Drought Response in Rice: The miRNA Story
Abstract
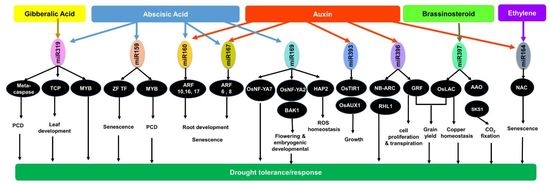
:1. Introduction
2. Hormonal Regulation of Drought
2.1. Abscisic Acid (ABA) in Drought Stress Signaling
2.2. Auxin in Drought Stress Signaling
3. The Role of miRNA in Growth and Development during Drought
4. The Role of miRNAs on Photosynthesis and Respiration during Drought
5. The Role of miRNAs in Stress Modulation
5.1. miRNAs and Senescence
5.2. miRNAs in Antioxidant Defense
5.3. miRNA in Osmotic Stress during Drought
6. Differences and Similarities in Drought Response between Rice and Other Plant Systems
6.1. Hormones Are Central to Drought Regulation
6.2. The Semi-Aquatic Rice Versus Terrestrial Plants
7. Conclusions and Future Prospects
Funding
Conflicts of Interest
References
- Baldrich, P.; Campo, S.; Wu, M.T.; Liu, T.T.; Hsing, Y.I.C.; Segundo, B.S. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 2015, 12, 847–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebert, L.F.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2018, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Djami-Tchatchou, A.T.; Sanan-Mishra, N.; Ntushelo, K.; Dubery, I.A. Functional roles of microRNAs in agronomically important plants—potential as targets for crop improvement and protection. Front. Plant Sci. 2017, 8, 378. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Pan, X.; Najeeb, U.; Tan, D.K.Y.; Fahad, S.; Zahoor, R.; Luo, H. Coping with drought: Stress and adaptive mechanisms, and management through cultural and molecular alternatives in cotton as vital constituents for plant stress resilience and fitness. Biol. Res. 2018, 51, 47. [Google Scholar] [CrossRef] [PubMed]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17, 1712. [Google Scholar] [CrossRef] [PubMed]
- Mittal, D.; Sharma, N.; Sharma, V.; Sopory, S.; Sanan-Mishra, N. Role of microRNAs in rice plant under salt stress. Ann. Biol. 2016, 168, 2–18. [Google Scholar] [CrossRef]
- Willmann, M.R.; Poethig, R.S. Conservation and evolution of miRNA regulatory programs in plant development. Curr. Opin. Plant Biol. 2007, 10, 503–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samad, A.F.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.; Zainal, Z.; Ismail, I. MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 2017, 8, 565. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, J.; Yan, J.; Gou, F.; Mao, Y.; Tang, G.; Botella, J.R.; Zhu, J.K. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl. Acad. Sci. USA 2017, 114, 5277–5282. [Google Scholar] [CrossRef] [Green Version]
- Pagliarani, C.; Vitali, M.; Ferrero, M.; Vitulo, N.; Incarbone, M.; Lovisolo, C.; Valle, G.; Schubert, A. The accumulation of miRNAs differentially modulated by drought stress is affected by grafting in grapevine. Plant Physiol. 2017, 173, 2180–2195. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Yi, F.; Xie, S.; Liu, Y.; Qi, X.; Yu, J. Genome-wide characterization of microRNA in foxtail millet (Setaria italica). BMC Plant Biol. 2013, 13, 212. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sandhu, N.; Dixit, S.; Yadav, S.; Swamy, B.; Shamsudin, N.A.A. Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice 2018, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Rahim, H.; Bhuiyan, M.; Lim, L.; Sabu, K.; Saad, A.; Azhar, M.; Wickneswari, R. Identification of quantitative trait loci for blast resistance in BC. Genet. Mol. Res. 2012, 11, 3277–3289. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zeng, T.; Xia, Q.; Qian, Q.; Yang, C.; Ding, Y.; Chen, L.; Wang, W. Unravelling miRNA regulation in yield of rice (Oryza sativa) based on differential network model. Sci. Rep. 2018, 8, 8498. [Google Scholar] [CrossRef] [PubMed]
- Balyan, S.; Kumar, M.; Mutum, R.D.; Raghuvanshi, U.; Agarwal, P.; Mathur, S.; Raghuvanshi, S. Identification of miRNA-mediated drought responsive multi-tiered regulatory network in drought tolerant rice, Nagina 22. Sci. Rep. 2017, 7, 15446. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Cai, Z.; Gan, S. Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 2004, 27, 521–549. [Google Scholar] [CrossRef]
- Yue, E.; Liu, Z.; Li, C.; Li, Y.; Liu, Q.; Xu, J.H. Overexpression of miR529a confers enhanced resistance to oxidative stress in rice (Oryza sativa L.). Plant Cell Rep. 2017, 36, 1171–1182. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cannon, C.H.; Cobb, G.P.; Anderson, T.A. Conservation and divergence of plant microRNA genes. Plant J. 2006, 46, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Daszkowska-Golec, A.; Szarejko, I. The molecular basis of ABA-mediated plant response to drought. In Abiotic Stress-Plant Responses and Applications in Agriculture; IntechOpen: London, UK, 2013. [Google Scholar]
- Khan, S.A.; Li, M.Z.; Wang, S.M.; Yin, H.J. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Pramanik, K.; Sharma, R.; Gantait, S.; Banerjee, J. In-silico study of biotic and abiotic stress-related transcription factor binding sites in the promoter regions of rice germin-like protein genes. PLoS ONE 2019, 14, e0211887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Yuan, L.J.; Shao, Y.; Du, W.; Yan, D.W.; Lu, Y.T. The disturbance of small RNA pathways enhanced abscisic acid response and multiple stress responses in Arabidopsis. Plant Cell Environ. 2008, 31, 562–574. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Shao, C.; Wang, H.; Chen, M. The regulatory activities of plant microRNAs: A more dynamic perspective. Plant Physiol. 2011, 157, 1583–1595. [Google Scholar] [CrossRef] [PubMed]
- Cheah, B.H.; Jadhao, S.; Vasudevan, M.; Wickneswari, R.; Nadarajah, K. Identification of functionally important microRNAs from rice inflorescence at heading stage of a qDTY4. 1-QTL bearing Near Isogenic Line under drought conditions. PLoS ONE 2017, 12, e0186382. [Google Scholar] [CrossRef] [PubMed]
- El-Kereamy, A.; Bi, Y.M.; Ranathunge, K.; Beatty, P.H.; Good, A.G.; Rothstein, S.J. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS ONE 2012, 7, e52030. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wen, H.; Teotia, S.; Du, Y.; Zhang, J.; Li, J.; Sun, H.; Tang, G.; Peng, T.; Zhao, Q. Suppression of microRNA159 impacts multiple agronomic traits in rice (Oryza sativa L.). BMC Plant Biol. 2017, 17, 215. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Micro RNAs and short-interfering RNAs in plants. J. Integr. Plant Biol. 2007, 49, 817–826. [Google Scholar] [CrossRef]
- Mohsenifard, E.; Ghabooli, M.; Mehri, N.; Bakhshi, B. Regulation of miR159 and miR396 mediated by Piriformospora indica confer drought tolerance in rice. J. Plant Mol. Breed. 2017, 5, 10–18. [Google Scholar]
- Reyes, J.L.; Chua, N.H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007, 49, 592–606. [Google Scholar] [CrossRef]
- Lee, J.H.; Terzaghi, W.; Deng, X.W. DWA3, an Arabidopsis DWD protein, acts as a negative regulator in ABA signal transduction. Plant Sci. 2011, 180, 352–357. [Google Scholar] [CrossRef]
- Bakhshi, B.; Fard, E.M.; Nikpay, N.; Ebrahimi, M.A.; Bihamta, M.R.; Mardi, M.; Salekdeh, G.H. MicroRNA signatures of drought signaling in rice root. PLoS ONE 2016, 11, e0156814. [Google Scholar] [CrossRef]
- Tian, C.; Zuo, Z.; Qiu, J.-L. Identification and characterization of ABA-responsive microRNAs in rice. J. Genet. Genom. 2015, 42, 393–402. [Google Scholar] [CrossRef]
- Xie, F.; Zhang, B. micro RNA evolution and expression analysis in polyploidized cotton genome. Plant Biotechnol. J. 2015, 13, 421–434. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, Y.C.; Wang, C.Y.; Luo, Y.C.; Huang, Q.J.; Chen, S.Y.; Zhou, H.; Qu, L.H.; Chen, Y.Q. Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett. 2009, 583, 723–728. [Google Scholar] [CrossRef] [Green Version]
- Pandey, B.; Gupta, O.P.; Pandey, D.M.; Sharma, I.; Sharma, P. Identification of new stress-induced microRNA and their targets in wheat using computational approach. Plant Signal. Behav. 2013, 8, e23932. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Able, A.J.; Able, J.A. SMARTER de-stressed cereal breeding. Trends Plant Sci. 2016, 21, 909–925. [Google Scholar] [CrossRef]
- Wei, L.; Zhang, D.; Xiang, F.; Zhang, Z. Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int. J. Plant Sci. 2009, 170, 979–989. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, H.; Zhang, Q.; Li, M.; Yan, M.; Wang, R.; Wang, L.; Welti, R.; Zhang, W.; Wang, X. Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 2009, 21, 2357–2377. [Google Scholar] [CrossRef]
- Ding, Y.; Tao, Y.; Zhu, C. Emerging roles of microRNAs in the mediation of drought stress response in plants. J. Exp. Bot. 2013, 64, 3077–3086. [Google Scholar] [CrossRef]
- Wen, M.; Xie, M.; He, L.; Wang, Y.; Shi, S.; Tang, T. Expression variations of miRNAs and mRNAs in rice (Oryza sativa). Genome Biol. Evol. 2016, 8, 3529–3544. [Google Scholar] [CrossRef]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef]
- Zhang, X.; Zou, Z.; Gong, P.; Zhang, J.; Ziaf, K.; Li, H.; Xiao, F.; Ye, Z. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 2011, 33, 403–409. [Google Scholar] [CrossRef]
- Wang, T.; Chen, L.; Zhao, M.; Tian, Q.; Zhang, W.H. Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genom. 2011, 12, 367. [Google Scholar] [CrossRef]
- Lee, D.K.; Kim, H.I.; Jang, G.; Chung, P.J.; Jeong, J.S.; Kim, Y.S.; Bang, S.W.; Jung, H.; Do Choi, Y.; Kim, J.K. The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci. 2015, 241, 199–210. [Google Scholar] [CrossRef]
- Macovei, A.; Gill, S.S.; Tuteja, N. microRNAs as promising tools for improving stress tolerance in rice. Plant Signal. Behav. 2012, 7, 1296–1301. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Liang, R.; Ge, L.; Li, W.; Xiao, H.; Lin, H.; Ruan, K.; Jin, Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007, 354, 585–590. [Google Scholar] [CrossRef]
- Gasparis, S.; Yanushevska, Y.; Nadolska-Orczyk, A. Bioinformatic identification and expression analysis of new microRNAs from wheat (Triticum aestivum L.). Acta Physiol. Plant. 2017, 39, 236. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef]
- Windels, D.; Bielewicz, D.; Ebneter, M.; Jarmolowski, A.; Szweykowska-Kulinska, Z.; Vazquez, F. miR393 is required for production of proper auxin signalling outputs. PLoS ONE 2014, 9, e95972. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, Y.; Liu, Z.; Kong, D.; Duan, M.; Luo, L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 2010, 61, 4157–4168. [Google Scholar] [CrossRef]
- Zhou, M.; Luo, H. Role of microRNA319 in creeping bentgrass salinity and drought stress response. Plant Signal. Behav. 2014, 9, 1375–1391. [Google Scholar] [CrossRef]
- Liu, D.; Song, Y.; Chen, Z.; Yu, D. Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis. Physiol. Plant. 2009, 136, 223–236. [Google Scholar] [CrossRef]
- Chaves, M.; Oliveira, M. Mechanisms underlying plant resilience to water deficits: Prospects for water-saving agriculture. J. Exp. Bot. 2004, 55, 2365–2384. [Google Scholar] [CrossRef]
- Sorin, C.; Bussell, J.D.; Camus, I.; Ljung, K.; Kowalczyk, M.; Geiss, G.; McKhann, H.; Garcion, C.; Vaucheret, H.; Sandberg, G. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 2005, 17, 1343–1359. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef]
- Huang, J.; Si, W.; Deng, Q.; Li, P.; Yang, S. Rapid evolution of avirulence genes in rice blast fungus Magnaporthe oryzae. BMC Genet. 2014, 15, 45. [Google Scholar] [CrossRef]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A review of auxin response factors (ARFs) in plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef]
- Bakhshi, B.; Salekdeh, G.H.; Bihamta, M.R.; Tohidfar, M. Characterization of Three Key MicroRNAs in Rice Root Architecture under Drought Stress using In silico Analysis and Quantitative Real-time PCR. Biosci. Biotechnol. Res. Asia 2014, 11, 555–565. [Google Scholar] [CrossRef]
- Gleeson, M.; Constantin, M.; Carroll, B.J.; Mitter, N. MicroRNAs as regulators of adventitious root development. J. Plant Biochem. Biotechnol. 2014, 23, 339–347. [Google Scholar] [CrossRef]
- Reed, J.W.; Wu, M.F.; Reeves, P.H.; Hodgens, C.; Yadav, V.; Hayes, S.; Pierik, R. Three Auxin Response Factors Promote Hypocotyl Elongation. Plant Physiol. 2018, 178, 864–875. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Feng, B.; Chen, T.; Zhang, X.; Tao, L.; Fu, G. Sugars, antioxidant enzymes and IAA mediate salicylic acid to prevent rice spikelet degeneration caused by heat stress. Plant Growth Regul. 2017, 83, 313–323. [Google Scholar] [CrossRef]
- Tian, G.W.; Mohanty, A.; Chary, S.N.; Li, S.; Paap, B.; Drakakaki, G.; Kopec, C.D.; Li, J.; Ehrhardt, D.; Jackson, D.; et al. High-Throughput Fluorescent Tagging of Full-Length Arabidopsis Gene Products in Planta. Plant Physiol. 2004, 135, 25–38. [Google Scholar] [CrossRef] [Green Version]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, S.; Zhu, C.; Chang, X.; Yue, C.; Wang, Z.; Lin, Y.; Lai, Z. Identification of drought-responsive miRNAs and physiological characterization of tea plant (Camellia sinensis L.) under drought stress. BMC Plant Biol. 2017, 17, 211. [Google Scholar] [CrossRef]
- Eckardt, N.A. MicroRNAs Regulate Auxin Homeostasis and Plant Development. Plant Cell 2005, 17, 1335–1338. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Xie, K.; Xiong, L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014, 65, 2119–2135. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar] [CrossRef]
- Liu, P.P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef]
- Yang, J.H.; Han, S.J.; Yoon, E.K.; Lee, W.S. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006, 34, 1892–1899. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Yu, Y.; Wang, C.Y.; Li, Z.Y.; Liu, Q.; Xu, J.; Liao, J.Y.; Wang, X.J.; Qu, L.H.; Chen, F. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Lee, B.H.; Ko, J.H.; Lee, S.; Lee, Y.; Pak, J.H.; Kim, J.H. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 2009, 151, 655–668. [Google Scholar] [CrossRef]
- Wei Xu, M.H.; Yuan, C.Y.; Wu, F.L.; Chen, S.B.; Xue, K. MicroRNA-139-5p inhibits cell proliferation and invasion by targeting insulin-like growth factor 1 receptor in human non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 3864. [Google Scholar]
- Liu, H.; Guo, S.; Xu, Y.; Li, C.; Zhang, Z.; Zhang, D.; Xu, S.; Zhang, C.; Chong, K. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 2014, 165, 160–174. [Google Scholar] [CrossRef]
- Zhang, C.; Bai, M.Y.; Chong, K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014, 33, 683–696. [Google Scholar] [CrossRef]
- Niu, Y.; Zhao, T.; Xu, X.; Li, J. Genome-wide identification and characterization of GRAS transcription factors in tomato (Solanum lycopersicum). PeerJ 2017, 5, 3955. [Google Scholar] [CrossRef]
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Rodriguez, R.E.; Mecchia, M.A.; Palatnik, J.F. Functional Specialization of the Plant miR396 Regulatory Network through Distinct MicroRNA–Target Interactions. PLoS Genet. 2012, 8, e1002419. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Ercoli, M.F.; Debernardi, J.M.; Breakfield, N.W.; Mecchia, M.A.; Sabatini, M.; Cools, T.; De Veylder, L.; Benfey, P.N.; Palatnik, J.F. MicroRNA miR396 Regulates the Switch between Stem Cells and Transit-Amplifying Cells in Arabidopsis Roots. Plant Cell 2015, 27, 3354–3366. [Google Scholar] [CrossRef]
- Nodine, M.D.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692. [Google Scholar] [CrossRef] [Green Version]
- Yu, N.; Cai, W.J.; Wang, S.; Shan, C.M.; Wang, L.J.; Chen, X.Y. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 2010, 22, 2322–2335. [Google Scholar] [CrossRef]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL 9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Gocal, G.F.; Sheldon, C.C.; Gubler, F.; Moritz, T.; Bagnall, D.J.; MacMillan, C.P.; Li, S.F.; Parish, R.W.; Dennis, E.S.; Weigel, D.; et al. GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis. Plant Physiol. 2001, 127, 1682–1693. [Google Scholar] [CrossRef]
- Liu, W.W.; Meng, J.; Cui, J.; Luan, Y.S. Characterization and Function of MicroRNA(∗)s in Plants. Front. Plant Sci. 2017, 8, 2200. [Google Scholar] [CrossRef]
- Alonso-Peral, M.M.; Li, J.; Li, Y.; Allen, R.S.; Schnippenkoetter, W.; Ohms, S.; White, R.G.; Millar, A.A. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol. 2010, 154, 757–771. [Google Scholar] [CrossRef]
- Xiong, L.; Wang, R.G.; Mao, G.; Koczan, J.M. Identification of Drought Tolerance Determinants by Genetic Analysis of Root Response to Drought Stress and Abscisic Acid. Plant Physiol. 2006, 142, 1065–1074. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef]
- Gutierrez, L.; Bussell, J.D.; Păcurar, D.I.; Schwambach, J.; Păcurar, M.; Bellini, C. Phenotypic Plasticity of Adventitious Rooting in Arabidopsis Is Controlled by Complex Regulation of AUXIN RESPONSE FACTOR Transcripts and MicroRNA Abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Do Choi, Y.; Kim, M.; Reuzeau, C.; Kim, J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Srivastava, A.K.; Pan, Y.; Bai, J.; Fang, J.; Shi, H.; Zhu, J.K. Knockdown of Rice MicroRNA166 Confers Drought Resistance by Causing Leaf Rolling and Altering Stem Xylem Development. Plant Physiol. 2018, 176, 2082–2094. [Google Scholar] [CrossRef] [Green Version]
- Ong, S.S.; Wickneswari, R. Characterization of microRNAs expressed during secondary wall biosynthesis in Acacia mangium. PLoS ONE 2012, 7, e49662. [Google Scholar] [CrossRef]
- Rubio-Somoza, I.; Weigel, D. MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 2011, 16, 258–264. [Google Scholar] [CrossRef]
- Boualem, A.; Laporte, P.; Jovanovic, M.; Laffont, C.; Plet, J.; Combier, J.P.; Niebel, A.; Crespi, M.; Frugier, F. MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 2008, 54, 876–887. [Google Scholar] [CrossRef]
- Hawker, N.P.; Bowman, J.L. Roles for Class III HD-Zip and KANADI Genes in Arabidopsis Root Development. Plant Physiol. 2004, 135, 2261–2270. [Google Scholar] [CrossRef] [Green Version]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 2012, 1819, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Grewal, R.K.; Saraf, S.; Deb, A.; Kundu, S. Differentially expressed microRNAs link cellular physiology to phenotypic changes in rice under stress conditions. Plant Cell Physiol. 2018, 59, 2143–2154. [Google Scholar] [CrossRef]
- Ferdous, J.; Hussain, S.S.; Shi, B.J. Role of microRNAs in plant drought tolerance. Plant Biotechnol. J. 2015, 13, 293–305. [Google Scholar] [CrossRef]
- Mutum, R.D.; Kumar, S.; Balyan, S.; Kansal, S.; Mathur, S.; Raghuvanshi, S. Identification of novel miRNAs from drought tolerant rice variety Nagina 22. Sci. Rep. 2016, 6, 30786. [Google Scholar] [CrossRef] [Green Version]
- Basu, S.; Ramegowda, V.; Kumar, A.; Pereira, A. Plant adaptation to drought stress. F1000Research 2016, 5. [Google Scholar] [CrossRef]
- Galle, A.; Florez-Sarasa, I.; Thameur, A.; de Paepe, R.; Flexas, J.; Ribas-Carbo, M. Effects of drought stress and subsequent rewatering on photosynthetic and respiratory pathways in Nicotiana sylvestris wild type and the mitochondrial complex I-deficient CMSII mutant. J. Exp. Bot. 2010, 61, 765–775. [Google Scholar] [CrossRef]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S. Plant drought stress: Effects, mechanisms and management. In Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2009; pp. 153–188. [Google Scholar]
- Berwal, M.K.; Goyal, P.; Chugh, L.; Kumar, R. Impact of Flag Leaf Removal on Grain Development and Nutrients Deposition in Pearl Millet Developing Grains. Vegetos 2017, 31. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, X.; Li, J.; Cai, H.; Deng, X.W.; Li, L. MicroRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell 2014, 26, 4933–4953. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, S.L.; Li, J.L.; Hu, X.H.; Wang, H.; Cao, X.L.; Xu, Y.J.; Zhao, Z.X.; Xiao, Z.Y.; Yang, N. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 2. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S.; et al. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [Green Version]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. MicroRNAs As Potential Targets for Abiotic Stress Tolerance in Plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef]
- Singh, I.; Smita, S.; Mishra, D.C.; Kumar, S.; Singh, B.K.; Rai, A. Abiotic Stress Responsive miRNA-Target Network and Related Markers (SNP, SSR) in Brassica juncea. Front. Plant Sci. 2017, 8, 1943. [Google Scholar] [CrossRef] [Green Version]
- Cheah, B.H.; Nadarajah, K.; Divate, M.D.; Wickneswari, R. Identification of four functionally important microRNA families with contrasting differential expression profiles between drought-tolerant and susceptible rice leaf at vegetative stage. BMC Genom. 2015, 16, 692. [Google Scholar] [CrossRef]
- Pan, L.; Zhao, H.; Yu, Q.; Bai, L.; Dong, L. miR397/Laccase gene mediated network improves tolerance to fenoxaprop-p-ethyl in Beckmannia syzigachne and Oryza sativa. Front. Plant Sci. 2017, 8, 879. [Google Scholar] [CrossRef]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef]
- Pomeranz, M.C.; Hah, C.; Lin, P.C.; Kang, S.G.; Finer, J.J.; Blackshear, P.J.; Jang, J.C. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 2010, 152, 151–165. [Google Scholar] [CrossRef]
- Lim, P.O.; Woo, H.R.; Nam, H.G. Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 2003, 8, 272–278. [Google Scholar] [CrossRef]
- Xu, X.; Bai, H.; Liu, C.; Chen, E.; Chen, Q.; Zhuang, J.; Shen, B. Genome-wide analysis of microRNAs and their target genes related to leaf senescence of rice. PLoS ONE 2014, 9, e114313. [Google Scholar] [CrossRef]
- Ma, N.; Ma, C.; Liu, Y.; Shahid, M.O.; Wang, C.; Gao, J. Petal senescence: A hormone view. J. Exp. Bot. 2018, 69, 719–732. [Google Scholar] [CrossRef]
- Cruz de Carvalho, M.H. Drought stress and reactive oxygen species: Production, scavenging and signaling. Plant Signal. Behav. 2008, 3, 156–165. [Google Scholar] [CrossRef]
- Loggini, B.; Scartazza, A.; Brugnoli, E.; Navari-Izzo, F. Antioxidative defense system, pigment composition, and photosynthetic efficiency in two wheat cultivars subjected to drought. Plant Physiol. 1999, 119, 1091–1100. [Google Scholar] [CrossRef]
- Mittler, R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002, 7, 405–410. [Google Scholar] [CrossRef]
- Li, T.; Li, H.; Zhang, Y.X.; Liu, J.Y. Identification and analysis of seven H2O2-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica). Nucleic Acids Res. 2011, 39, 2821–2833. [Google Scholar] [CrossRef]
- Kantar, M.; Lucas, S.J.; Budak, H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta 2011, 233, 471–484. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Andrés-Colás, N.; Carrió-Seguí, A.; Abdel-Ghany, S.E.; Pilon, M.; Peñarrubia, L. Expression of the Intracellular COPT3-Mediated Cu Transport Is Temporally Regulated by the TCP16 Transcription Factor. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Csiszár, J.; Gallé, Á.; Horváth, E.; Dancsó, P.; Gombos, M.; Váry, Z.; Erdei, L.; Györgyey, J.; Tari, I. Different peroxidase activities and expression of abiotic stress-related peroxidases in apical root segments of wheat genotypes with different drought stress tolerance under osmotic stress. Plant Physiol. Biochem. 2012, 52, 119–129. [Google Scholar] [CrossRef]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2014, 5, 771. [Google Scholar] [CrossRef]
- Slama, I.; Abdelly, C.; Bouchereau, A.; Flowers, T.; Savouré, A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann. Bot. 2015, 115, 433–447. [Google Scholar] [CrossRef] [Green Version]
- Roelofs, D.; Aarts, M.; Schat, H.; Van Straalen, N. Functional ecological genomics to demonstrate general and specific responses to abiotic stress. Funct. Ecol. 2008, 22, 8–18. [Google Scholar] [CrossRef]
- Crowe, J.H.; Crowe, L.M. Preservation of mammalian cells-learning nature’s tricks. Nat. Biotechnol. 2000, 18, 145–146. [Google Scholar] [CrossRef]
- Richards, A.B.; Krakowka, S.; Dexter, L.B.; Schmid, H.; Wolterbeek, A.P.; Waalkens-Berendsen, D.H.; Shigoyuki, A.; Kurimoto, M. Trehalose: A review of properties, history of use and human tolerance, and results of multiple safety studies. Food Chem. Toxicol. 2002, 40, 871–898. [Google Scholar] [CrossRef]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.E.; Stitt, M.; Schmid, M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 2013, 339, 704–707. [Google Scholar] [CrossRef]
- Eastmond, P.J.; Van Dijken, A.J.; Spielman, M.; Kerr, A.; Tissier, A.F.; Dickinson, H.G.; Jones, J.D.; Smeekens, S.C.; Graham, I.A. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 2002, 29, 225–235. [Google Scholar] [CrossRef]
- Kaplan, F.; Guy, C.L. β-Amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004, 135, 1674–1684. [Google Scholar] [CrossRef]
- Peng, T.; Sun, H.; Du, Y.; Zhang, J.; Li, J.; Liu, Y.; Zhao, Y.; Zhao, Q. Characterization and expression patterns of microRNAs involved in rice grain filling. PLoS ONE 2013, 8, e54148. [Google Scholar] [CrossRef]
- Sengupta, A.; Chakraborty, M.; Saha, J.; Gupta, B.; Gupta, K. Polyamines: Osmoprotectants in plant abiotic stress adaptation. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Springer: Berlin/Heidelberg, Germany, 2016; pp. 97–127. [Google Scholar]
- Minocha, R.; Majumdar, R.; Minocha, S.C. Polyamines and abiotic stress in plants: A complex relationship1. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef]
- Umate, P.; Tuteja, R. Architectures of the unique domains associated with the DEAD-box helicase motif. Cell Cycle 2010, 9, 4228–4235. [Google Scholar] [CrossRef] [Green Version]
- Macovei, A.; Tuteja, N. Different expression of miRNAs targeting helicases in rice in response to low and high dose rate γ-ray treatments. Plant Signal. Behav. 2013, 8, e25128. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, M.; Zhou, H.; Li, M.; Huang, L.; Yin, X.; Zhao, G.; Lin, F.; Xia, X.; Xu, G. OsSGL, a Novel DUF1645 Domain-Containing Protein, Confers Enhanced Drought Tolerance in Transgenic Rice and Arabidopsis. Front. Plant Sci. 2016, 7, 2001. [Google Scholar] [CrossRef] [Green Version]
- Akdogan, G.; Tufekci, E.D.; Uranbey, S.; Unver, T. miRNA-based drought regulation in wheat. Funct. Integr. Genom. 2016, 16, 221–233. [Google Scholar] [CrossRef]
- Barrera-Figueroa, B.E.; Gao, L.; Diop, N.N.; Wu, Z.; Ehlers, J.D.; Roberts, P.A.; Close, T.J.; Zhu, J.K.; Liu, R. Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol. 2011, 11, 127. [Google Scholar] [CrossRef]
- Jajic, I.; Sarna, T.; Strzalka, K. Senescence, stress, and reactive oxygen species. Plants 2015, 4, 393–411. [Google Scholar] [CrossRef]
- Sade, N.; del Mar Rubio-Wilhelmi, M.; Umnajkitikorn, K.; Blumwald, E. Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 2017, 69, 845–853. [Google Scholar] [CrossRef]
- Pirasteh-Anosheh, H.; Saed-Moucheshi, A.; Pakniyat, H.; Pessarakli, M. Stomatal responses to drought stress. In Water Stress and Crop Plants; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016; pp. 24–40. [Google Scholar]
- Jung, J.H.; Park, C.M. MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 2007, 225, 1327–1338. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef]
- Shuai, P.; Liang, D.; Zhang, Z.; Yin, W.; Xia, X. Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genom. 2013, 14, 233. [Google Scholar] [CrossRef]
- Kumar, A.; Dixit, S.; Ram, T.; Yadaw, R.; Mishra, K.; Mandal, N. Breeding high-yielding drought-tolerant rice: Genetic variations and conventional and molecular approaches. J. Exp. Bot. 2014, 65, 6265–6278. [Google Scholar] [CrossRef]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [Green Version]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nadarajah, K.; Kumar, I.S. Drought Response in Rice: The miRNA Story. Int. J. Mol. Sci. 2019, 20, 3766. https://doi.org/10.3390/ijms20153766
Nadarajah K, Kumar IS. Drought Response in Rice: The miRNA Story. International Journal of Molecular Sciences. 2019; 20(15):3766. https://doi.org/10.3390/ijms20153766
Chicago/Turabian StyleNadarajah, Kalaivani, and Ilakiya Sharanee Kumar. 2019. "Drought Response in Rice: The miRNA Story" International Journal of Molecular Sciences 20, no. 15: 3766. https://doi.org/10.3390/ijms20153766