Morphological, Transcriptomic and Hormonal Characterization of Trimonoecious and Subandroecious Pumpkin (Cucurbita maxima) Suggests Important Roles of Ethylene in Sex Expression
Abstract
:1. Introduction
2. Results
2.1. Flower Phenotype
2.2. RNA-Seq of Pumpkin
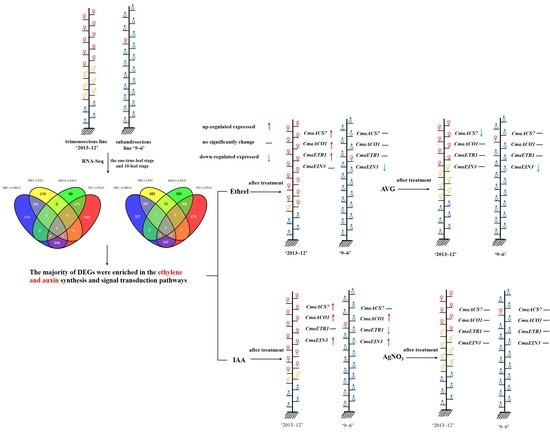
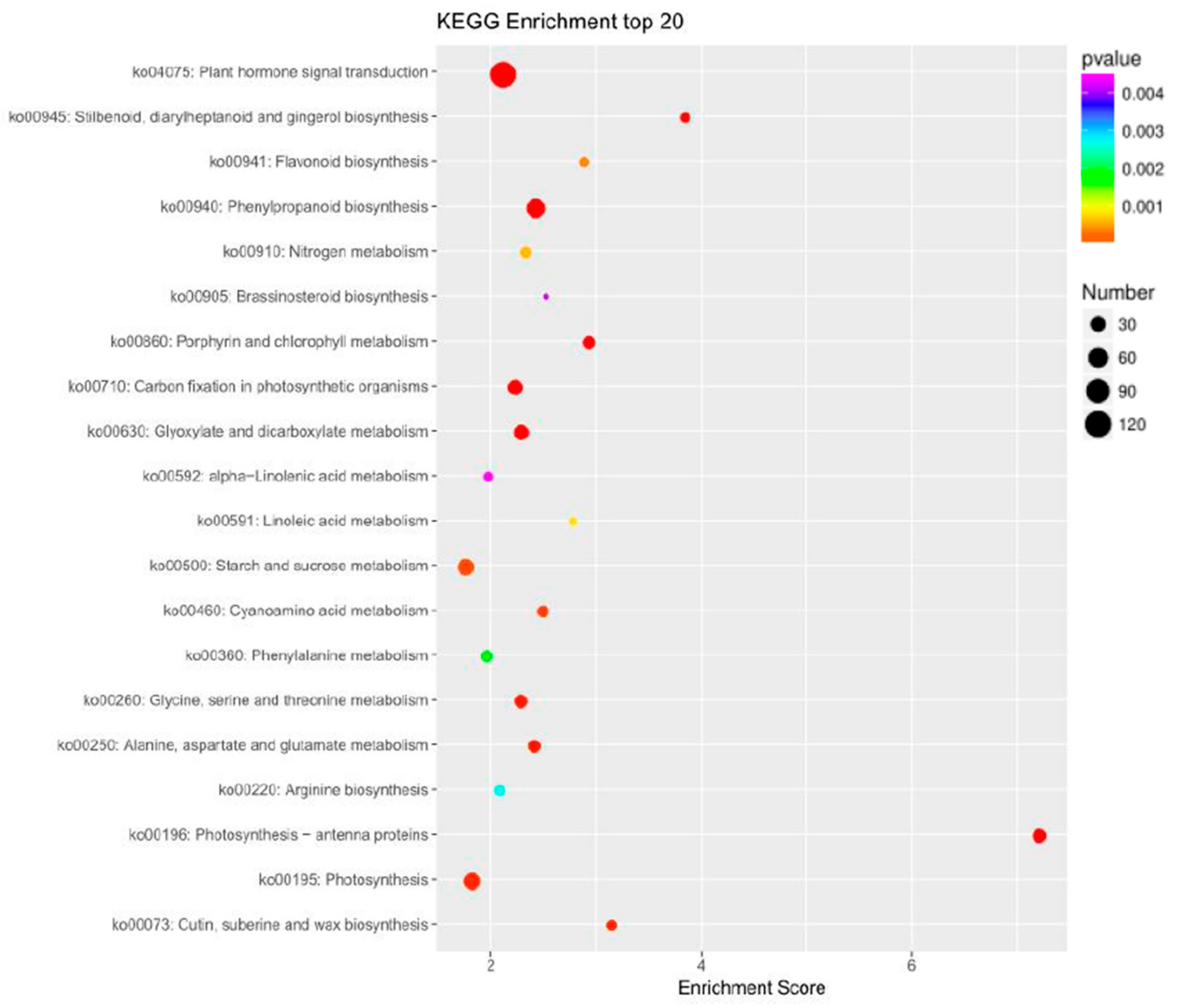
2.3. Differential Expression Analysis and Functional Annotation
2.4. Hormone Genes Associated with Sex Expression
2.5. Plant Phenotypes after Chemical Treatments
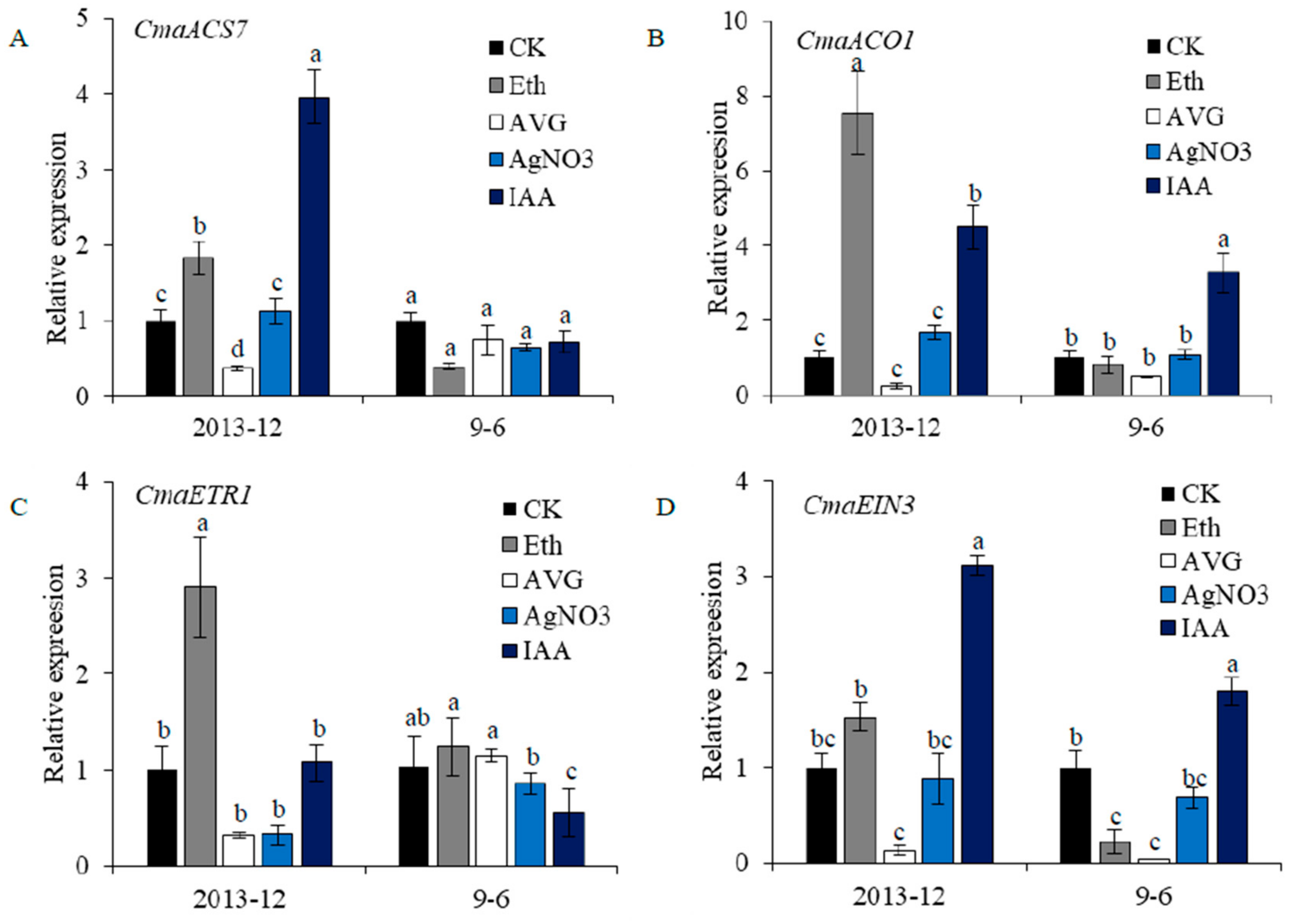
2.6. Gene Expression under Chemical Treatment
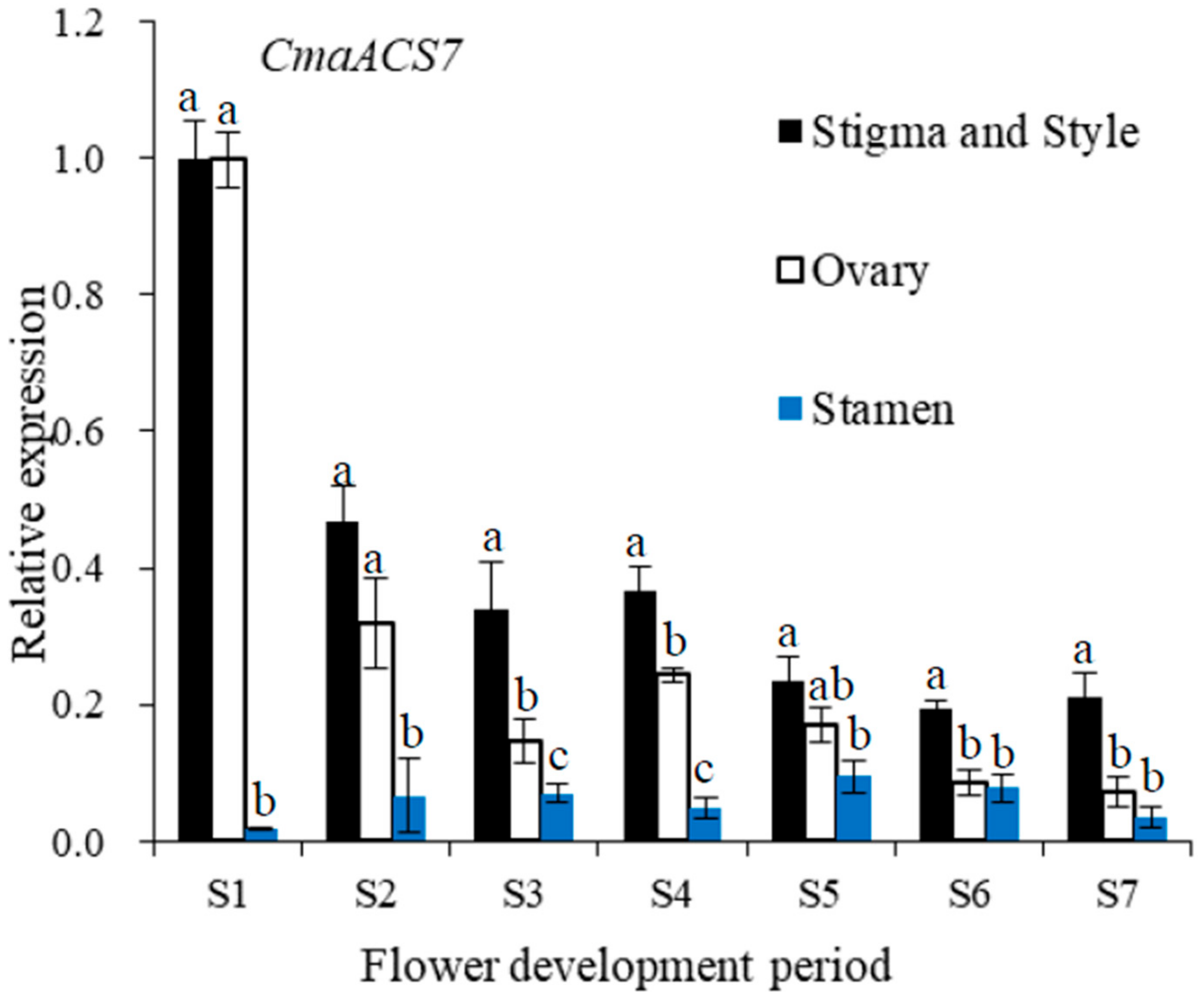
2.7. CmaACS7 Expression at Different Flower Development Stages
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Extraction and RNA-Seq Processing
4.3. RNA-Seq Assembly, Annotation and Transcriptome Sequence Analysis
4.4. Quantitative RT-PCR Analysis
4.5. Plant Hormone Treatments
4.6. Gene expression at Different Stages of Flower Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | 1-aminocyclopropane-1-carboxylate |
| ACO | 1-aminocyclopropane-1-carboxylate oxidase |
| ACS | 1-aminocyclopropane-1-carboxylate synthetase |
| ERF | Ethylene-responsive factor |
| EIN | Ethylene- insensitive |
| WEI | Weak ethylene insensitive |
| CTR | Constitutive triple response |
| RNA-seq | RNA sequencing |
| C. maxima | Cucurbita maxima |
| C. pepo | Cucurbita pepo |
| C. moschata | Cucurbita moschata |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| FPKM | Fragments per kilobase of exon per million fragments mapped |
| bp | Base pairs |
| DEGs | Differentially expressed genes |
| MAPK | Mitogen-activated protein kinase |
| LS1 | One-true-leaf stage |
| LS10 | 10-leaf stage |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| PCA | Principal component analysis |
| AVG | Aminoethoxyvinyl glycine |
| IAA | Indoleacetic acid |
References
- Ekaterina, P.; Holly, A.L.; Sue, A.H.; Rebecca, G. Effect of modified endogenous ethylene production on sex expression, bisexual flower development and fruit production in melon (Cucumis melo L.). Sex. Plant Reprod. 2005, 18, 131–142. [Google Scholar]
- Kater, M.M.; Franken, J.; Carney, K.J.; Colombo, L.; Angenent, G.C. Sex determination in the monoecious species cucumber is confined to specific floral whorls. Plant Cell 2001, 13, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Fujii, N.; Miyazawa, Y.; Yamasaki, S.; Matsuura, S.; Mizusawa, H.; Fujita, Y.; Takahashi, H. Correlation between development of female flower buds and expression of the CS-ACS2 gene in cucumber plants. J. Exp. Bot. 2007, 58, 2897–2907. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, S.; Liu, S.; Pan, J.; Zhang, Z.; Tao, Q.; Shi, Q.; Jia, Z.; Zhang, W.; Chen, H.; et al. Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics 2009, 182, 1381–1385. [Google Scholar] [CrossRef] [PubMed]
- Boualem, A.; Troadec, C.; Camps, C.; Lemhemdi, A.; Morin, H.; Sari, M.; Fraenkel-Zagouri, R.; Kovalski, I.; Dogimont, C.; Perl-Treves, R.; et al. A cucurbit androecygene reveals how unisexual flowers develop and dioecy emerges. Plant Genetics 2015, 350, 688–691. [Google Scholar]
- Zhang, Z.; Mao, L.; Chen, H.; Bu, F.; Li, G.; Sun, J.; Li, S.; Sun, H.; Jiao, C.; Blakely, R.; et al. Genome-wide mapping of structural variations reveals a copy number variant that determines reproductive morphology in cucumber. Plant Cell 2015, 27, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.F.; Hoffman, N.E. Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 2003, 35, 155–189. [Google Scholar] [CrossRef]
- Boualem, A.; Fergany, M.; Fernandez, R.; Troadec, C.; Martin, A.; Morin, H.; Sari, M.A.; Collin, F.; Flowers, J.M.; Pitrat, M.; et al. A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 2008, 321, 836–838. [Google Scholar] [CrossRef]
- Boualem, A.; Troadec, C.; Kovalski, I.; Sari, M.A.; Perl-Treves, R.; Bendahmane, A. A conserved ethylene biosynthesis enzyme leads to andromonoecy in two Cucumis species. PLoS ONE 2009, 4, e6144. [Google Scholar] [CrossRef]
- Martin, A.; Troadec, C.; Boualem, A.; Rajab, M.; Fernandez, R.; Morin, H.; Pitrat, M.; Dogimont, C.; Bendahmane, A. A transposon-induced epigenetic change leads to sex determination in melon. Nature 2009, 461, 1135–1138. [Google Scholar] [CrossRef]
- Adnane, B.; Afef, L.; Marie-Agnes, S.; Sarah, P.; Christelle, T.; Fadi, A.; Ilknur, S.; Nebahat, S.; Catherine, D.; Abdelhafid, B. The androandroecious sex determination gene predates the separation of Cucumis and Citrullus Genera. PLoS ONE 2016, 11, e0155444. [Google Scholar]
- Ji, G.; Zhang, J.; Zhang, H.; Sun, H.; Gong, G.; Shi, J.; Tian, S.; Guo, S.; Ren, Y.; Shen, H.; et al. Mutation in the gene encoding 1-aminocyclopropane-1-carboxylate synthase 4 (CitACS4) led to andromonoecy in watermelon. J. Integr. Plant Biol. 2016, 9, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Knopf, R.R.; Trebitsh, T. The female-specific Cs-ACS1G gene of cucumber. A case of gene duplication and recombination between the non-sex-specific 1-aminocyclopropane-1-carboxylate synthase gene and a branched-chain amino acid transaminase gene. Plant Cell Physiol. 2006, 47, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, J.; Li, S.; Cui, Q.; Zhang, H.; Xin, F.; Wang, H.; Lin, T.; Gao, D.; Wang, S.; et al. An ACC oxidase gene essential for cucumber carpel development. Mol. Plant. 2016, 9, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Fujii, N.; Takahashi, H. The ethylene-regulated expression of CS-ETR2 and CS-ERS genes in cucumber plants and their possible involvement with sex expression in flowers. Plant Cell Physiol. 2000, 41, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, F.; Duan, Q.; Han, T.; Xu, Z.; Bai, S. Ethylene perception is involved in female cucumber flower development. Plant J. 2010, 61, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Tao, Q.; Niu, H.; Wang, Z.; Zhang, W.; Wang, H.; Wang, S.; Zhang, X.; Li, Z. Ethylene responsive factor ERF110 mediates ethylene-regulated transcription of a sex determination-related orthologous gene in two Cucumis species. J. Exp. Bot. 2018, 25, 2953–2965. [Google Scholar] [CrossRef]
- Pan, J.; Wang, G.; Wen, H.; Du, H.; Lian, H.; He, H.; Pan, J.; Cai, R. Differential gene expression caused by the F and M Loci provides insight into ethylene-mediated female flower differentiation in cucumber. Front Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Peñaranda, A.; Payan, M.C.; Garrido, D.; Gómez, P.; Jamilena, M. Production of fruits with attached flowers in zucchini squash is correlated with the arrest of maturation of female flowers. J. Hortic. Sci. Biotech. 2007, 82, 579–584. [Google Scholar] [CrossRef]
- Kubicki, B. Androecious strains of Cucurbita pepo L. Genet. Pol. 1970, 11, 45–51. [Google Scholar]
- Dossey, B.F.; Bemis, W.P.; Scheerens, J.C. Genetic control of gynoecy in the buffalo gourd. J. Hered. 1981, 72, 355–356. [Google Scholar] [CrossRef]
- Manzano, S.; Dominguez, V.J.; Garrido, D.; Gómez, P.; Jamilena, M. A recessive gene conferring ethylene insensitivity and androecy in Cucurbita pepo. Cucurbitaceae Ixth Eucarpia Meeting on Genetics and breeding of Cucurbitaceae. INRA 2008, 563–567. [Google Scholar]
- Manzano, S.; Martınez, C.; Domınguez, V.; Avalos, E.; Garrido, D.; Gómez, P.; Jamilena, M. A major gene conferring reduced ethylene sensitivity and maleness in Cucurbita pepo. J. Plant Growth Regul. 2010, 29, 73–80. [Google Scholar] [CrossRef]
- Manzano, S.; Martınez, C.; Gomez, P.; Garrido, D.; Jamilena, M. Cloning and characterisation of two CTR1-like genes in Cucurbita pepo: Regulation of their expression during male and female flower development. Sex Plant Reprod. 2010, 23, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Manzano, S.; Martınez, C.; Megías, Z.; Garrido, D.; Picó, B.; Jamilena, M. Involvement of ethylene biosynthesis and signalling in the transition from male to female flowering in the monoecious Cucurbita pepo. J. Plant Growth Regul. 2013, 32, 789–798. [Google Scholar] [CrossRef]
- Manzano, S.; Martínez, C.; García, J.M.; Megías, Z.; Jamilena, M. Involvement of ethylene in sex expression and female flower development in watermelon (Citrullus lanatus). Plant Physiol. Biochem. 2014, 85, 96–104. [Google Scholar] [CrossRef]
- Yang, W. Mapping and candidate gene analysis of strong female characteristic in Cucurbita maxima Duch. Master’s Thesis, Hebei University of Engineering, Hebei, China, December 2016. [Google Scholar]
- Blanca, J.; Cañizares, J.; Roig, C.; Ziarsolo, P.; Nuez, F.; Picó, B. Transcriptome characterization and high throughput SSRs and SNPs discovery in Cucurbita pepo (Cucurbitaceae). BMC Genom. 2011, 12, 104. [Google Scholar] [CrossRef]
- Esteras, C.; Gómez, P.; Monforte, A.J.; Blanca, J.; Vicente-Dólera, N.; Roig, C.; Nuez, F.; Picó, B. High-throughput SNP genotyping in Cucurbita pepo for map construction and quantitative trait loci mapping. BMC Genom. 2012, 13, 80–101. [Google Scholar] [CrossRef]
- Wu, T.; Luo, S.; Wang, R.; Zhong, Y.; Xu, X.; Lin, Y.; He, X.; Sun, B.; Huang, H. The first Illumina-based de novo transcriptome sequencing and analysis of pumpkin (Cucurbita moschata Duch.) and SSR marker development. Mol. Breeding 2014, 34, 1437–1447. [Google Scholar] [CrossRef]
- Summers, C.F.; Gulliford, C.M.; Carlson, C.H.; Lillis, J.A.; Carlson, M.O.; Cadle-Davidson, L.; Gent, D.H.; Smart, C.D. Identification of genetic variation between obligate plant pathogens Pseudoperonospora cubensis and P. humuli using RNA sequencing and genotyping-by-sequencing. PLoS ONE 2015, 10, e0143665. [Google Scholar] [CrossRef]
- Wyatt, L.E.; Strickler, S.R.; Mueller, L.A.; Mazourek, M. An acorn squash (Cucurbita pepo ssp. ovifera) fruit and seed transcriptome as a resource for the study of fruit traits in Cucurbita. Hortic. Res. 2015, 2. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, L.E.; Strickler, S.R.; Mueller, L.A.; Mazourek, M. Comparative analysis of Cucurbita pepo metabolism throughout fruit development in acorn squash and oilseed pumpkin. Hortic. Res. 2016, 3, 16045. [Google Scholar] [CrossRef] [PubMed]
- Xanthopoulou, A.; Ganopoulos, I.; Psomopoulos, F.; Manioudaki, M.; Moysiadis, T.; Kapazoglou, A.; Osathanunkul, M.; Michailidou, S.; Kalivas, A.; Tsaftaris, A.; et al. De novo comparative transcriptome analysis of genes involved in fruit morphology of pumpkin cultivars with extreme size difference and development of EST-SSR markers. Gene 2017, 622, 50–66. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, F.; Rosales, R.; Palma, F.; Manzano, S.; Canizares, J.; Jamilena, M.; Garrido, D. Transcriptomic changes in Cucurbita pepo fruit after cold storage: Differential response between two cultivars contrasting in chilling sensitivity. BMC Genom. 2018, 19, 125. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.L.; Chen, B.H.; Chen, X.J.; Guo, Y.Y.; Yang, H.L.; Li, X.Z.; Wang, G.Y. Transcriptome profiling of pumpkin (Cucurbita moschata Duch.) leaves infected with powdery mildew. PLoS ONE 2018, 13, e0190175. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yang, X.; Wang, Y.; Xu, W.; Cui, C.; Qu, S. Content changes of endogenous hormones and polyamine during flower development in pumpkin. China Sciencepap. 2016, 11, 2096–2099. [Google Scholar]
- Guo, S.; Zheng, Y.; Joung, Y.; Liu, S.; Zhang, Z.; Crasta, O.; Sobral, B.W.; Xu, Y.; Huang, S.; Fei, Z. Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genom. 2010, 11, 384. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Qin, Z.; Zhou, X.; Feng, Z.; Du, Y. Transcriptome profile analysis of floral sex determination in cucumber. J. Plant Physiol. 2010, 167, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Sheng, Y.; Luan, F.; Ma, H.; Liu, S. RNA-Seq transcriptome profiling reveals differentially expressed genes involved in sex expression in melon. Crop Sci. 2016, 55, 1686–1695. [Google Scholar] [CrossRef]
- Yamasaki, S.; Fujii, N.; Matsuura, S.; Mizusawa, H.; Takahashi, H. The M locus and ethylene-controlled sex determination in andromonoecious cucumber plants. Plant Cell Physiol. 2001, 42, 608–619. [Google Scholar] [CrossRef]
- Martínez, C.; Manzano, S.; Megías, Z.; Barrera, A.; Boualem, A.; Garrido, D.; Bendahmane, A.; Jamilena, M. Molecular and functional characterization of CpACS27A gene reveals its involvement in monoecy instability and other associated traits in squash (Cucurbita pepo L.). Planta 2014, 239, 1201–1215. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wei, B.; Liu, A.; Zhang, J.; Feng, H. Female flowers were induced by ethephon in andromonecious Cucumis melo L. China Veg. 2010, 4, 67–70. (In Chinese) [Google Scholar]
- Rudich, J.; Halevy, A.; Kedar, N. Increase in femaleness of three cucurbits by treatment with ethrel, an ethylene releasing compound. Planta 1969, 86, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shi, J.; Ji, G.; Zhang, H.; Gong, G.; Guo, S.; Ren, Y.; Fan, J.; Tian, S.; Xu, Y. Modulation of sex expression in four forms of watermelon by gibberellin, ethephone and silver nitrate. Hortic. Plant J. 2017, 3, 91–100. [Google Scholar] [CrossRef]
- Li, Z.; Wang, S.; Tao, Q.; Pan, J.; Si, L.; Gong, Z.; Cai, R. A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L.). J. Exp. Bot. 2012, 63, 4475–4484. [Google Scholar] [CrossRef]
- Martínez, C.; Manzano, S.; Megías, Z.; Garrido, D.; Picó, B.; Jamilena, M. Involvement of ethylene biosynthesis and signaling in fruit set and early fruit development in zucchini squash (Cucurbita pepo L.). BMC Plant Biol. 2013, 13, 139. [Google Scholar] [CrossRef]
- Tatsuki, M.; Nakajima, N.; Fujii, H.; Shimada, T.; Nakano, M.; Hayashi, K.; Hayama, H.; Yoshioka, H.; Nakamura, Y. Increased levels of IAA are required for system 2 ethylene synthesis causing fruit softening in peach (Prunus persica L. Batsch). J. Exp. Bot. 2013, 64, 1049–1059. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Yoo, S.D.; Sheen, J. Differential regulation of EIN3 stability by glucose and ethylene signaling in plants. Nature 2003, 425, 521–525. [Google Scholar] [CrossRef]
- Guo, H.W.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCF (EBFl/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef]
- Potuschak, T.; Lechner, E.; Parmentier, Y.; Yanagisawa, S.; Grava, S.; Koncz, C.; Genschik, P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell 2003, 115, 679–689. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Fields, C.J.; Goto, N.; Heuer, M.L.; Rice, P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010, 38, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 14–26. [Google Scholar] [CrossRef] [PubMed]

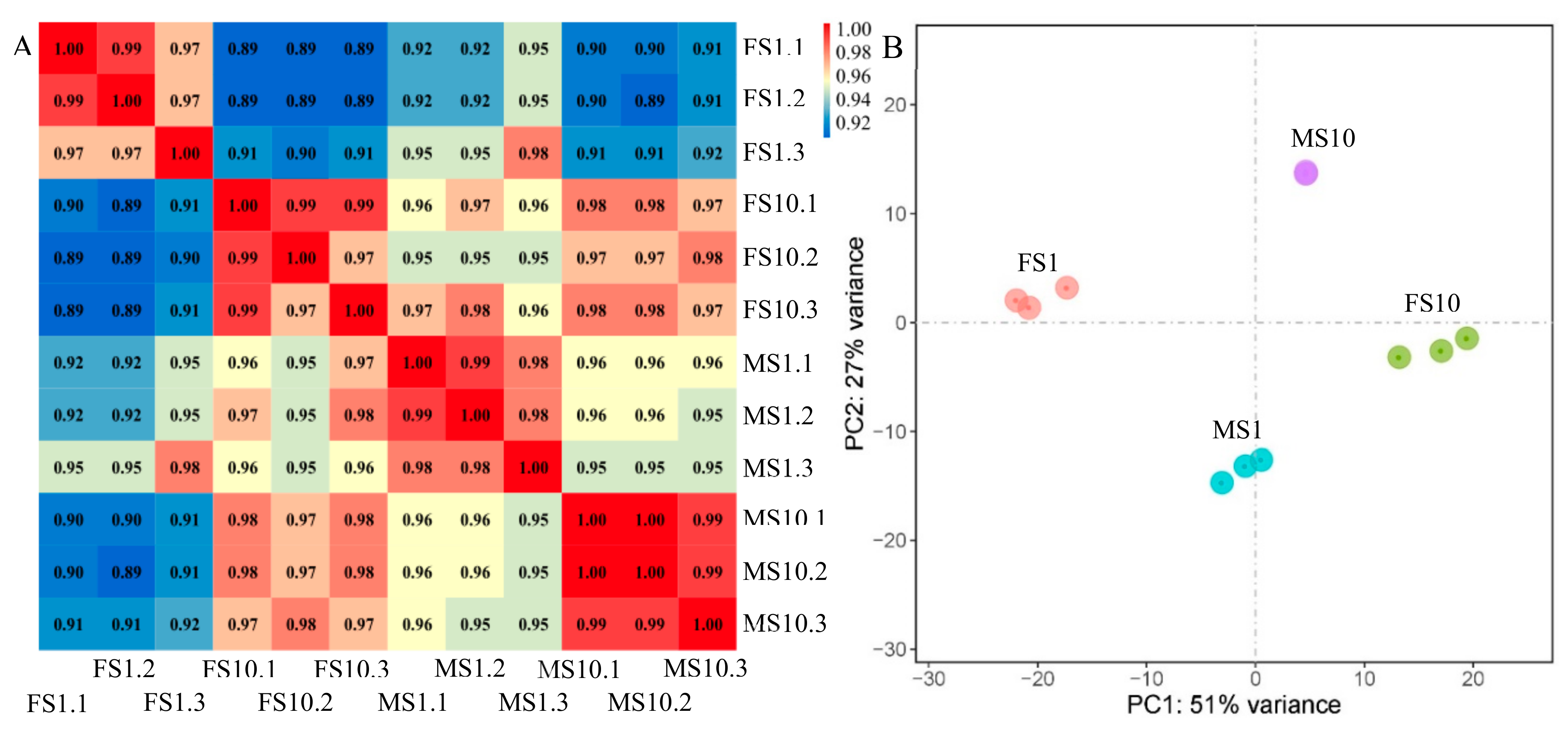
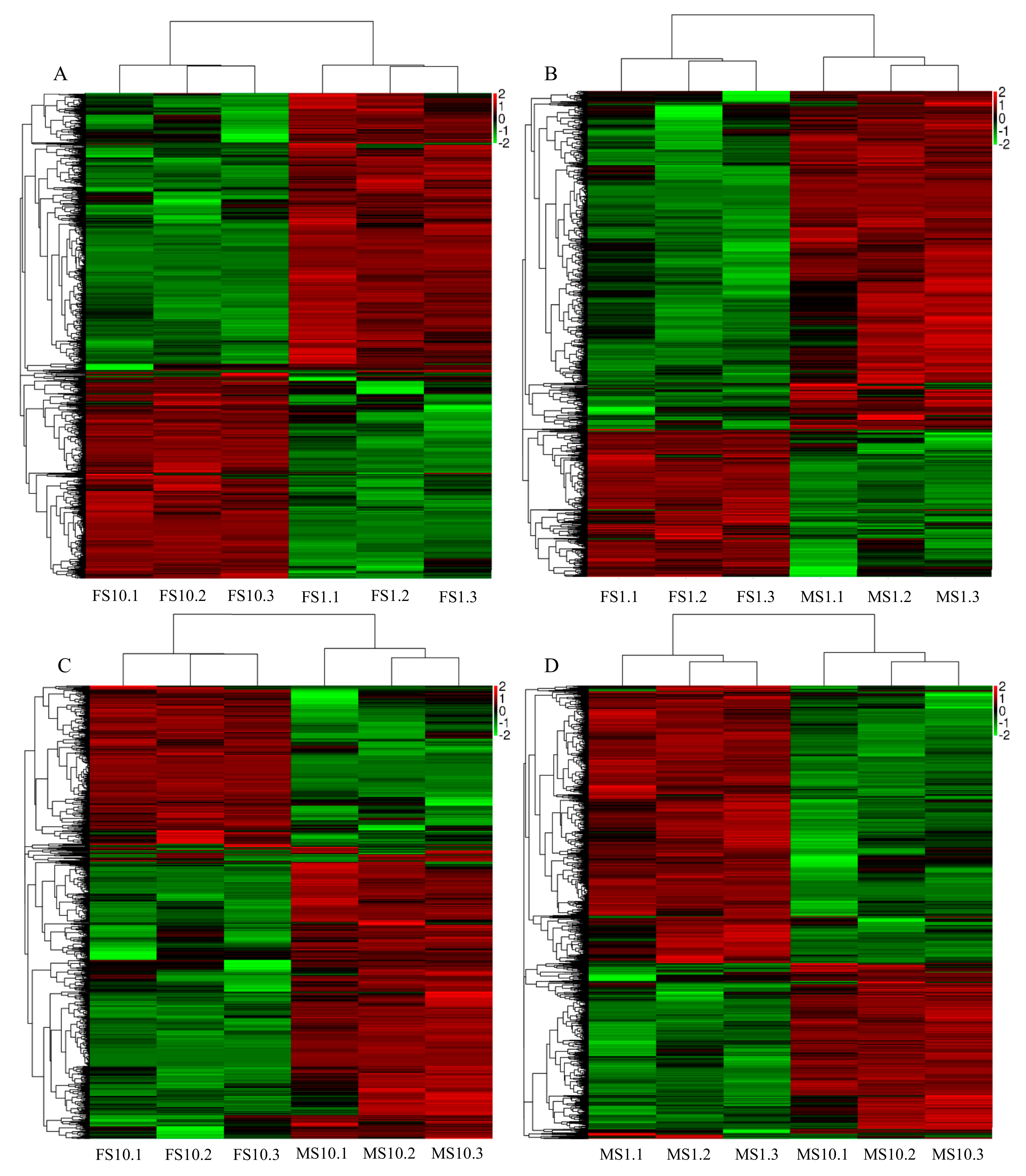



| Sample ID | Raw Reads (Mb) | Raw Bases (Gb) | Clean Reads (Mb) | Clean Bases (Gb) | Valid Bases (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|
| MS1.1 | 38.21 | 4.78 | 37.06 | 4.63 | 96.95 | 94.29 | 45 |
| MS1.2 | 43.52 | 5.44 | 42.17 | 5.27 | 96.86 | 94.57 | 45 |
| MS1.3 | 46.69 | 5.84 | 45.39 | 5.67 | 97.16 | 94.60 | 45 |
| MS10.1 | 46.31 | 5.79 | 44.96 | 5.62 | 97.05 | 94.55 | 45 |
| MS10.2 | 43.91 | 5.49 | 42.54 | 5.32 | 96.84 | 94.47 | 45 |
| MS10.3 | 42.03 | 5.25 | 40.70 | 5.09 | 96.81 | 94.28 | 45 |
| FS1.1 | 48.34 | 6.04 | 46.86 | 5.86 | 96.90 | 94.49 | 45 |
| FS1.2 | 45.03 | 5.63 | 43.67 | 5.46 | 96.95 | 94.51 | 45 |
| FS1.3 | 47.38 | 5.92 | 45.86 | 5.73 | 96.75 | 94.46 | 45 |
| FS10.1 | 43.06 | 5.38 | 41.86 | 5.23 | 97.17 | 94.71 | 45 |
| FS10.2 | 38.27 | 4.78 | 36.83 | 4.6 | 96.22 | 93.89 | 44.5 |
| FS10.3 | 42.65 | 5.33 | 41.33 | 5.16 | 96.87 | 94.60 | 44.5 |
| Gene ID | FS1 VS FS10 log2FoldChange | MS10 VS FS10 log2FoldChange | GO Term |
|---|---|---|---|
| CmaCh03G005620 | 1.14 | 1.18 | 1-aminocyclopropane-1-carboxylate oxidase |
| CmaCh02G000660 | 2.75 | 0.26 | 1-aminocyclopropane-1-carboxylate oxidase |
| CmaCh01G009990 | −1.43 | −0.16 | negative regulation of ethylene-activated signaling pathway, auxin efflux transmembrane transporter activity, auxin homeostasis, auxin polar transport, auxin-activated signaling pathway |
| CmaCh01G016830 | −1.25 | −0.44 | negative regulation of ethylene-activated signaling pathway, regulation of abscisic acid-activated signaling pathway, cytokinin-activated signaling pathway |
| CmaCh02G014030 | 1.82 | 1.32 | ethylene-activated signaling pathway, cytokinin-activated signaling pathway |
| CmaCh04G006970 | 2.74 | 1.42 | ethylene-activated signaling pathway |
| CmaCh04G002620 | 1.87 | 0.15 | ethylene-activated signaling pathway |
| CmaCh04G012360 | 1.69 | 0.44 | ethylene-activated signaling pathway |
| CmaCh04G020120 | 2.35 | 0.07 | ethylene-activated signaling pathway |
| CmaCh04G008070 | −1.66 | −0.36 | ethylene-activated signaling pathway |
| CmaCh04G015210 | 1.81 | 0.74 | ethylene-activated signaling pathway |
| CmaCh04G004940 | Inf | Inf | ethylene-activated signaling pathway |
| CmaCh05G004890 | 1.08 | 0.3 | ethylene-activated signaling pathway, cytokinin-activated signaling pathway |
| CmaCh05G001690 | 1.40 | 0.10 | ethylene-activated signaling pathway |
| CmaCh07G005340 | 2.45 | 0.70 | ethylene-activated signaling pathway |
| CmaCh09G004610 | 1.31 | 0.42 | ethylene-activated signaling pathway |
| CmaCh09G005960 | −3.13 | −0.82 | negative regulation of ethylene-activated signaling pathway, gibberellin biosynthetic process |
| CmaCh09G008010 | 2.02 | 1.57 | response to jasmonic acid and salicylic acid |
| CmaCh10G006590 | 1.34 | 0.04 | ethylene-activated signaling pathway |
| CmaCh11G006380 | 1.38 | 0.49 | ethylene-activated signaling pathway |
| CmaCh11G008140 | 1.01 | 0.11 | ethylene-activated signaling pathway |
| CmaCh11G019380 | 1.17 | 0.42 | negative regulation of ethylene-activated signaling pathway |
| CmaCh11G005470 | 1.09 | 0.20 | ethylene-activated signaling pathway |
| CmaCh17G008890 | 2.32 | 0.86 | ethylene-activated signaling pathway |
| CmaCh19G010510 | 1.53 | 0.12 | negative regulation of ethylene-activated signaling pathway |
| Treatment | Female Flowers Per Plant % | First Female Flower Node | Bisexual Flowers Per Plant % | |||
|---|---|---|---|---|---|---|
| 2013–12 | 9–6 | 2013–12 | 9–6 | 2013–12 | 9–6 | |
| Ethrel | 72.5 ± 3.5a | 5 ± 1a | 5.9 ± 0.4c | 13.7 ± 1.5a | 3 ± 1.5b | 0 |
| IAA | 67.5 ± 5.5a | 4.5 ± 1.5a | 6.9 ± 1.5c | 14.6 ± 2.3a | 5.5 ± 1b | 0 |
| AVG | 37 ± 3.5c | 2 ± 1a | 12.5 ± 0.8a | 17.1 ± 1.2a | 28.5 ± 7.5a | 0 |
| AgNO3 | 33.5 ± 0.5c | 2 ± 1.2a | 13.7 ± 0.9a | 17 ± 1.6a | 33.5 ± 4a | 0 |
| control | 51 ± 5b | 4 ± 0.8a | 10.1 ± 1.6b | 15.4 ± 1.2a | 27.5 ± 5a | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Yan, C.; Zou, B.; Wang, C.; Xu, W.; Cui, C.; Qu, S. Morphological, Transcriptomic and Hormonal Characterization of Trimonoecious and Subandroecious Pumpkin (Cucurbita maxima) Suggests Important Roles of Ethylene in Sex Expression. Int. J. Mol. Sci. 2019, 20, 3185. https://doi.org/10.3390/ijms20133185
Wang Y, Yan C, Zou B, Wang C, Xu W, Cui C, Qu S. Morphological, Transcriptomic and Hormonal Characterization of Trimonoecious and Subandroecious Pumpkin (Cucurbita maxima) Suggests Important Roles of Ethylene in Sex Expression. International Journal of Molecular Sciences. 2019; 20(13):3185. https://doi.org/10.3390/ijms20133185
Chicago/Turabian StyleWang, Yunli, Chundong Yan, Bingxue Zou, Chaojie Wang, Wenlong Xu, Chongshi Cui, and Shuping Qu. 2019. "Morphological, Transcriptomic and Hormonal Characterization of Trimonoecious and Subandroecious Pumpkin (Cucurbita maxima) Suggests Important Roles of Ethylene in Sex Expression" International Journal of Molecular Sciences 20, no. 13: 3185. https://doi.org/10.3390/ijms20133185