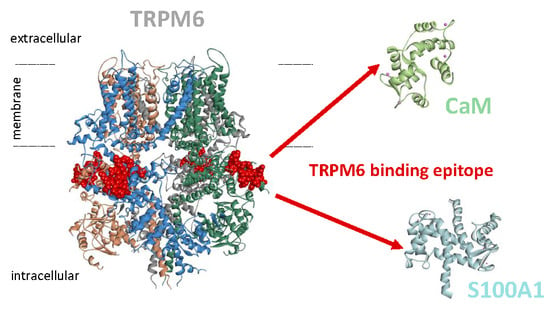
TRPM6 N-Terminal CaM- and S100A1-Binding Domains
Abstract
:1. Introduction
2. Results
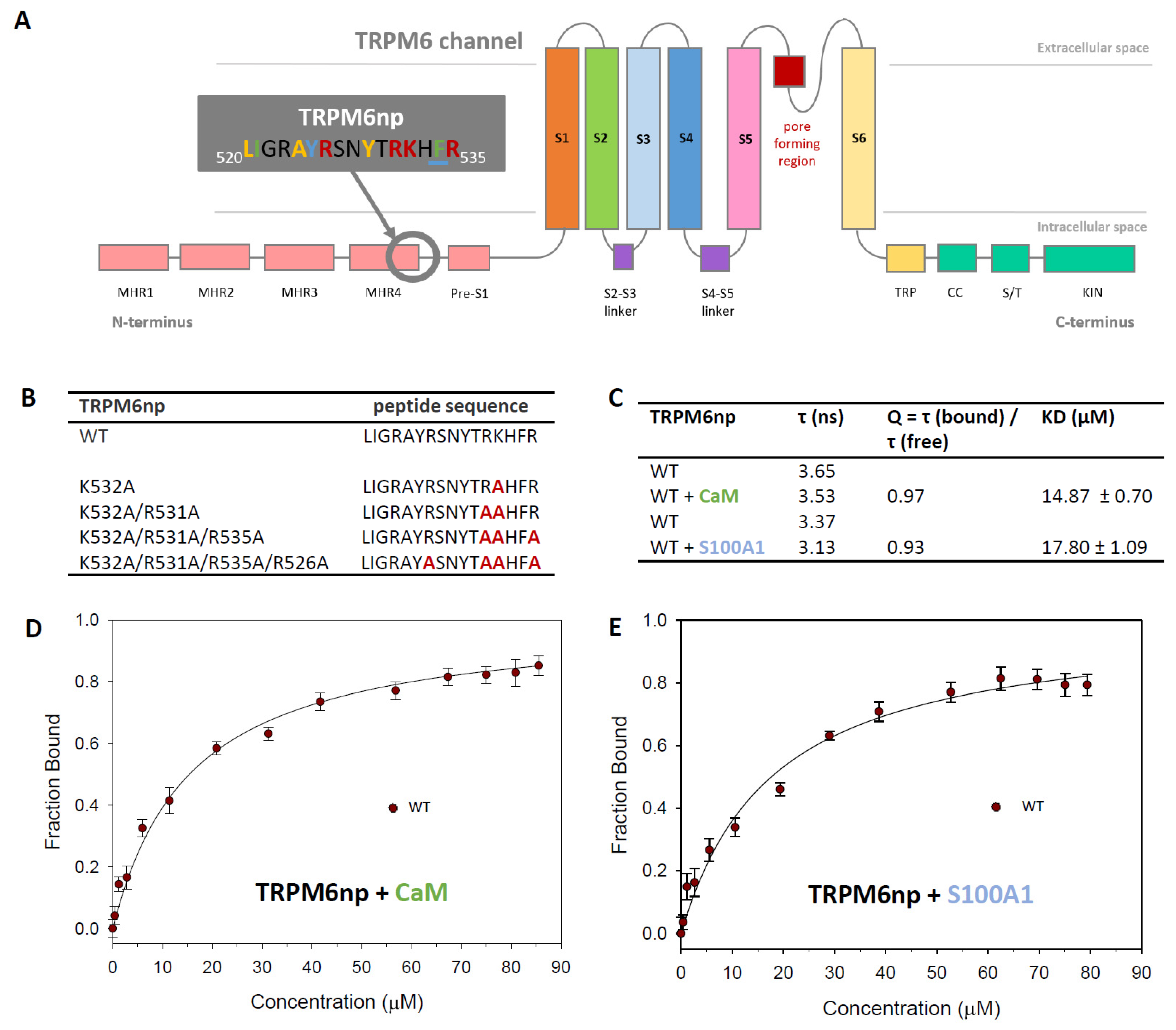
2.1. The Identification of TRPM6np Binding Domains for CaM and S100A1
2.2. CaM- and S100A1-Binding Domains of the TRPM6 N-Termini Overlap
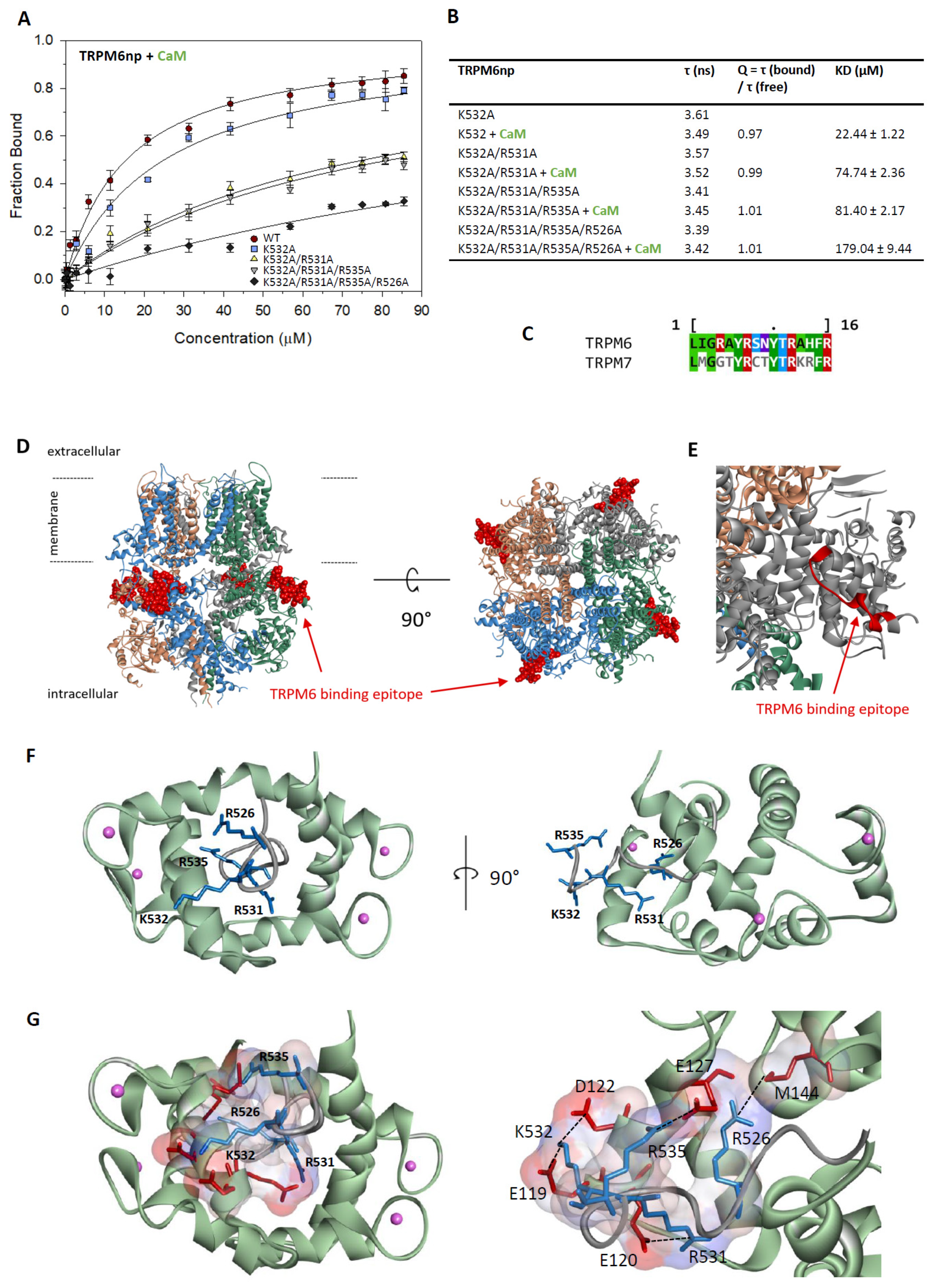
2.3. Characterisation of the TRPM6np/CaM Complex
2.4. Characterisation of the TRPM6np/S100A1 Complex
2.5. TRPM6np Molecular Modelling and CaM and S100A1 Docking
3. Discussion
4. Methods
4.1. TRPM6-Binding Domain Design and Synthesis
4.2. Expression and Purification of CaM and S100A1
4.3. Steady-State Fluorescence Anisotropy Measurements
4.4. Lifetime Experiments
4.5. Model Building
4.6. Ligand Docking
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ATP | adenosine trisphosphate |
| CaM | calmodulin |
| CBPs | calcium-binding proteins |
| Cryo-EM | Cryogenic Electron Microscopy |
| IDPs | intrinsically disordered proteins |
| PIP2 | phosphatidylinositol 4, 5- bisphosphate |
| RyR | rhyanodine receptor |
| S100A1 | S100 calcium-binding protein A1 |
| TRP | transient receptor potential channel |
| TRPM | transient receptor potential channel melastatin |
| TRPM6np | transient receptor potential channel melastatin 6 N-terminal peptide |
| TRPs | transient receptor potential channels |
| TRPV | transient receptor potential channels |
| WT | wild type |
References
- Kraft, R.; Harteneck, C. The mammalian melastatin-related transient receptor potential cation channels: An overview. Pflug. Arch. 2005, 451, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Voets, T.; Nilius, B.; Hoefs, S.; van der Kemp, A.W.; Droogmans, G.; Bindels, R.J.; Hoenderop, J.G. TRPM6 forms the Mg2+ influx channel involved in intestinal and renal Mg2+ absorption. J. Biol. Chem. 2004, 279, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Fonfria, E.; Murdock, P.R.; Cusdin, F.S.; Benham, C.D.; Kelsell, R.E.; McNulty, S. Tissue distribution profiles of the human TRPM cation channel family. J. Recept Signal Transduct Res. 2006, 26, 159–178. [Google Scholar] [CrossRef] [PubMed]
- Groenestege, W.M.; Hoenderop, J.G.; van den Heuvel, L.; Knoers, N.; Bindels, R.J. The epithelial Mg2+ channel transient receptor potential melastatin 6 is regulated by dietary Mg2+ content and estrogens. J. Am. Soc. Nephrol. 2006, 17, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.R.; Lin, M.J.; McIntosh, L.S.; Sham, J.S. Functional expression of transient receptor potential melastatin-and vanilloid-related channels in pulmonary arterial and aortic smooth muscle. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2006, L1267–L1276. [Google Scholar] [CrossRef] [PubMed]
- Mandinova, A.; Atar, D.; Schafer, B.; Spiess, M.; Aebi, U.; Heizmann, C.W. Distinct subcellular localization of calcium binding S100 proteins in human smooth muscle cells and their relocation in response to rises in intracellular calcium. J. Cell Sci. 1998, 111, 2043–2054. [Google Scholar] [PubMed]
- Bagchi, I.C.; Huang, Q.; Means, A.R. Identification of amino acids essential for calmodulin binding and activation of smooth muscle myosin light chain kinase. J. Biol. Chem. 1992, 267, 3024–3029. [Google Scholar] [PubMed]
- Marston, S.B.; Fraser, I.; Huber, P.; Pritchard, K.; Gusev, N.B.; Torok, K. Location of two contact sites between human smooth muscle caldesmon and Ca (2+)-calmodulin. J. Biol. Chem. 1994, 269, 8134–8139. [Google Scholar] [PubMed]
- Paunier, L.; Radde, I.C.; Kooh, S.W.; Conen, P.E.; Fraser, D.D.D. Primary hypomagnesemia with secondary hypocalcemia in an infant. Pediatrics 1968, 41, 385–402. [Google Scholar]
- Schlingmann, K.P.; Weber, S.; Peters, M.; Niemann Nejsum, L.; Vitzthum, H.; Klingel, K.; Kratz, M.; Haddad, E.; Ristoff, E.; Dinour, D.; et al. Hypomagnesemia with secondary hypocalcemia is caused by mutations in TRPM6, a new member of the TRPM gene family. Nat. Genet. 2002, 31, 166–170. [Google Scholar] [CrossRef]
- Walder, R.Y.; Landau, D.; Meyer, P.; Shalev, H.; Tsolia, M.; Borochowitz, Z.; Boettger, M.B.; Beck, G.E.; Englehardt, R.K.; Carmi, R.; et al. Mutation of TRPM6 causes familial hypomagnesemia with secondary hypocalcemia. Nat. Genet. 2002, 31, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Sanz, N.; Fernandez-Carvajal, A.; Morenilla-Palao, C.; Planells-Cases, R.; Fajardo-Sanchez, E.; Fernandez-Ballester, G.; Ferrer-Montiel, A. Identification of a tetramerization domain in the C terminus of the vanilloid receptor. J. Neurosci. 2004, 24, 5307–5314. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Jiang, J.; Yue, L. Functional characterization of homo- and heteromeric channel kinases TRPM6 and TRPM7. J. Gen. Physiol. 2006, 127, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yu, H.; Huang, J.; Faouzi, M.; Schmitz, C.; Penner, R.; Fleig, A. The TRPM6 kinase domain determines the Mg.ATP sensitivity of TRPM7/M6 heteromeric ion channels. J. Biol. Chem. 2014, 289, 5217–5227. [Google Scholar] [CrossRef] [PubMed]
- Chubanov, V.; Waldegger, S.; Mederos y Schnitzler, M.; Vitzthum, H.; Sassen, M.C.; Seyberth, H.W.; Konrad, M.; Gudermann, T. Disruption of TRPM6/TRPM7 complex formation by a mutation in the TRPM6 gene causes hypomagnesemia with secondary hypocalcemia. Proc. Natl. Acad. Sci. USA 2004, 101, 2894–2899. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Cao, E.; Julius, D.; Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 2013, 504, 107–112. [Google Scholar] [CrossRef]
- Moiseenkova-Bell, V.Y.; Stanciu, L.A.; Serysheva, I.; Tobe, B.J.; Wensel, T.G. Structure of TRPV1 channel revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 2008, 105, 7451–7455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Cao, E.; Julius, D.; Cheng, Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 2016, 534, 347–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Autzen, H.E.; Myasnikov, A.G.; Campbell, M.G.; Asarnow, D.; Julius, D.; Cheng, Y. Structure of the human TRPM4 ion channel in a lipid nanodisc. Science 2018, 359, 228–232. [Google Scholar] [CrossRef] [Green Version]
- Winkler, P.A.; Huang, Y.; Sun, W.; Du, J.; Lu, W. Electron cryo-microscopy structure of a human TRPM4 channel. Nature 2017, 552, 200–204. [Google Scholar] [CrossRef]
- Chubanov, V.; Mittermeier, L.; Gudermann, T. TRPM7 reflected in Cryo-EMirror. Cell Calcium 2018, 76, 129–131. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Li, Z.; Li, J.; Hulse, R.E.; Santa-Cruz, A.; Valinsky, W.C.; Abiria, S.A.; Krapivinsky, G.; Zhang, J.; Clapham, D.E. Structure of the mammalian TRPM7, a magnesium channel required during embryonic development. Proc. Natl. Acad. Sci. USA 2018, 115, E8201–E8210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owsianik, G.; D’Hoedt, D.; Voets, T.; Nilius, B. Structure-function relationship of the TRP channel superfamily. Rev. Physiol. Biochem. Pharmacol. 2006, 156, 61–90. [Google Scholar] [PubMed]
- Clapham, D.E. TRP channels as cellular sensors. Nature 2003, 426, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Harteneck, C. Proteins modulating TRP channel function. Cell Calcium 2003, 33, 303–310. [Google Scholar] [CrossRef]
- Obukhov, A.; Schultz, G.; Lückhoff, A. Regulation of heterologously expressed transient receptor potential-like channels by calcium ions. Neuroscience 1998, 85, 487–495. [Google Scholar] [CrossRef]
- Zhu, M.X. Multiple roles of calmodulin and other Ca 2+-binding proteins in the functional regulation of TRP channels. Pflügers Arch. 2005, 451, 105–115. [Google Scholar] [CrossRef]
- Kinoshita-Kawada, M.; Tang, J.; Xiao, R.; Kaneko, S.; Foskett, J.K.; Zhu, M.X. Inhibition of TRPC5 channels by Ca 2+-binding protein 1 in Xenopus oocytes. Pflügers Arch. 2005, 450, 345–354. [Google Scholar] [CrossRef]
- Jirku, M.; Lansky, Z.; Bednarova, L.; Sulc, M.; Monincova, L.; Majer, P.; Vyklicky, L.; Vondrasek, J.; Teisinger, J.; Bousova, K. The characterization of a novel S100A1 binding site in the N-terminus of TRPM1. Int. J. Biochem. Cell Biol. 2016, 78, 186–193. [Google Scholar] [CrossRef]
- Holakovska, B.; Grycova, L.; Jirku, M.; Sulc, M.; Bumba, L.; Teisinger, J. Calmodulin and S100A1 protein interact with N terminus of TRPM3 channel. J. Biol. Chem. 2012, 287, 16645–16655. [Google Scholar] [CrossRef]
- Bousova, K.; Herman, P.; Vecer, J.; Bednarova, L.; Monincova, L.; Majer, P.; Vyklicky, L.; Vondrasek, J.; Teisinger, J. Shared CaM- and S100A1-binding epitopes in the distal TRPM4 N terminus. FEBS J. 2018, 285, 599–613. [Google Scholar] [CrossRef] [PubMed]
- Tong, Q.; Zhang, W.; Conrad, K.; Mostoller, K.; Cheung, J.Y.; Peterson, B.Z.; Miller, B.A. Regulation of the transient receptor potential channel TRPM2 by the Ca2+ sensor calmodulin. J. Biol. Chem. 2006, 281, 9076–9085. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Sun, B.; Du, J.; Yang, W.; Chen, H.C.; Overton, J.D.; Runnels, L.W.; Yue, L. Phosphatidylinositol 4,5-bisphosphate (PIP(2)) controls magnesium gatekeeper TRPM6 activity. Sci. Rep. 2011, 1, 146. [Google Scholar] [CrossRef] [PubMed]
- Park, E.Y.J.; Baik, J.Y.; Kwak, M.; So, I. The role of calmodulin in regulating calcium-permeable PKD2L1 channel activity. Korean J. Physiol. Pharmacol. 2019, 23, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Emery, E.C.; Diakogiannaki, E.; Gentry, C.; Psichas, A.; Habib, A.M.; Bevan, S.; Fischer, M.J.; Reimann, F.; Gribble, F.M. Stimulation of GLP-1 secretion downstream of the ligand-gated ion channel TRPA1. Diabetes 2015, 64, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Dang, S.; van Goor, M.K.; Asarnow, D.; Wang, Y.; Julius, D.; Cheng, Y.; van der Wijst, J. Structural insight into TRPV5 channel function and modulation. Proc. Natl. Acad. Sci. USA 2019, 116, 8869–8878. [Google Scholar] [CrossRef] [Green Version]
- Hasan, R.; Zhang, X. Ca(2+) Regulation of TRP Ion Channels. Int. J. Mol. Sci. 2018, 19, 1256. [Google Scholar] [CrossRef]
- Babu, Y.S.; Sack, J.S.; Greenhough, T.J.; Bugg, C.E.; Means, A.R.; Cook, W.J.J.J. Three-dimensional structure of calmodulin. Nature 1985, 315, 37–40. [Google Scholar] [CrossRef]
- Barbato, G.; Ikura, M.; Kay, L.E.; Pastor, R.W.; Bax, A.A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: The central helix is flexible. Biochemistry 1992, 31, 5269–5278. [Google Scholar] [CrossRef]
- Babu, Y.S.; Bugg, C.E.; Cook, W.J.J. Structure of calmodulin refined at 2.2 A resolution. J. Mol. Biol. 1988, 204, 191–204. [Google Scholar] [CrossRef]
- Chin, D.; Means, A.R.R. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000, 10, 322–328. [Google Scholar] [CrossRef]
- Singh, A.K.; McGoldrick, L.L.; Twomey, E.C.; Sobolevsky, A.I.I. Mechanism of calmodulin inactivation of the calcium-selective TRP channel TRPV6. Sci. Adv. 2018, 4. [Google Scholar] [CrossRef] [PubMed]
- Hasan, R.; Leeson-Payne, A.T.; Jaggar, J.H.; Zhang, X. Calmodulin is responsible for Ca(2+)-dependent regulation of TRPA1 Channels. Sci. Rep. 2017, 7, 45098. [Google Scholar] [CrossRef]
- Bate, N.; Caves, R.E.; Skinner, S.P.; Goult, B.T.; Basran, J.; Mitcheson, J.S.; Vuister, G.W. A novel mechanism for calmodulin-dependent inactivation of transient receptor potential vanilloid 6. Biochemistry 2018, 57, 2611–2622. [Google Scholar] [CrossRef] [PubMed]
- Prosser, B.L.; Wright, N.T.; Hernandez-Ochoa, E.O.; Varney, K.M.; Liu, Y.; Olojo, R.O.; Zimmer, D.B.; Weber, D.J.; Schneider, M.F. S100A1 binds to the calmodulin-binding site of ryanodine receptor and modulates skeletal muscle excitation-contraction coupling. J. Biol. Chem. 2008, 283, 5046–5057. [Google Scholar] [CrossRef] [PubMed]
- Wright, N.T.; Varney, K.M.; Ellis, K.C.; Markowitz, J.; Gitti, R.K.; Zimmer, D.B.; Weber, D.J. The three-dimensional solution structure of Ca(2+)-bound S100A1 as determined by NMR spectroscopy. J. Mol. Biol. 2005, 353, 410–426. [Google Scholar] [CrossRef]
- Bily, J.; Grycova, L.; Holendova, B.; Jirku, M.; Janouskova, H.; Bousova, K.; Teisinger, J. Characterization of the S100A1 protein binding site on TRPC6 C-terminus. PLoS ONE 2013, 8, e62677. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.-Y.; Procko, E.; Gaudet, R. Distinct properties of Ca2+–calmodulin binding to N-and C-terminal regulatory regions of the TRPV1 channel. J. Gen. Physiol. 2012, 140, 541–555. [Google Scholar] [CrossRef]
- Yap, K.L.; Kim, J.; Truong, K.; Sherman, M.; Yuan, T.; Ikura, M. Calmodulin target database. J. Struct. Funct. Genom. 2000, 1, 8–14. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Brown, N.P.; Leroy, C.; Sander, C. MView: A web-compatible database search or multiple alignment viewer. Bioinformatics 1998, 14, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255. [Google Scholar] [CrossRef] [PubMed]
- Fallon, J.L.; Halling, D.B.; Hamilton, S.L.; Quiocho, F.A. Structure of calmodulin bound to the hydrophobic IQ domain of the cardiac Cav1. 2 calcium channel. Structure 2005, 13, 1881–1886. [Google Scholar] [CrossRef] [PubMed]
- Wright, N.T.; Prosser, B.L.; Varney, K.M.; Zimmer, D.B.; Schneider, M.F.; Weber, D.J. S100A1 and calmodulin compete for the same binding site on ryanodine receptor. J. Biol. Chem. 2008, 283, 26676–26683. [Google Scholar] [CrossRef] [PubMed]
- Grycova, L.; Holendova, B.; Lansky, Z.; Bumba, L.; Jirku, M.; Bousova, K.; Teisinger, J. Ca2+ Binding protein S100A1 competes with calmodulin and PIP2 for binding site on the C-terminus of the TPRV1 receptor. ACS Chem. Neurosci. 2014, 6, 386–392. [Google Scholar] [CrossRef]
- Prosser, B.L.; Hernández-Ochoa, E.O.; Schneider, M.F. S100A1 and calmodulin regulation of ryanodine receptor in striated muscle. Cell Calcium 2011, 50, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Whicher, J.R.; MacKinnon, R. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science 2016, 353, 664–669. [Google Scholar] [CrossRef]
- López-Romero, A.E.; Hernández-Araiza, I.; Torres-Quiroz, F.; Tovar-Y-Romo, L.B.; Islas, L.D.; Rosenbaum, T. TRP ion channels: Proteins with conformational flexibility. Channels 2019, 13, 207–226. [Google Scholar] [CrossRef] [Green Version]
- Jirku, M.; Bumba, L.; Bednarova, L.; Kubala, M.; Sulc, M.; Franek, M.; Vyklicky, L.; Vondrasek, J.; Teisinger, J.; Bousova, K. Characterization of the part of N-terminal PIP2 binding site of the TRPM1 channel. Biophys. Chem. 2015, 207, 135–142. [Google Scholar] [CrossRef]
- Neshich, G.; Togawa, R.C.; Mancini, A.L.; Kuser, P.R.; Yamagishi, M.E.; Pappas, G.; Torres, W.V.; e Campos, T.F.; Ferreira, L.L.; Luna, F.M. STING Millennium: A web-based suite of programs for comprehensive and simultaneous analysis of protein structure and sequence. Nucleic Acids Res. 2003, 31, 3386–3392. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins: Struct. Funct. Bioinform. 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vajda, S.; Yueh, C.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Kozakov, D. New additions to the C lus P ro server motivated by CAPRI. Proteins: Struct. Funct. Bioinform. 2017, 85, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Biovia, D.S. Discovery Studio Modeling Environment. Release: 2017; Dassault Systèmes: San Diego, CA, USA, 2016. [Google Scholar]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zouharova, M.; Herman, P.; Hofbauerová, K.; Vondrasek, J.; Bousova, K. TRPM6 N-Terminal CaM- and S100A1-Binding Domains. Int. J. Mol. Sci. 2019, 20, 4430. https://doi.org/10.3390/ijms20184430
Zouharova M, Herman P, Hofbauerová K, Vondrasek J, Bousova K. TRPM6 N-Terminal CaM- and S100A1-Binding Domains. International Journal of Molecular Sciences. 2019; 20(18):4430. https://doi.org/10.3390/ijms20184430
Chicago/Turabian StyleZouharova, Monika, Petr Herman, Kateřina Hofbauerová, Jiri Vondrasek, and Kristyna Bousova. 2019. "TRPM6 N-Terminal CaM- and S100A1-Binding Domains" International Journal of Molecular Sciences 20, no. 18: 4430. https://doi.org/10.3390/ijms20184430