Effective Treatment of Metastatic Melanoma by Combining MAPK and PI3K Signaling Pathway Inhibitors
Abstract
1. Introduction
2. Results
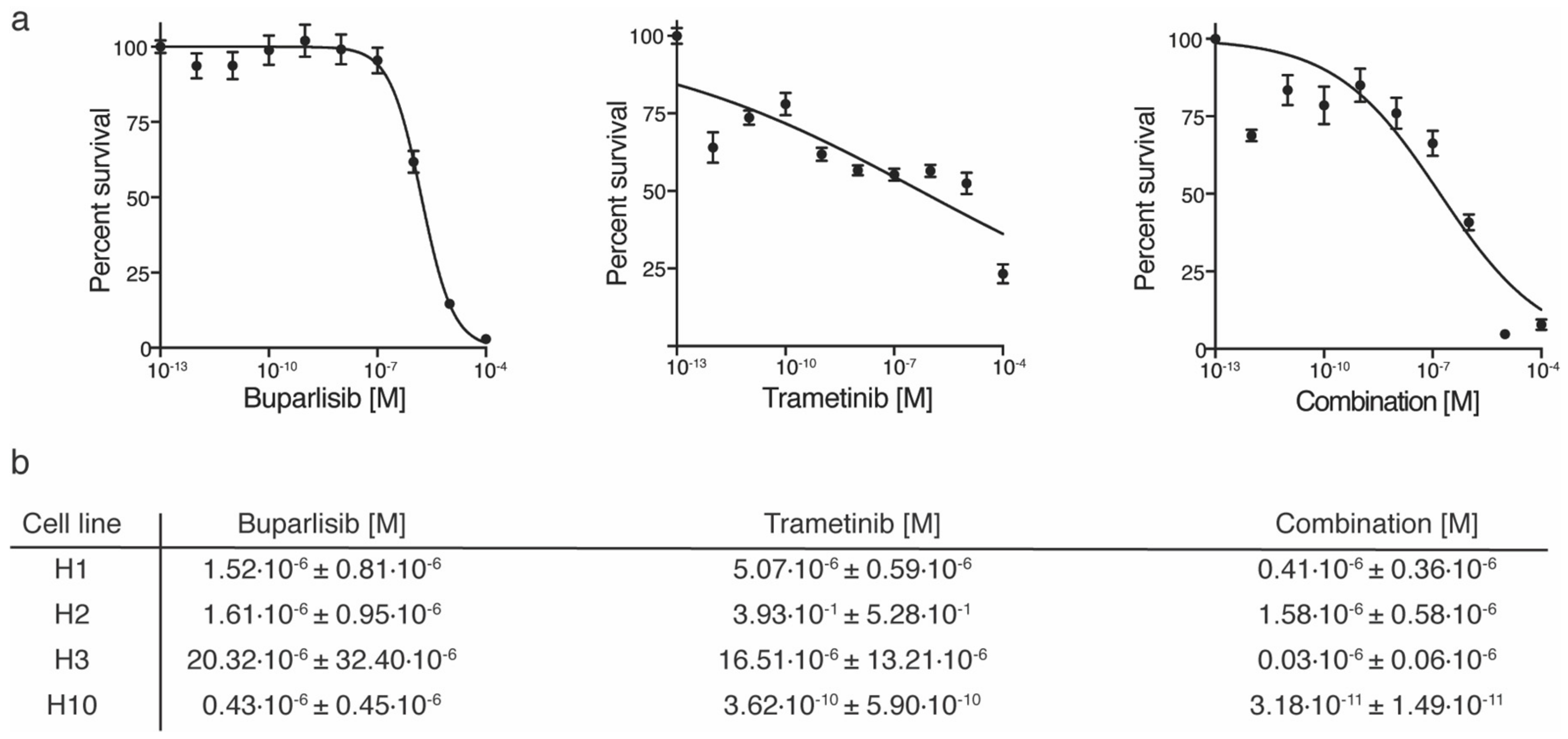
2.1. Combined Trametinib and Buparlisib Treatment Is More Effective Than Monotherapies In Vitro
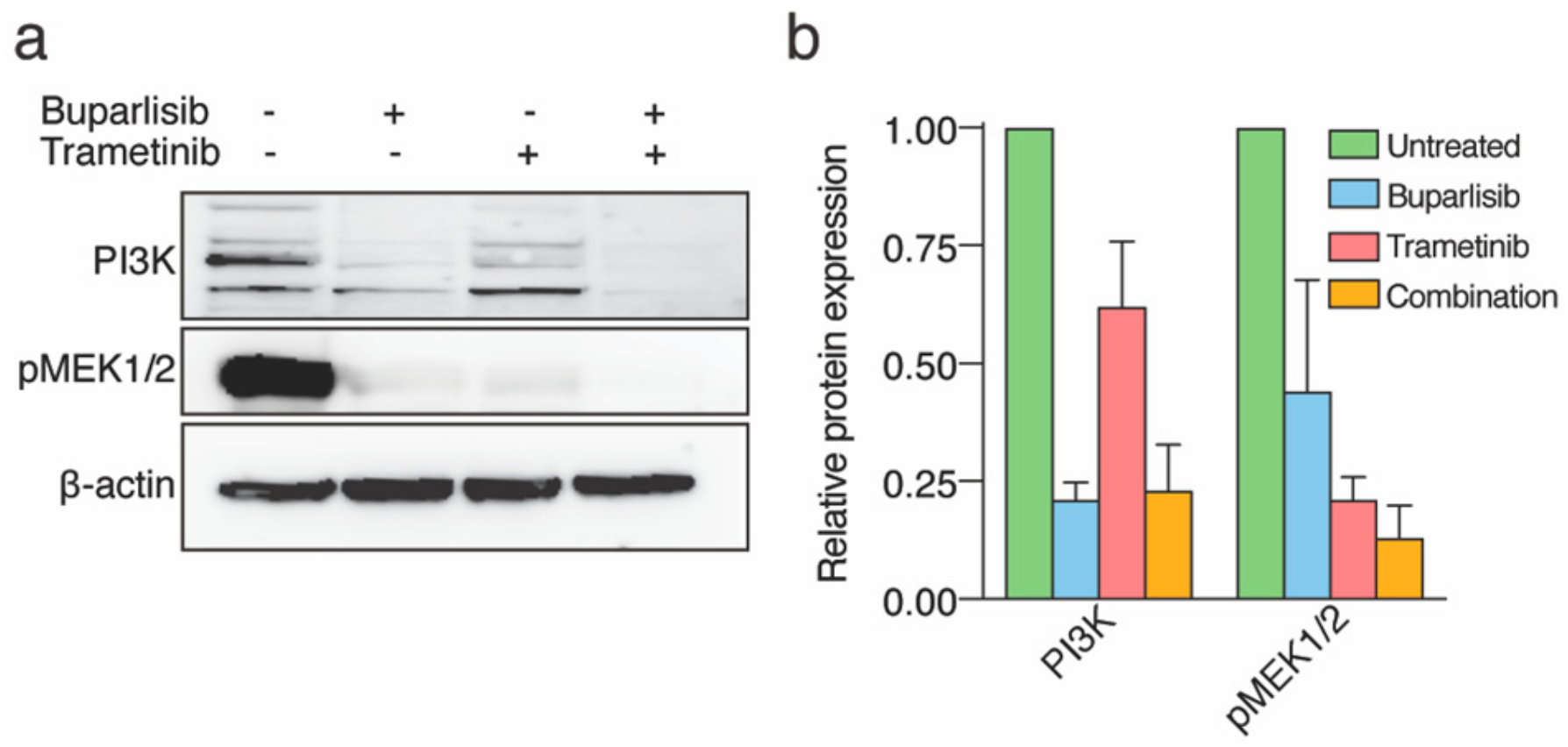
2.2. Treatment with Buparlisib and Trametinib Decreases Target Protein Expressions
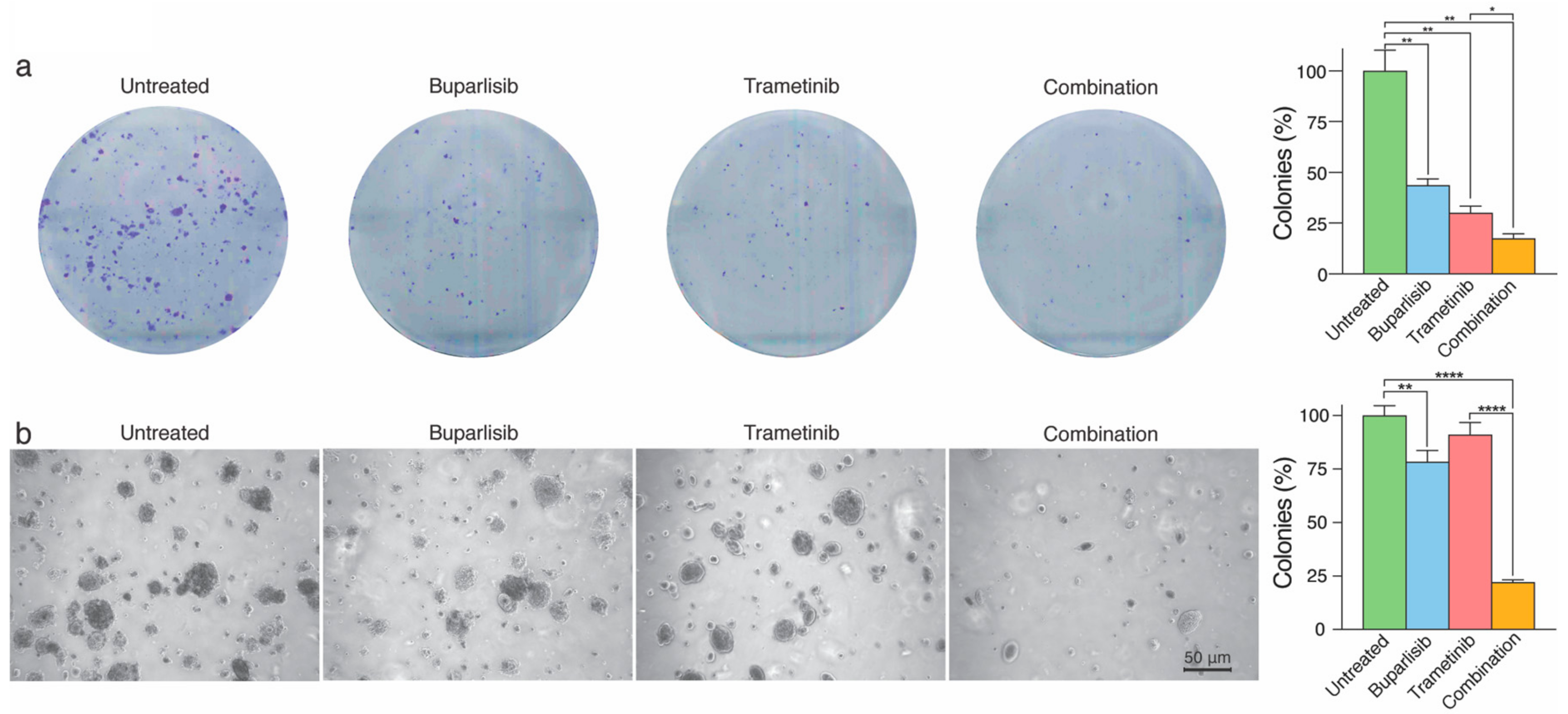
2.3. Combined Treatment Inhibits 2D and 3D Colony Formation More Effectively Than Single Drug Treatment
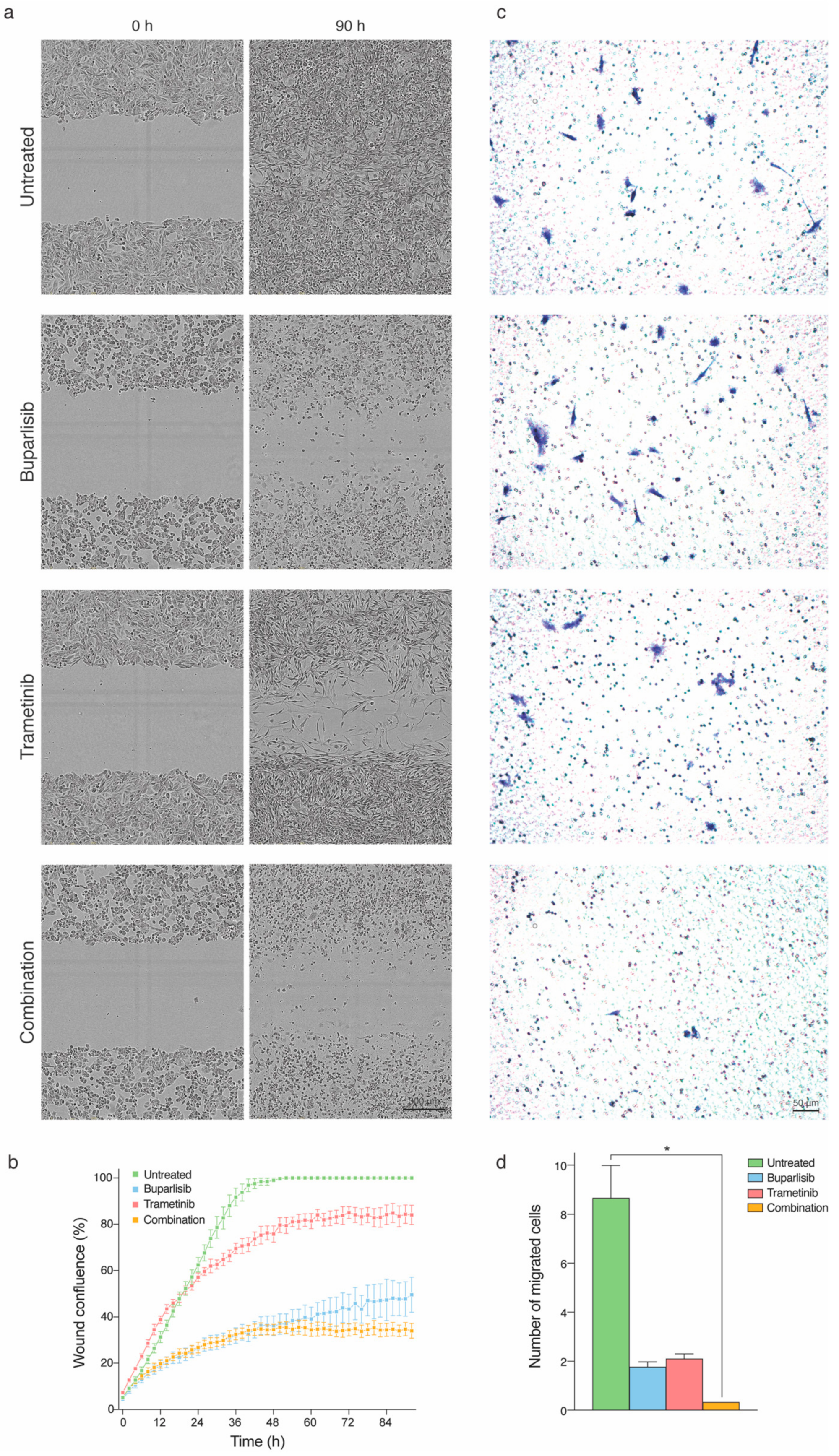
2.4. Tumor Cell Migration and Directional Cell Migration towards a Chemo-Attractant Is Hampered by Combination Treatment
2.5. Melanoma Cell Morphology Is Altered upon Combination Treatment
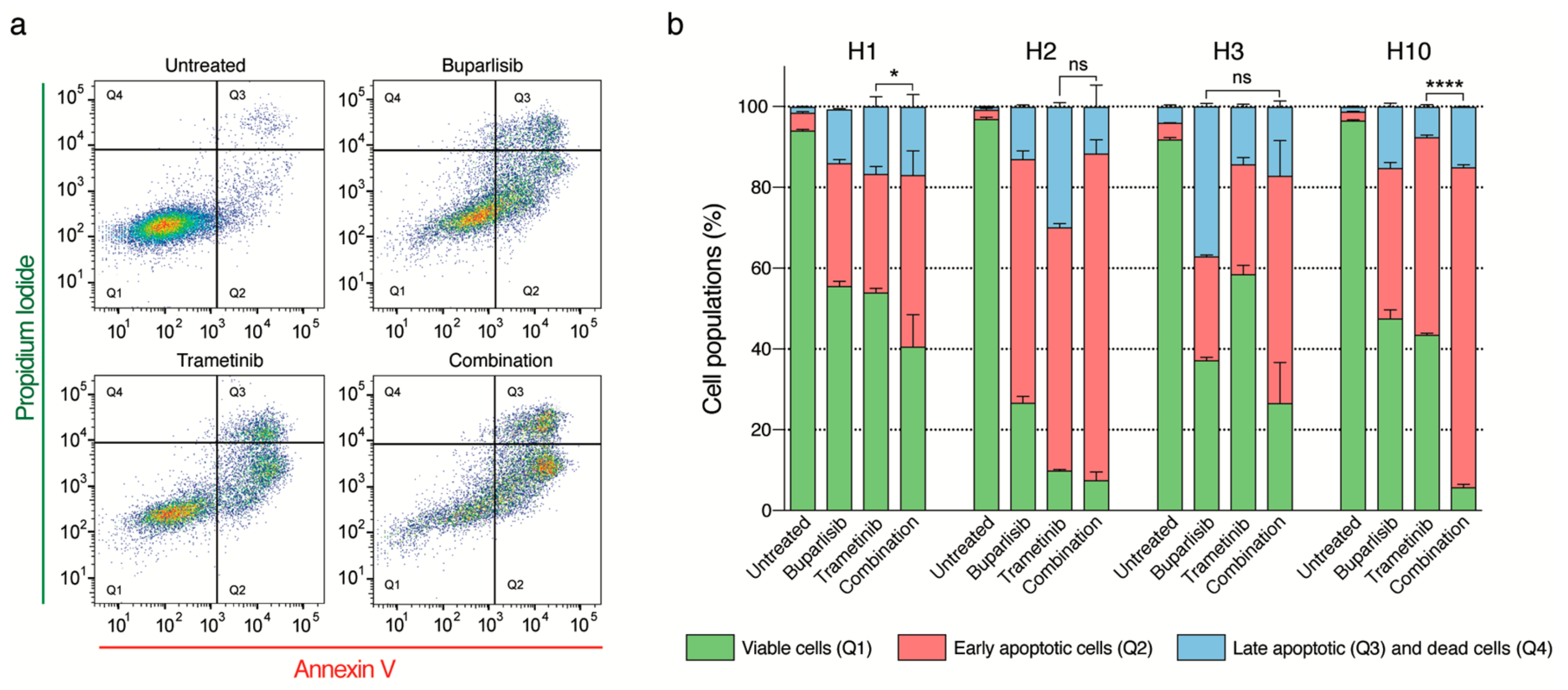
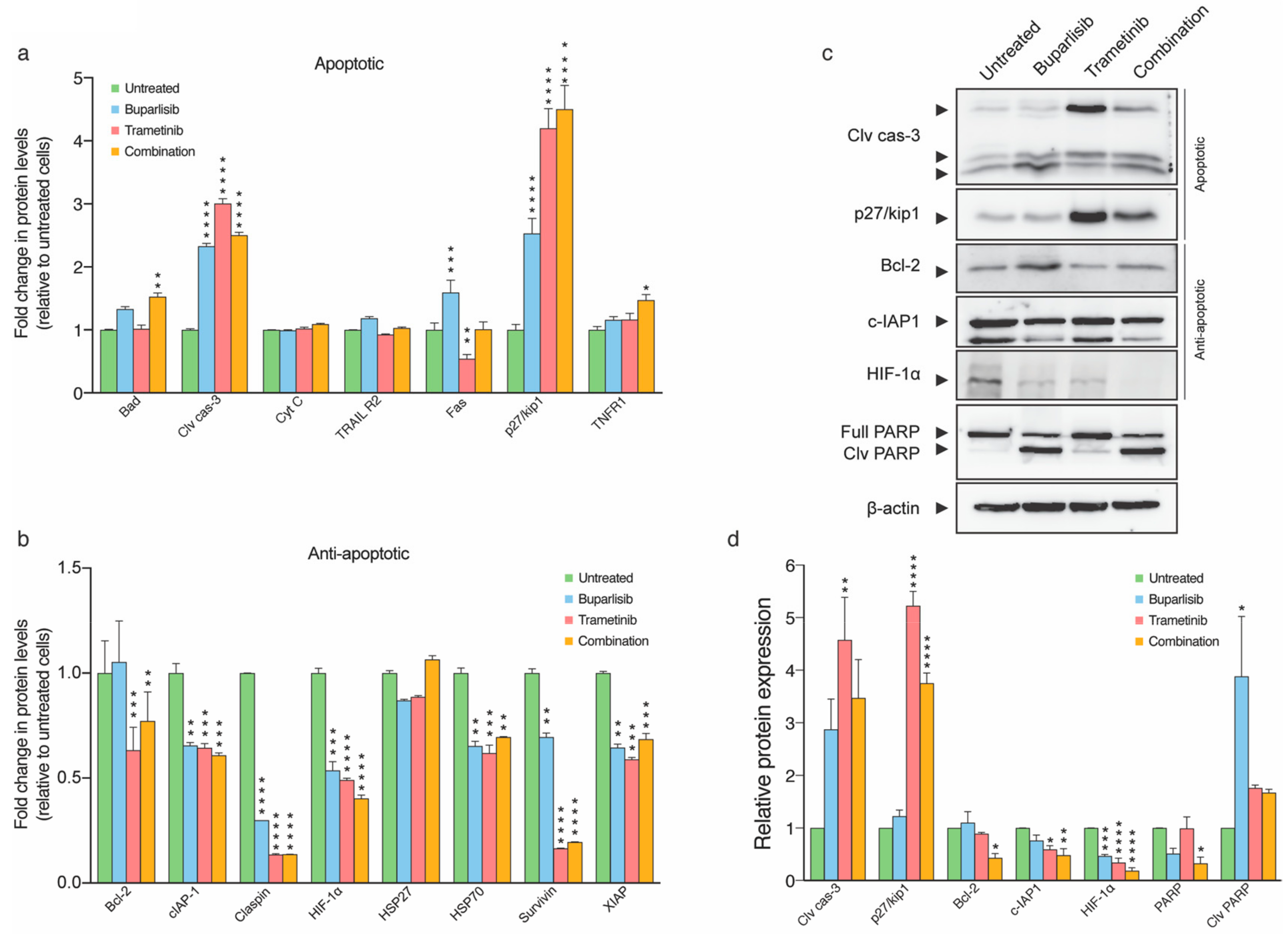
2.6. Combined Treatment Induces More Apoptosis Than Single Drug Treatment
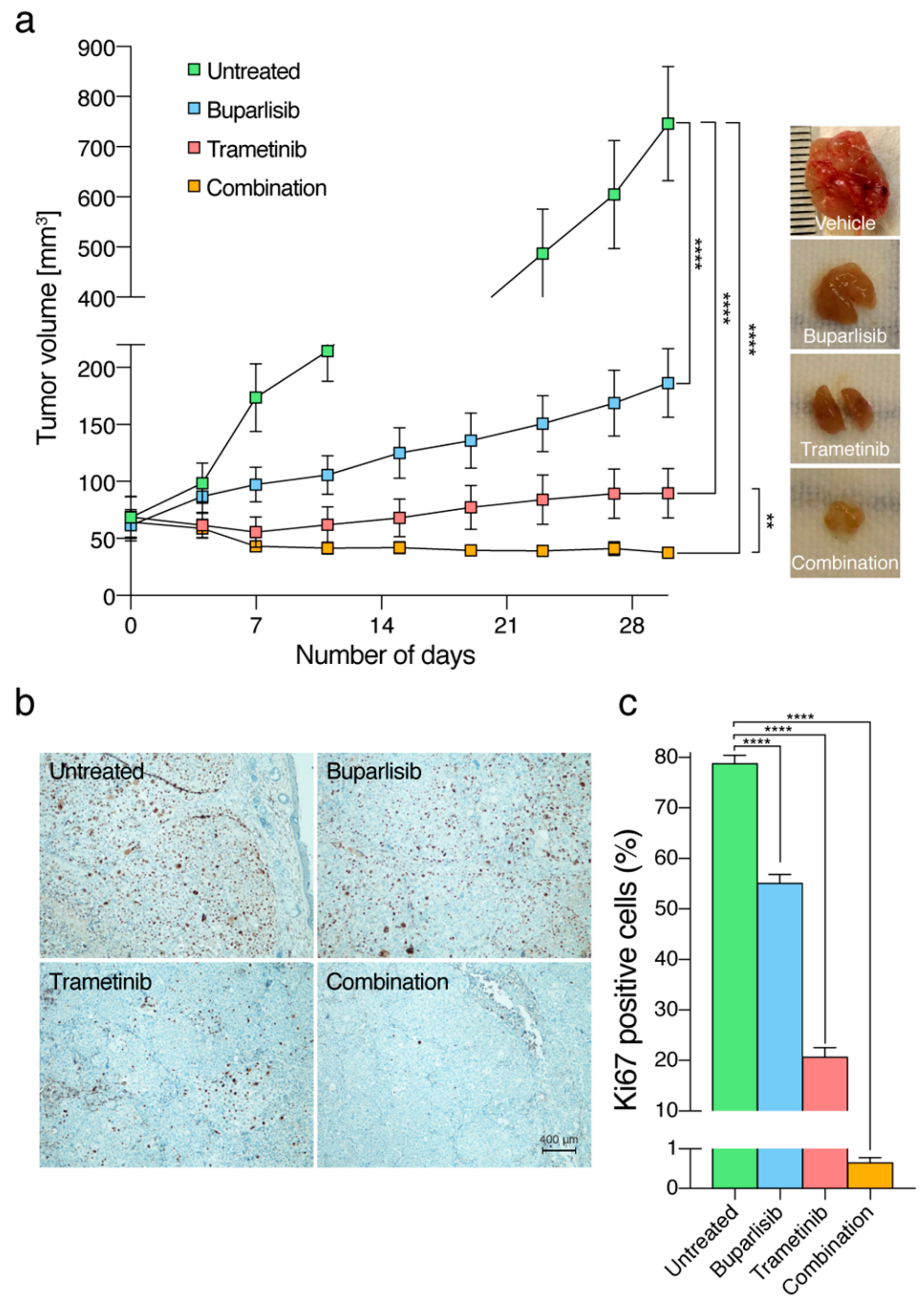
2.7. The Combinatorial Approach Is More Efficacious Than Single Treatments In Vivo
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Drugs
4.3. Animals
4.4. Cell Viability Assay
4.5. Clonogenic Assays
4.6. Cell Migration
4.7. Apoptosis Assay
4.8. Apoptosis Array
4.9. Western Blot Analysis
4.10. In Vivo Experiments
4.11. Histology and Immunohistochemistry
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BBB | Blood-brain barrier |
| BRAF | v-Raf murine sarcoma viral oncogene homolog |
| EMT | Epithelial-mesenchymal transition |
| ERK | Extracellular signal-regulated kinase |
| FDA | Food and Drug Administration |
| MAPK | Mitogen activated protein kinase |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MEK | Mitogen-activated protein kinase |
| NOD/SCID | Non-obese diabetic/severe compromised immunodeficiency |
| NRAS | Neuroblastoma RAS viral oncogene homolog |
| NSCLC | Non-small-cell lung cancer |
| PI | Propidium iodide |
| PI3K | Phosphoinositide 3-kinase |
| PTEN | Phosphatase and tensin homolog |
| STR | Short tandem repeat |
| TNFR1 | Tumor necrosis factor receptor 1 |
References
- Marks, R. Epidemiology of melanoma. Clin. Exp. Dermatol. 2000, 25, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Abate-Daga, D.; Ramello, M.C.; Smalley, I.; Forsyth, P.A.; Smalley, K.S.M. The biology and therapeutic management of melanoma brain metastases. Biochem. Pharmacol. 2018, 153, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Sundstrom, T.; Espedal, H.; Harter, P.N.; Fasmer, K.E.; Skaftnesmo, K.O.; Horn, S.; Hodneland, E.; Mittelbronn, M.; Weide, B.; Beschorner, R.; et al. Melanoma brain metastasis is independent of lactate dehydrogenase A expression. Neuro. Oncol. 2015, 17, 1374–1385. [Google Scholar] [CrossRef] [PubMed]
- Berghoff, A.S.; Preusser, M. New developments in brain metastases. Ther. Adv. Neurol. Disord. 2018, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Daphu, I.; Horn, S.; Stieber, D.; Varughese, J.; Spriet, E.; Dale, H.; Skaftnesmo, K.; Bjerkvig, R.; Thorsen, F. In Vitro Treatment of Melanoma Brain Metastasis by Simultaneously Targeting the MAPK and PI3K Signaling Pathways. Int. J. Mol. Sci. 2014, 15, 8773–8794. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghi, A.; Spandidos, D.A.; McCubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Fischer, G.M.; Vashisht Gopal, Y.N.; McQuade, J.L.; Peng, W.; DeBerardinis, R.J.; Davies, M.A. Metabolic strategies of melanoma cells: Mechanisms, interactions with the tumor microenvironment, and therapeutic implications. Pigment Cell Melanoma Res. 2018, 31, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Schittek, B.; Busch, S.; Garbe, C.; Smalley, K.; Satyamoorthy, K.; Li, G.; Herlyn, M. The RAS/RAF/MEK/ERK and PI3K/AKT signaling pathways present molecular targets for the effective treatment of advanced melanoma. Front. Biosci. 2005, 10, 2986–3001. [Google Scholar] [CrossRef] [PubMed]
- Isola, A.L.; Eddy, K.; Chen, S. Biology, Therapy and Implications of Tumor Exosomes in the Progression of Melanoma. Cancers (Basel) 2016, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.A. The role of the PI3K-AKT pathway in melanoma. Cancer J. 2012, 18, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Brastianos, P.K.; Carter, S.L.; Santagata, S.; Cahill, D.P.; Taylor-Weiner, A.; Jones, R.T.; Van Allen, E.M.; Lawrence, M.S.; Horowitz, P.M.; Cibulskis, K.; et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov. 2015, 5, 1164–1177. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Tardell, C.; Higgins, B.; Packman, K.; Boylan, J.F.; Niu, H. BRAFV600E negatively regulates the AKT pathway in melanoma cell lines. PLoS ONE 2012, 7, e42598. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Greger, J.G.; Eastman, S.D.; Zhang, V.; Bleam, M.R.; Hughes, A.M.; Smitheman, K.N.; Dickerson, S.H.; Laquerre, S.G.; Liu, L.; Gilmer, T.M. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol. Cancer Ther. 2012, 11, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Gilmartin, A.G.; Bleam, M.R.; Groy, A.; Moss, K.G.; Minthorn, E.A.; Kulkarni, S.G.; Rominger, C.M.; Erskine, S.; Fisher, K.E.; Yang, J.; et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 2011, 17, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Fung, C.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Hyman, J.; Shahheydari, H.; Tembe, V.; Thompson, J.F.; Saw, R.P.; et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 2014, 5, 5694. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, K.T.; Robert, C.; Hersey, P.; Nathan, P.; Garbe, C.; Milhem, M.; Demidov, L.V.; Hassel, J.C.; Rutkowski, P.; Mohr, P.; et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N. Engl. J. Med. 2012, 367, 107–114. [Google Scholar] [CrossRef]
- Yap, T.A.; Bjerke, L.; Clarke, P.A.; Workman, P. Drugging PI3K in cancer: Refining targets and therapeutic strategies. Curr. Opin. Pharmacol. 2015, 23, 98–107. [Google Scholar] [CrossRef]
- Engelman, J.A. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat. Rev. Cancer 2009, 9, 550–562. [Google Scholar] [CrossRef] [PubMed]
- Maira, S.M.; Pecchi, S.; Huang, A.; Burger, M.; Knapp, M.; Sterker, D.; Schnell, C.; Guthy, D.; Nagel, T.; Wiesmann, M.; et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol. Cancer Ther. 2012, 11, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, S.M.; Kleylein-Sohn, J.; Gaulis, S.; Kauffmann, A.; Blommers, M.J.; Kazic-Legueux, M.; Laborde, L.; Hattenberger, M.; Stauffer, F.; Vaxelaire, J.; et al. Characterization of the mechanism of action of the pan class I PI3K inhibitor NVP-BKM120 across a broad range of concentrations. Mol. Cancer Ther. 2012, 11, 1747–1757. [Google Scholar] [CrossRef] [PubMed]
- Ando, Y.; Inada-Inoue, M.; Mitsuma, A.; Yoshino, T.; Ohtsu, A.; Suenaga, N.; Sato, M.; Kakizume, T.; Robson, M.; Quadt, C.; et al. Phase I dose-escalation study of buparlisib (BKM120), an oral pan-class I PI3K inhibitor, in Japanese patients with advanced solid tumors. Cancer Sci. 2014, 105, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, J.; Vultur, A.; Lee, J.T.; Somasundaram, R.; Fukunaga-Kalabis, M.; Cipolla, A.K.; Wubbenhorst, B.; Xu, X.; Gimotty, P.A.; Kee, D.; et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 2010, 18, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.N.; Kong, L.R.; Sim, W.J.; Ng, H.C.; Ong, W.R.; Thiery, J.P.; Huynh, H.; Goh, B.C. Combinatorial treatment using targeted MEK and SRC inhibitors synergistically abrogates tumor cell growth and induces mesenchymal-epithelial transition in non-small-cell lung carcinoma. Oncotarget 2015, 6, 29991–30005. [Google Scholar] [CrossRef]
- Speranza, M.C.; Nowicki, M.O.; Behera, P.; Cho, C.F.; Chiocca, E.A.; Lawler, S.E. BKM-120 (Buparlisib): A Phosphatidyl-Inositol-3 Kinase Inhibitor with Anti-Invasive Properties in Glioblastoma. Sci. Rep. 2016, 6, 20189. [Google Scholar] [CrossRef]
- Cordaro, F.G.; De Presbiteris, A.L.; Camerlingo, R.; Mozzillo, N.; Pirozzi, G.; Cavalcanti, E.; Manca, A.; Palmieri, G.; Cossu, A.; Ciliberto, G.; et al. Phenotype characterization of human melanoma cells resistant to dabrafenib. Oncol. Rep. 2017, 38, 2741–2751. [Google Scholar] [CrossRef]
- Bedard, P.L.; Tabernero, J.; Janku, F.; Wainberg, Z.A.; Paz-Ares, L.; Vansteenkiste, J.; Van Cutsem, E.; Perez-Garcia, J.; Stathis, A.; Britten, C.D.; et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin. Cancer Res. 2015, 21, 730–738. [Google Scholar] [CrossRef]
- Wee, S.; Jagani, Z.; Xiang, K.X.; Loo, A.; Dorsch, M.; Yao, Y.M.; Sellers, W.R.; Lengauer, C.; Stegmeier, F. PI3K pathway activation mediates resistance to MEK inhibitors in KRAS mutant cancers. Cancer Res. 2009, 69, 4286–4293. [Google Scholar] [CrossRef]
- Sinnberg, T.; Lasithiotakis, K.; Niessner, H.; Schittek, B.; Flaherty, K.T.; Kulms, D.; Maczey, E.; Campos, M.; Gogel, J.; Garbe, C.; et al. Inhibition of PI3K-AKT-mTOR signaling sensitizes melanoma cells to cisplatin and temozolomide. J. Investig. Dermatol. 2009, 129, 1500–1515. [Google Scholar] [CrossRef] [PubMed]
- Niessner, H.; Schmitz, J.; Tabatabai, G.; Schmid, A.M.; Calaminus, C.; Sinnberg, T.; Weide, B.; Eigentler, T.K.; Garbe, C.; Schittek, B.; et al. PI3K Pathway Inhibition Achieves Potent Antitumor Activity in Melanoma Brain Metastases In Vitro and In Vivo. Clin. Cancer Res. 2016, 22, 5818–5828. [Google Scholar] [CrossRef] [PubMed]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology (Bethesda) 2017, 32, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Lintz, M.; Munoz, A.; Reinhart-King, C.A. The Mechanics of Single Cell and Collective Migration of Tumor Cells. J. Biomech. Eng. 2017, 139, 021005. [Google Scholar] [CrossRef] [PubMed]
- Justus, C.R.; Leffler, N.; Ruiz-Echevarria, M.; Yang, L.V. In vitro cell migration and invasion assays. J. Vis. Exp. 2014, e51046. [Google Scholar]
- Zhao, Y.; Ge, C.C.; Wang, J.; Wu, X.X.; Li, X.M.; Li, W.; Wang, S.S.; Liu, T.; Hou, J.Z.; Sun, H.; et al. MEK inhibitor, PD98059, promotes breast cancer cell migration by inducing beta-catenin nuclear accumulation. Oncol. Rep. 2017, 38, 3055–3063. [Google Scholar] [CrossRef] [PubMed]
- Mabuchi, S.; Ohmichi, M.; Kimura, A.; Hisamoto, K.; Hayakawa, J.; Nishio, Y.; Adachi, K.; Takahashi, K.; Arimoto-Ishida, E.; Nakatsuji, Y.; et al. Inhibition of phosphorylation of BAD and Raf-1 by Akt sensitizes human ovarian cancer cells to paclitaxel. J. Biol. Chem. 2002, 277, 33490–33500. [Google Scholar] [CrossRef]
- Huang, Q.; Li, F.; Liu, X.; Li, W.; Shi, W.; Liu, F.F.; O’Sullivan, B.; He, Z.; Peng, Y.; Tan, A.C.; et al. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radiotherapy. Nat. Med. 2011, 17, 860–866. [Google Scholar] [CrossRef]
- Motti, M.L.; Califano, D.; Troncone, G.; De Marco, C.; Migliaccio, I.; Palmieri, E.; Pezzullo, L.; Palombini, L.; Fusco, A.; Viglietto, G. Complex regulation of the cyclin-dependent kinase inhibitor p27kip1 in thyroid cancer cells by the PI3K/AKT pathway: Regulation of p27kip1 expression and localization. Am. J. Pathol. 2005, 166, 737–749. [Google Scholar] [CrossRef]
- Haydn, J.M.; Hufnagel, A.; Grimm, J.; Maurus, K.; Schartl, M.; Meierjohann, S. The MAPK pathway as an apoptosis enhancer in melanoma. Oncotarget 2014, 5, 5040–5053. [Google Scholar] [CrossRef]
- Siddiqa, A.; Long, L.M.; Li, L.; Marciniak, R.A.; Kazhdan, I. Expression of HER-2 in MCF-7 breast cancer cells modulates anti-apoptotic proteins Survivin and Bcl-2 via the extracellular signal-related kinase (ERK) and phosphoinositide-3 kinase (PI3K) signalling pathways. BMC Cancer 2008, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shi, Y.; Giranda, V.L.; Luo, Y. Inhibition of the phosphatidylinositol 3-kinase/Akt pathway sensitizes MDA-MB468 human breast cancer cells to cerulenin-induced apoptosis. Mol. Cancer Ther. 2006, 5, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.; Klinger, V.; Keksel, C.; Bonigut, V.; Kiefer, D.; Caspers, J.; Walther, J.; Wos-Maganga, M.; Weickhardt, S.; Rohn, G.; et al. Inhibition of the PI3K but not the MEK/ERK pathway sensitizes human glioma cells to alkylating drugs. Cancer Cell Int. 2018, 18, 69. [Google Scholar] [CrossRef] [PubMed]
- Powis, G.; Kirkpatrick, L. Hypoxia inducible factor-1alpha as a cancer drug target. Mol. Cancer Ther. 2004, 3, 647–654. [Google Scholar] [PubMed]
- Ramos, A.M.; Fernandez, C.; Amran, D.; Sancho, P.; de Blas, E.; Aller, P. Pharmacologic inhibitors of PI3K/Akt potentiate the apoptotic action of the antileukemic drug arsenic trioxide via glutathione depletion and increased peroxide accumulation in myeloid leukemia cells. Blood 2005, 105, 4013–4020. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, M.; Andrulis, M.; Stuhmer, T.; Muller, E.; Hofmann, C.; Steinbrunn, T.; Heimberger, T.; Schraud, H.; Kressmann, S.; Einsele, H.; et al. The PI3K/Akt signaling pathway regulates the expression of Hsp70, which critically contributes to Hsp90-chaperone function and tumor cell survival in multiple myeloma. Haematologica 2013, 98, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Horinaka, M.; Shinnoh, M.; Yoshioka, T.; Miki, T.; Sakai, T. A novel HDAC inhibitor OBP-801 and a PI3K inhibitor LY294002 synergistically induce apoptosis via the suppression of survivin and XIAP in renal cell carcinoma. Int. J. Oncol. 2013, 43, 1080–1086. [Google Scholar] [CrossRef]
- Altieri, D.C. Survivin, cancer networks and pathway-directed drug discovery. Nat. Rev. Cancer 2008, 8, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Boucher, M.J.; Morisset, J.; Vachon, P.H.; Reed, J.C.; Laine, J.; Rivard, N. MEK/ERK signaling pathway regulates the expression of Bcl-2, Bcl-X(L), and Mcl-1 and promotes survival of human pancreatic cancer cells. J. Cell Biochem. 2000, 79, 355–369. [Google Scholar] [CrossRef]
- Volm, M.; Koomagi, R. Hypoxia-inducible factor (HIF-1) and its relationship to apoptosis and proliferation in lung cancer. Anticancer Res. 2000, 20, 1527–1533. [Google Scholar]
- Huang, J.; Lyu, H.; Wang, J.; Liu, B. MicroRNA regulation and therapeutic targeting of survivin in cancer. Am. J. Cancer Res. 2015, 5, 20–31. [Google Scholar] [PubMed]
- Boulares, A.H.; Yakovlev, A.G.; Ivanova, V.; Stoica, B.A.; Wang, G.; Iyer, S.; Smulson, M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. J. Biol. Chem. 1999, 274, 22932–22940. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Yoshida, T.; Kurachi, R.; Kakegawa, J.; Hori, Y.; Nanayama, T.; Hayakawa, K.; Abe, H.; Takagi, K.; Matsuzaki, Y.; et al. Identification of JTP-70902, a p15(INK4b)-inductive compound, as a novel MEK1/2 inhibitor. Cancer Sci. 2007, 98, 1809–1816. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.S.; Flaherty, K.T.; Infante, J.R.; Falchook, G.S.; Kefford, R.; Daud, A.; Hamid, O.; Gonzalez, R.; Kudchadkar, R.R.; Lawrence, D.P.; et al. Updated safety and efficacy results from a phase I/II study of the oral BRAF inhibitor dabrafenib (GSK2118436) combined with the oral MEK 1/2 inhibitor trametinib (GSK1120212) in patients with BRAFi-naive metastatic melanoma. J. Clin. Oncol. 2012, 30, 8510. [Google Scholar]
- Bissig-Choisat, B.; Kettlun-Leyton, C.; Legras, X.D.; Zorman, B.; Barzi, M.; Chen, L.L.; Amin, M.D.; Huang, Y.H.; Pautler, R.G.; Hampton, O.A.; et al. Novel patient-derived xenograft and cell line models for therapeutic testing of pediatric liver cancer. J. Hepatol. 2016, 65, 325–333. [Google Scholar] [CrossRef] [PubMed]
- De Gooijer, M.C.; Zhang, P.; Buil, L.C.M.; Citirikkaya, C.H.; Thota, N.; Beijnen, J.H.; van Tellingen, O. Buparlisib is a brain penetrable pan-PI3K inhibitor. Sci. Rep. 2018, 8, 10784. [Google Scholar] [CrossRef] [PubMed]
- Vaidhyanathan, S.; Mittapalli, R.K.; Sarkaria, J.N.; Elmquist, W.F. Factors influencing the CNS distribution of a novel MEK-1/2 inhibitor: Implications for combination therapy for melanoma brain metastases. Drug Metab. Dispos. 2014, 42, 1292–1300. [Google Scholar] [CrossRef] [PubMed]
- Eichler, A.F.; Chung, E.; Kodack, D.P.; Loeffler, J.S.; Fukumura, D.; Jain, R.K. The biology of brain metastases-translation to new therapies. Nat. Rev. Clin. Oncol. 2011, 8, 344–356. [Google Scholar] [CrossRef]
- Bischof, K.; Knappskog, S.; Hjelle, S.M.; Stefansson, I.; Woie, K.; Salvesen, H.B.; Gjertsen, B.T.; Bjorge, L. Influence of p53 Isoform Expression on Survival in High-Grade Serous Ovarian Cancers. Sci. Rep. 2019, 9, 5244. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nymark Aasen, S.; Parajuli, H.; Hoang, T.; Feng, Z.; Stokke, K.; Wang, J.; Roy, K.; Bjerkvig, R.; Knappskog, S.; Thorsen, F. Effective Treatment of Metastatic Melanoma by Combining MAPK and PI3K Signaling Pathway Inhibitors. Int. J. Mol. Sci. 2019, 20, 4235. https://doi.org/10.3390/ijms20174235
Nymark Aasen S, Parajuli H, Hoang T, Feng Z, Stokke K, Wang J, Roy K, Bjerkvig R, Knappskog S, Thorsen F. Effective Treatment of Metastatic Melanoma by Combining MAPK and PI3K Signaling Pathway Inhibitors. International Journal of Molecular Sciences. 2019; 20(17):4235. https://doi.org/10.3390/ijms20174235
Chicago/Turabian StyleNymark Aasen, Synnøve, Himalaya Parajuli, Tuyen Hoang, Zichao Feng, Krister Stokke, Jiwei Wang, Kislay Roy, Rolf Bjerkvig, Stian Knappskog, and Frits Thorsen. 2019. "Effective Treatment of Metastatic Melanoma by Combining MAPK and PI3K Signaling Pathway Inhibitors" International Journal of Molecular Sciences 20, no. 17: 4235. https://doi.org/10.3390/ijms20174235
APA StyleNymark Aasen, S., Parajuli, H., Hoang, T., Feng, Z., Stokke, K., Wang, J., Roy, K., Bjerkvig, R., Knappskog, S., & Thorsen, F. (2019). Effective Treatment of Metastatic Melanoma by Combining MAPK and PI3K Signaling Pathway Inhibitors. International Journal of Molecular Sciences, 20(17), 4235. https://doi.org/10.3390/ijms20174235





