Chemical and Transcriptomic Analysis of Cuticle Lipids under Cold Stress in Thellungiella salsuginea
Abstract
1. Introduction
2. Results
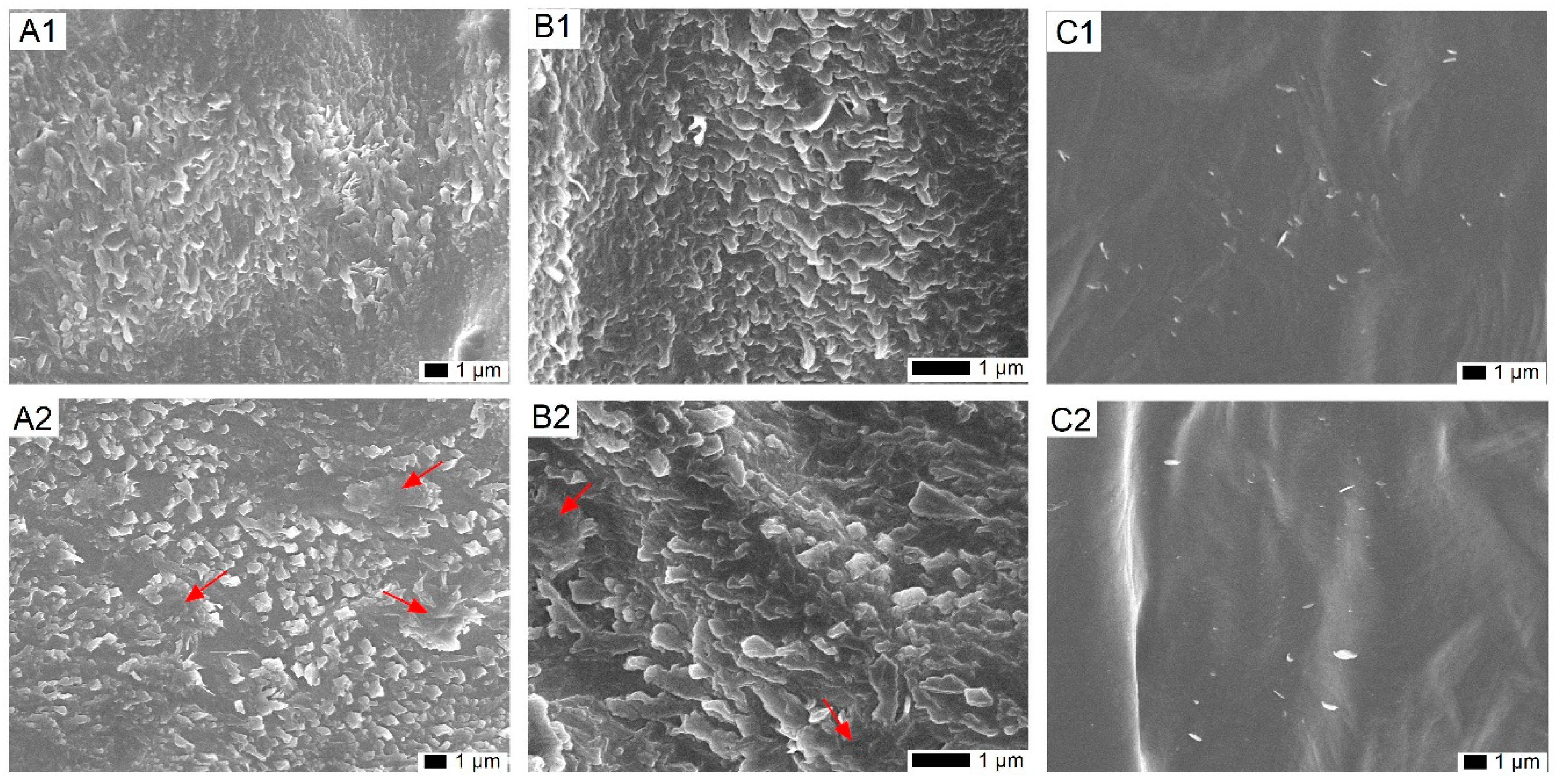
2.1. Impact of Cold on the Epicuticular Wax Structure on Leaves of T. salsuginea
2.2. Impact of Cold Treatment on Cuticular Wax of T. salsuginea
2.3. Impact of Cold Treatment on Cutin Monomers of T. salsuginea
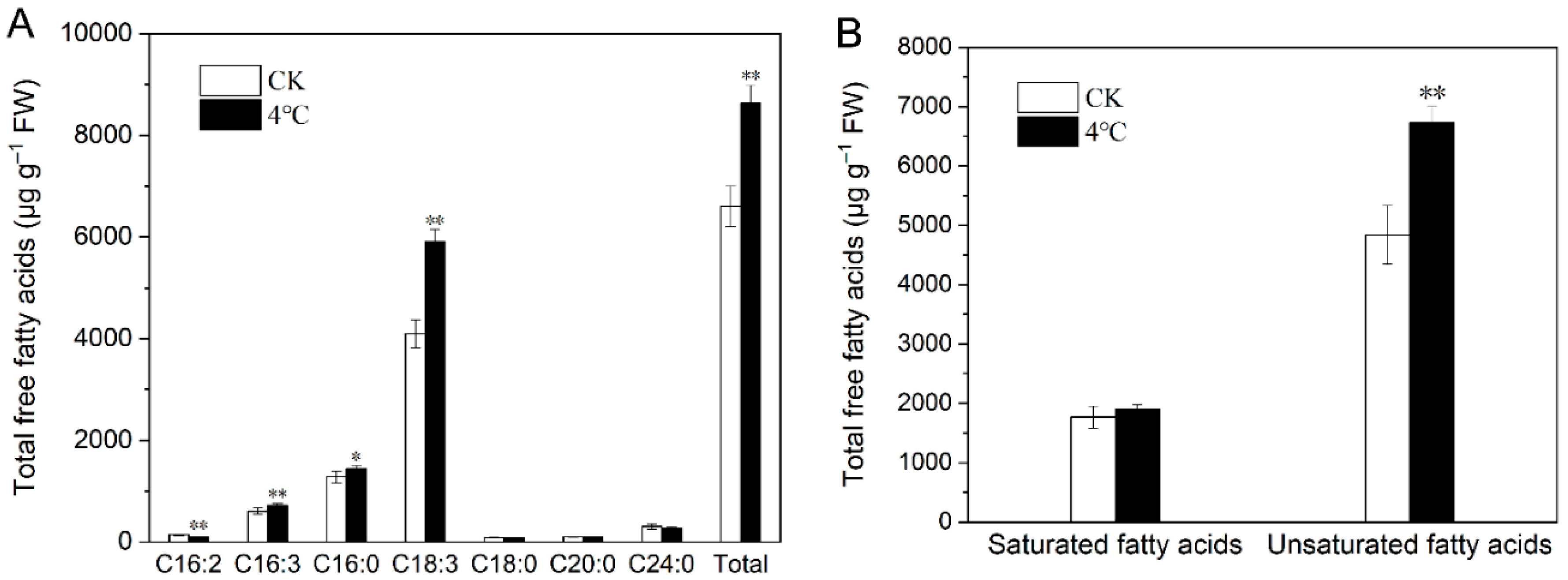
2.4. Impact of Cold Stress on Total Fatty Acids in T. salsuginea
2.5. Transcriptome Sequencing and Reads Mapping
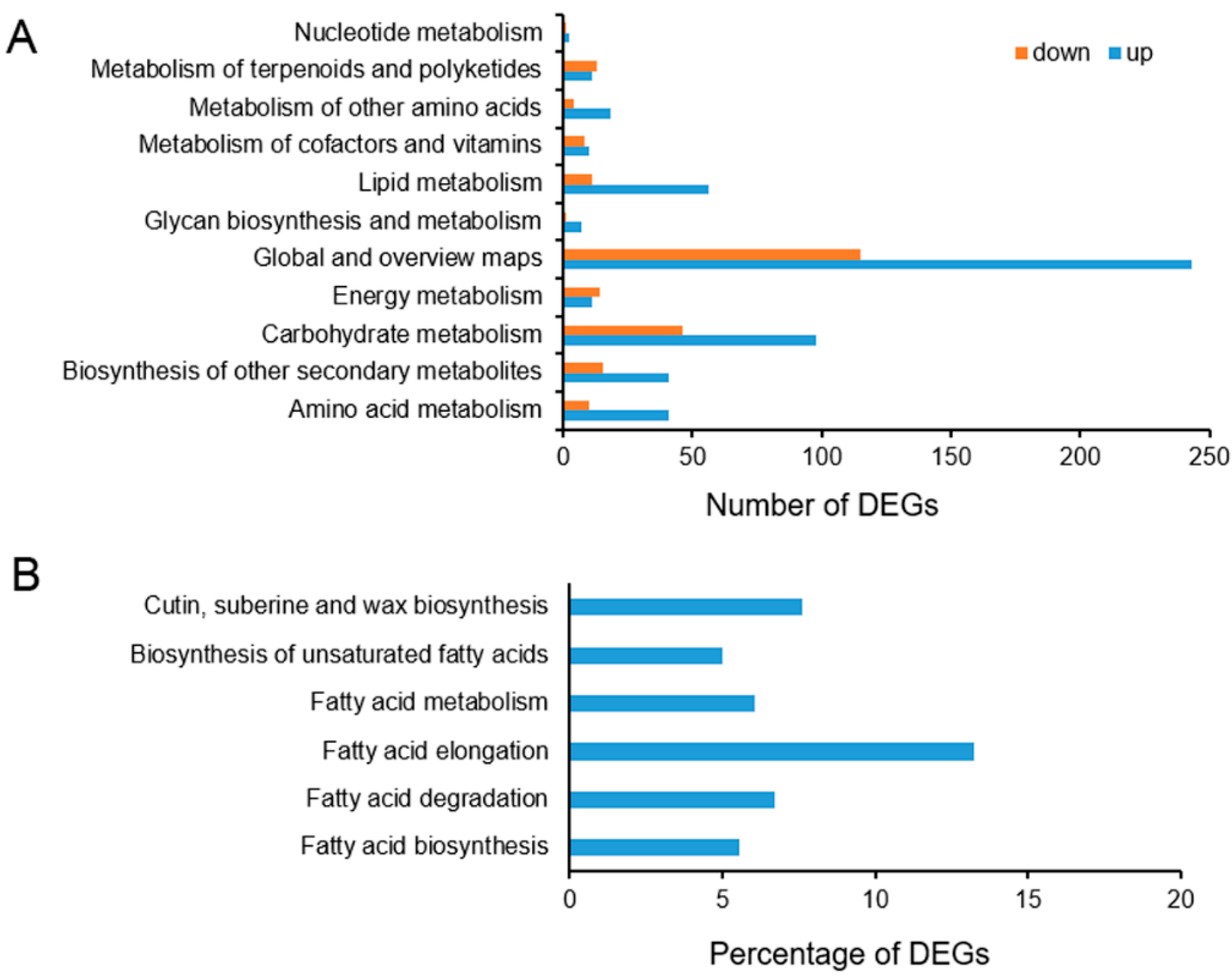
2.6. Functional Annotation and Classification of the Differentially Expressed Genes (DEGs)
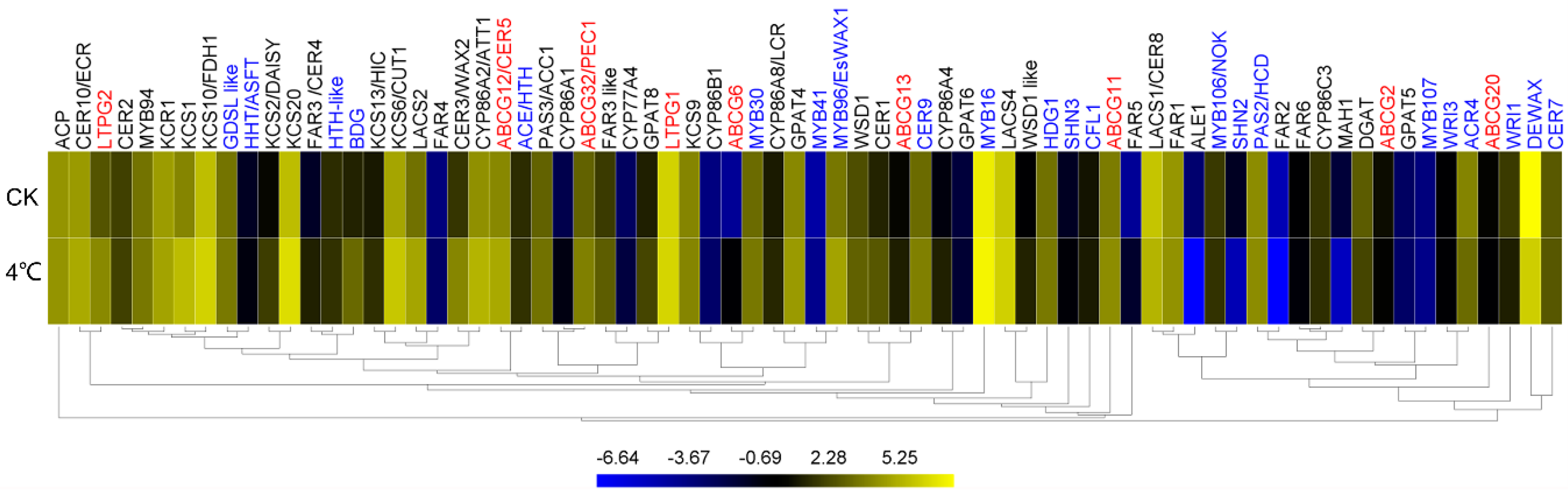
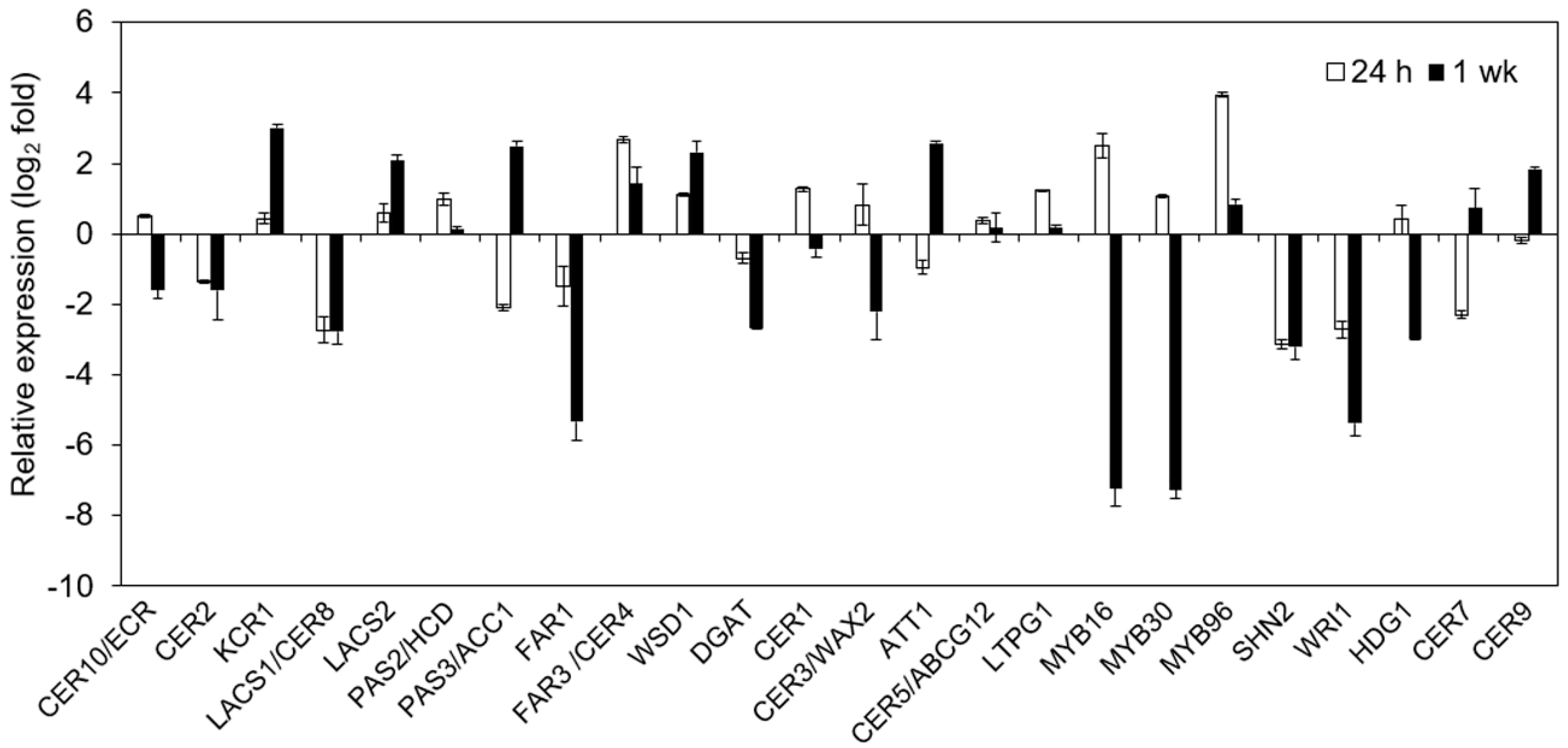
2.7. Impact of Cold on Cuticle-Associated Gene Expression
3. Discussion
4. Materials and Methods
4.1. Plant Growth Condition and Low-Temperature Treatment
4.2. Scanning Electron Microscopy
4.3. Analysis of Surface Cuticular Wax Composition
4.4. Analysis of Cutin Monomer Composition
4.5. Determination of Total Fatty Acid Profiles
4.6. RNA Extraction, Library Preparation, and Transcriptome Sequencing
4.7. Reads Mapping and Gene Quantification
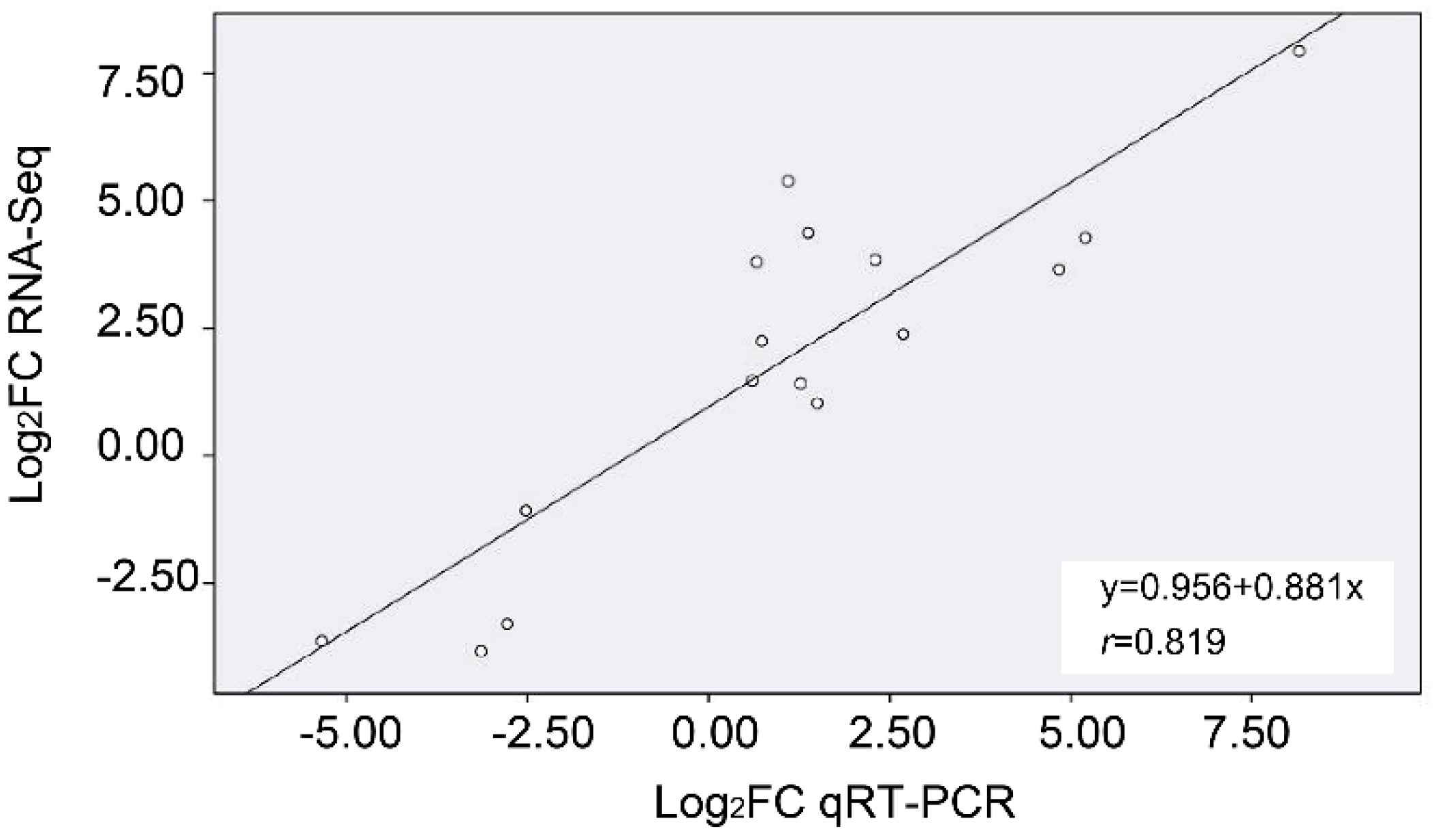
4.8. qRT-PCR
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BSTFA | N,O-bis(trimethylsilyl)trifluoroacetamide |
| DEG | Differential expressed gene |
| FID | Flame ionization detector |
| FPKM | Fragment per kilobase of exon model per million mapped reads |
| GC | Gas chromatography |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| qRT-PCR | Quantitative reverse transcription RT-PCR |
| SEM | Scanning electron microscopy |
References
- Hirabayashi, Y.; Mahendran, R.; Koirala, S.; Konoshima, L.; Yamazaki, D.; Watanabe, S.; Kim, H.; Kanae, S. Global flood risk under climate change. Nat. Clim. Chang. 2013, 3, 816. [Google Scholar] [CrossRef]
- Deng, J.M. Advances of studies on plant freezing-tolerance mechanism: Freezing tolerance gene expression and its function. Chin. Bull. Bot. 2001, 18, 521–530. [Google Scholar]
- Lee, Y.P.; Babakov, A.; de Boer, B.; Zuther, E.; Hincha, D.K. Comparison of freezing tolerance, compatible solutes and polyamines in geographically diverse collections of Thellungiella sp. and Arabidopsis thaliana accessions. BMC Plant Biol. 2012, 12, 131. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Li, H.G.; Wang, J.J.; Su, Y.; Wang, H.L.; Feng, C.H.; Yang, Y.; Niu, M.X.; Liu, C.; Yin, W.; et al. PeSTZ1, a C2H2-type zinc finger transcription factor from populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2. Plant Biotechnol. J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef]
- Zhang, M.; Ye, J.; Xu, Q.; Feng, Y.; Yuan, X.; Yu, H.; Wang, Y.; Wei, X.; Yang, Y. Genome-wide association study of cold tolerance of Chinese indica rice varieties at the bud burst stage. Plant Cell Rep. 2018, 37, 529–539. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, S.; Dong, J.; Yang, T.; Mao, X.; Liu, Q.; Wang, X.; Liu, B. A novel functional gene associated with cold tolerance at the seedling stage in rice. Plant Biotechnol. J. 2017, 15, 1141–1148. [Google Scholar] [CrossRef]
- Haake, V.; Cook, D.; Riechmann, J.L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef]
- Ding, Y.; Jia, Y.; Shi, Y.; Zhang, X.; Song, C.; Gong, Z.; Yang, S. OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 2018, 37, e98228. [Google Scholar] [CrossRef]
- Jenks, M.A.; Eigenbrode, S.D.; Lemieux, B. Cuticular waxes of Arabidopsis. Arab. Book/Am. Soc. Plant Biol. 2002, 1, e0016. [Google Scholar] [CrossRef] [PubMed]
- Riederer, M. Thermodynamics of the water permeability of plant cuticles: Characterization of the polar pathway. J. Exp. Bot. 2006, 57, 2937–2942. [Google Scholar] [CrossRef] [PubMed]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef] [PubMed]
- Jetter, R.; Kunst, L.; Samuels, A.L. Composition of plant cuticular waxes. Biol. Plant Cuticle 2006, 23, 145–181. [Google Scholar]
- Yang, X.; Zhao, H.; Kosma, D.K.; Tomasi, P.; Dyer, J.M.; Li, R.; Liu, X.; Wang, Z.; Parsons, E.P.; Jenks, M.A.; et al. The acyl desaturase CER17 is involved in producing wax unsaturated primary alcohols and cutin monomers. Plant Physiol. 2017, 173, 1109–1124. [Google Scholar] [CrossRef]
- Pollard, M.; Beisson, F.; Li, Y.; Ohlrogge, J.B. Building lipid barriers: Biosynthesis of cutin and suberin. Trends Plant Sci. 2008, 13, 236–246. [Google Scholar] [CrossRef]
- Shepherd, T.; Wynne Griffiths, D. The effects of stress on plant cuticular waxes. New Phytol. 2006, 171, 469–499. [Google Scholar] [CrossRef]
- Ladaniya, M.S. Physico-chemical, respiratory and fungicide residue changes in wax coated mandarin fruit stored at chilling temperature with intermittent warming. J. Food Sci. Technol. 2011, 48, 150–158. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Broeckling, C.D.; Sumner, L.W.; Wang, Z.Y. Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 2007, 64, 265–278. [Google Scholar] [CrossRef]
- Amid, A.; Lytovchenko, A.; Fernie, A.R.; Warren, G.; Thorlby, G.J. The sensitive to freezing3 mutation of Arabidopsis thaliana is a cold-sensitive allele of homomeric acetyl-CoA carboxylase that results in cold-induced cuticle deficiencies. J. Exp. Bot. 2012, 63, 5289–5299. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.; Gao, H.; Chen, H.; Wu, W.; Fang, X. Changes in cuticular wax composition of two blueberry cultivars during fruit ripening and postharvest cold storage. J. Agric. Food Chem. 2018, 66, 2870–2876. [Google Scholar] [CrossRef]
- Inan, G.; Zhang, Q.; Li, P.; Wang, Z.; Cao, Z.; Zhang, H.; Zhang, C.; Quist, T.M.; Goodwin, S.M.; Zhu, J.; et al. Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 2004, 135, 1718–1737. [Google Scholar] [CrossRef] [PubMed]
- Teusink, R.S.; Rahman, M.; Bressan, R.A.; Jenks, M.A. Cuticular waxes on Arabidopsis thaliana close relatives Thellungiella halophila and Thellungiella parvula. Int. J. Plant Sci. 2002, 163, 309–315. [Google Scholar] [CrossRef]
- Taji, T.; Seki, M.; Satou, M.; Sakurai, T.; Kobayashi, M.; Ishiyama, K.; Narusaka, Y.; Narusaka, M.; Zhu, J.K.; Shinozaki, K. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray. Plant Physiol. 2004, 135, 1697–1709. [Google Scholar] [CrossRef] [PubMed]
- Amtmann, A. Learning from evolution: Thellungiella generates new knowledge on essential and critical components of abiotic stress tolerance in plants. Mol. Plant 2009, 2, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Guevara, D.R.; Champigny, M.J.; Tattersall, A.; Dedrick, J.; Wong, C.E.; Li, Y.; Labbe, A.; Ping, C.L.; Wang, Y.; Nuin, P.; et al. Transcriptomic and metabolomic analysis of Yukon Thellungiella plants grown in cabinets and their natural habitat show phenotypic plasticity. BMC Plant Biol. 2012, 12, 175. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Feng, J.; Lu, S.; Lohrey, G.T.; An, H.; Zhou, Y.; Jenks, M.A. Leaf cuticular lipids on the Shandong and Yukon ecotypes of saltwater cress, Eutrema salsugineum, and their response to water deficiency and impact on cuticle permeability. Physiol. Plant. 2014, 151, 446–458. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Chen, N.; Song, B.; He, J.; Zhou, Y.; Jenks, M.A.; Xu, X. Compositional and transcriptomic analysis associated with cuticle lipid production on rosette and inflorescence stem leaves in the extremophyte Thellungiella salsuginea. Physiol. Plant. 2019, 165, 584–603. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Zhang, Z.; Wang, J.Y.; Oh, D.H.; Dassanayake, M.; Liu, B.; Huang, Q.; Sun, H.X.; Xia, R.; Wu, Y.; et al. Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. USA 2012, 109, 12219–12224. [Google Scholar] [CrossRef] [PubMed]
- Kunst, L.; Samuels, A.L. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 2003, 42, 51–80. [Google Scholar] [CrossRef]
- Rowland, O.; Zheng, H.; Hepworth, S.R.; Lam, P.; Jetter, R.; Kunst, L. CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 2006, 142, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Aarts, M.G.; Keijzer, C.J.; Stiekema, W.J.; Pereira, A. Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 1995, 7, 2115–2127. [Google Scholar] [PubMed]
- Chen, X.; Goodwin, S.M.; Boroff, V.L.; Liu, X.; Jenks, M.A. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 2003, 15, 1170–1185. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Domergue, F.; Pascal, S.; Jetter, R.; Renne, C.; Faure, J.D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Joubes, J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 2012, 24, 3106–3118. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Song, C.; Wang, X. Investigation on response mechanism of epicuticular wax on Arabidopsis thaliana under cold stress. Sci. Agric. Sin. 2014, 47, 252–261. [Google Scholar]
- Wang, M.; Wang, Y.; Wu, H.; Xu, J.; Li, T.; Hegebarth, D.; Jetter, R.; Chen, L.; Wang, Z. Three TaFAR genes function in the biosynthesis of primary alcohols and the response to abiotic stresses in Triticum aestivum. Sci. Rep. 2016, 6, 25008. [Google Scholar] [CrossRef] [PubMed]
- Cameron, K.D.; Teece, M.A.; Smart, L.B. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol. 2006, 140, 176–183. [Google Scholar] [CrossRef]
- Szafranek, B.M.; Synak, E.E. Cuticular waxes from potato (Solanum tuberosum) leaves. Phytochemistry 2006, 67, 80–90. [Google Scholar] [CrossRef]
- Kim, K.S.; Park, S.H.; Jenks, M.A. Changes in leaf cuticular waxes of sesame (Sesamum indicum L.) plants exposed to water deficit. J. Plant Physiol. 2007, 164, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Park, S.H.; Kim, D.K.; Jenks, M.A. Influence of water deficit on leaf cuticular waxes of soybean (Glycine max [L.] Merr.). Int. J. Plant Sci. 2007, 168, 307–316. [Google Scholar] [CrossRef]
- Kosma, D.K.; Bourdenx, B.; Bernard, A.; Parsons, E.P.; Lu, S.; Joubes, J.; Jenks, M.A. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 2009, 151, 1918–1929. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xiao, L.; Feng, J.; Chen, N.; Chen, Y.; Song, B.; Xue, K.; Shi, S.; Zhou, Y.; Jenks, M.A. Cuticle lipids on heteromorphic leaves of Populus euphratica Oliv. growing in riparian habitats differing in available soil moisture. Physiol. Plant. 2016, 158, 318–330. [Google Scholar] [CrossRef]
- Hooker, T.S.; Millar, A.A.; Kunst, L. Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol. 2002, 129, 1568–1580. [Google Scholar] [CrossRef]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Park, G.C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef]
- Guo, L.; Yang, H.; Zhang, X.; Yang, S. Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 2013, 64, 1755–1767. [Google Scholar] [CrossRef]
- Xiao, F.; Mark Goodwin, S.; Xiao, Y.; Sun, Z.; Baker, D.; Tang, X.; Jenks, M.A.; Zhou, J.M. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004, 23, 2903–2913. [Google Scholar] [CrossRef]
- Wang, Q.; Guan, X.; Zenghui, H.U.; Cunfuamp, L.U.; Leng, P. Relationship between cold tolerance and leaf structure of the three species of Sedum. Chin. J. Appl. Environ. Biol. 2013, 19, 280–285. [Google Scholar] [CrossRef]
- Riederer, M.; Schreiber, L. Protecting against water loss: Analysis of the barrier properties of plant cuticles. J. Exp. Bot. 2001, 52, 2023–2032. [Google Scholar] [CrossRef]
- Bessire, M.; Borel, S.; Fabre, G.; Carraca, L.; Efremova, N.; Yephremov, A.; Cao, Y.; Jetter, R.; Jacquat, A.C.; Metraux, J.P.; et al. A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 2011, 23, 1958–1970. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.M.; Jenks, M.A. Plant cuticle function as a barrier to water loss. Plant Abiotic Stress 2007, 14–36. [Google Scholar]
- Steponkus, P.L.; Uemura, M.; Joseph, R.A.; Gilmour, S.J.; Thomashow, M.F. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 95, 14570–14575. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Browse, J. Cold Comfort Farm: The acclimation of plants to freezing temperatures. Plant Cell Environ. 2000, 23, 893–902. [Google Scholar] [CrossRef]
- Uemura, M.; Warren, G.; Steponkus, P.L. Freezing sensitivity in the sfr4 mutant of Arabidopsis is due to low sugar content and is manifested by loss of osmotic responsiveness. Plant Physiol. 2003, 131, 1800–1807. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Broeckling, C.D.; Blancaflor, E.B.; Sledge, M.K.; Sumner, L.W.; Wang, Z.Y. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 2005, 42, 689–707. [Google Scholar] [CrossRef] [PubMed]
- Bonaventure, G.; Beisson, F.; Ohlrogge, J.; Pollard, M. Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: Occurrence of octadeca-cis-6, cis-9-diene-1,18-dioate as the major component. Plant J. 2004, 40, 920–930. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book/Am. Soc. Plant Biol. 2013, 11, e0161. [Google Scholar] [CrossRef]
- Molina, I.; Bonaventure, G.; Ohlrogge, J.; Pollard, M. The lipid polyester composition of Arabidopsis thaliana and Brassica napus seeds. Phytochemistry 2006, 67, 2597–2610. [Google Scholar] [CrossRef]
- Lee, S.B.; Jung, S.J.; Go, Y.S.; Kim, H.U.; Kim, J.K.; Cho, H.J.; Park, O.K.; Suh, M.C. Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 2009, 60, 462–475. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Tarazona, S.; Garcã-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034-1. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Sample Name | CK_1 | CK_2 | CK_3 | 4 °C_1 | 4 °C_2 | 4 °C_3 |
|---|---|---|---|---|---|---|
| Raw Data Size (bp) | 1,197,620,550 | 1,162,182,850 | 1,199,812,800 | 1,140,096,000 | 1,178,295,300 | 1,188,882,800 |
| Raw Reads Number | 23,952,411 | 23,243,657 | 23,996,256 | 22,801,920 | 23,565,906 | 23,777,656 |
| Clean Data Size (bp) | 1,195,529,000 | 1,159,996,950 | 1,197,116,750 | 1,137,586,350 | 1,176,087,850 | 1,186,401,500 |
| Clean Reads Number | 23,910,580 | 23,199,939 | 23,942,335 | 22,751,727 | 23,521,757 | 23,728,030 |
| Clean Data Rate (%) | 99.82 | 99.81 | 99.77 | 99.77 | 99.81 | 99.79 |
| Clean Reads Q20 (%) | 96.80 | 96.20 | 96.70 | 96.00 | 95.70 | 96.50 |
| Total Mapped Reads (%) (reference genome/gene) | 97.12/89.86 | 96.89/89.85 | 97.32/90.35 | 96.90/90.73 | 96.87/90.59 | 97.17/90.96 |
| Unique Match (%) (reference genome/gene) | 79.91/75.68 | 77.53/75.42 | 79.88/75.52 | 78.31/78.78 | 79.52/78.56 | 80.76/78.71 |
| Multiposition Match (%) (reference genome/gene) | 17.21/14.19 | 19.36/14.43 | 17.44/14.84 | 18.59/11.95 | 17.35/12.03 | 16.41/12.24 |
| Total Unmapped Reads (%) (reference genome/gene) | 2.88/10.14 | 3.11/10.15 | 2.68/9.65 | 3.10/9.27 | 3.14/9.41 | 2.83/9.04 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Tang, S.; Yang, D.; Chen, Y.; Ling, L.; Zou, Y.; Zhou, M.; Xu, X. Chemical and Transcriptomic Analysis of Cuticle Lipids under Cold Stress in Thellungiella salsuginea. Int. J. Mol. Sci. 2019, 20, 4519. https://doi.org/10.3390/ijms20184519
He J, Tang S, Yang D, Chen Y, Ling L, Zou Y, Zhou M, Xu X. Chemical and Transcriptomic Analysis of Cuticle Lipids under Cold Stress in Thellungiella salsuginea. International Journal of Molecular Sciences. 2019; 20(18):4519. https://doi.org/10.3390/ijms20184519
Chicago/Turabian StyleHe, Junqing, Shuai Tang, Di Yang, Yue Chen, Ludi Ling, Yanli Zou, Minqi Zhou, and Xiaojing Xu. 2019. "Chemical and Transcriptomic Analysis of Cuticle Lipids under Cold Stress in Thellungiella salsuginea" International Journal of Molecular Sciences 20, no. 18: 4519. https://doi.org/10.3390/ijms20184519
APA StyleHe, J., Tang, S., Yang, D., Chen, Y., Ling, L., Zou, Y., Zhou, M., & Xu, X. (2019). Chemical and Transcriptomic Analysis of Cuticle Lipids under Cold Stress in Thellungiella salsuginea. International Journal of Molecular Sciences, 20(18), 4519. https://doi.org/10.3390/ijms20184519





