Increased FLYWCH1 Expression is Negatively Correlated with Wnt/β-catenin Target Gene Expression in Acute Myeloid Leukemia Cells
Abstract
:1. Introduction
2. Results
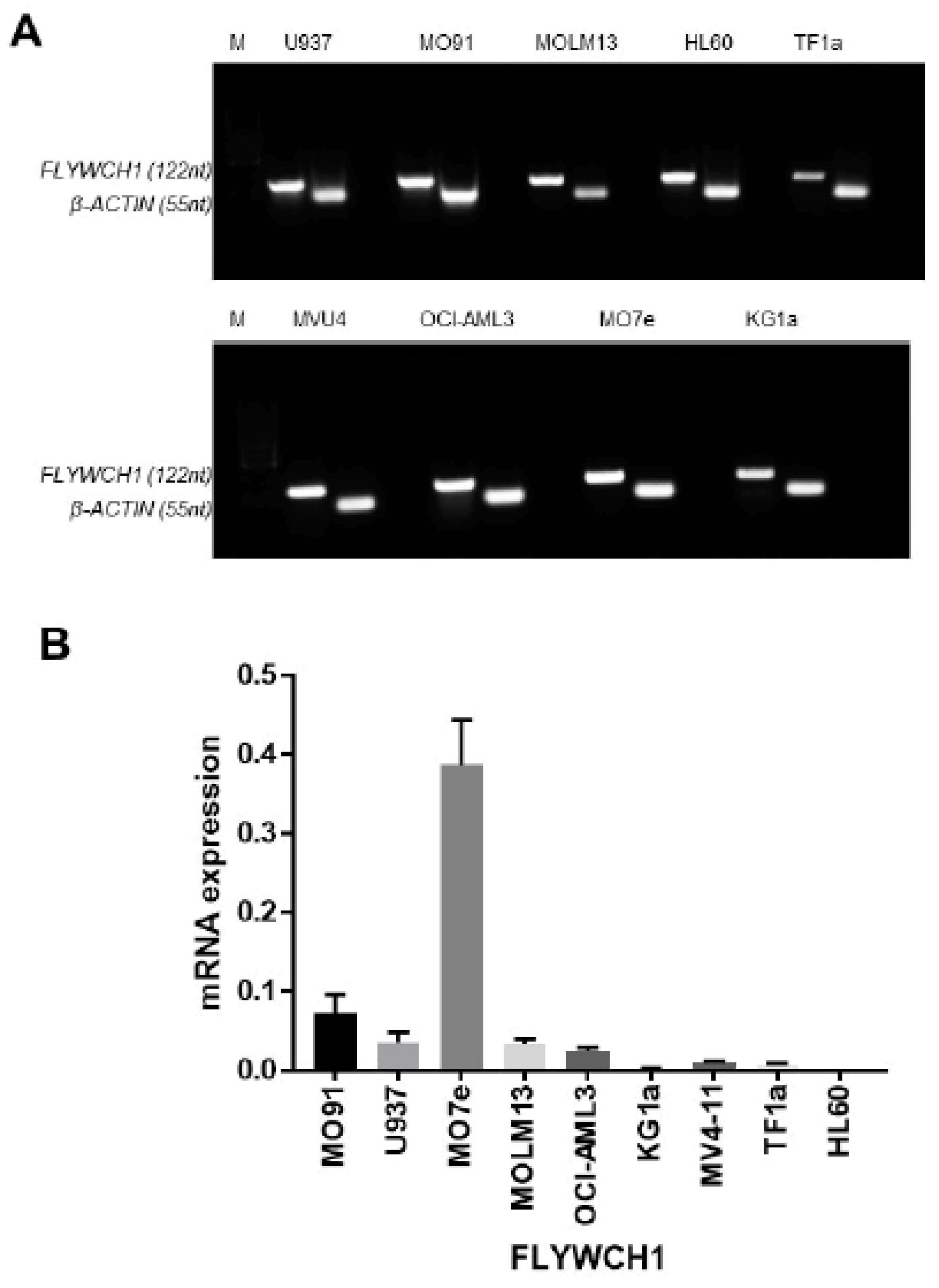
2.1. FLYWCH1 mRNA was Differentially Expressed in the AML Cell Lines
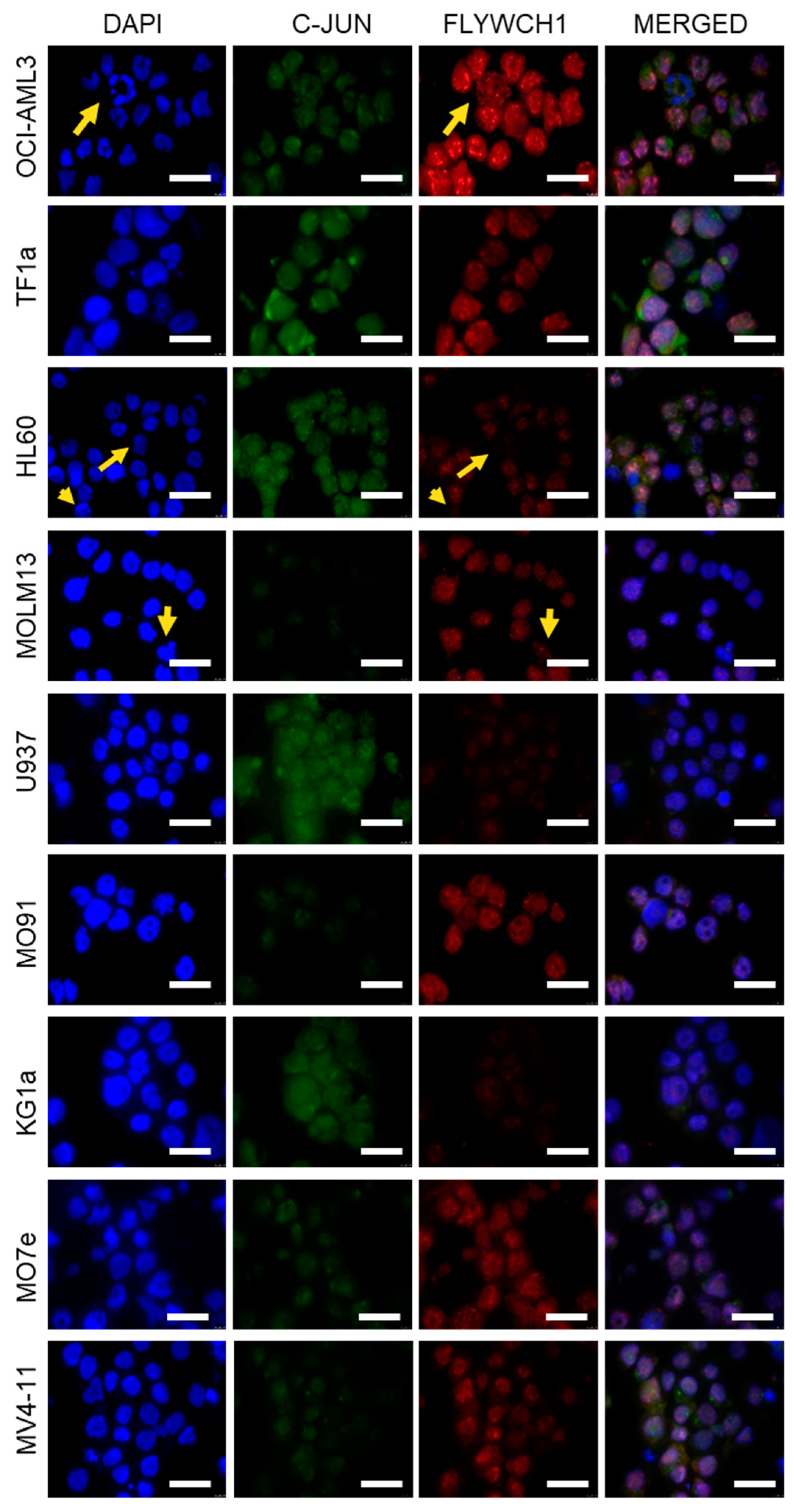
2.2. Immunofluorescence Staining Indicated Differential FLYWCH1 Protein Expression Levels in AML Cell Lines
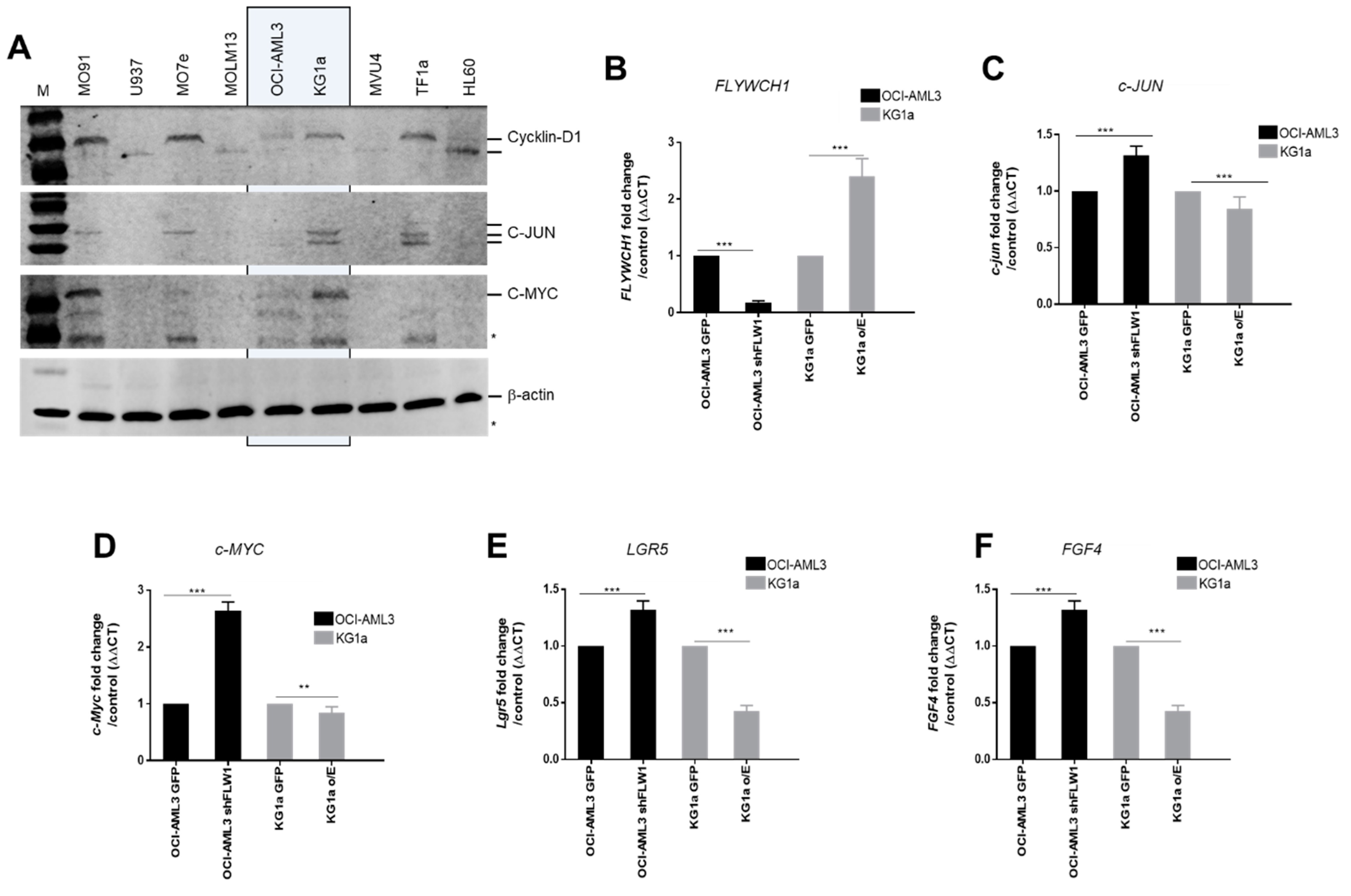
2.3. FLYWCH1 Represses the Expression of the β-catenin Target Genes
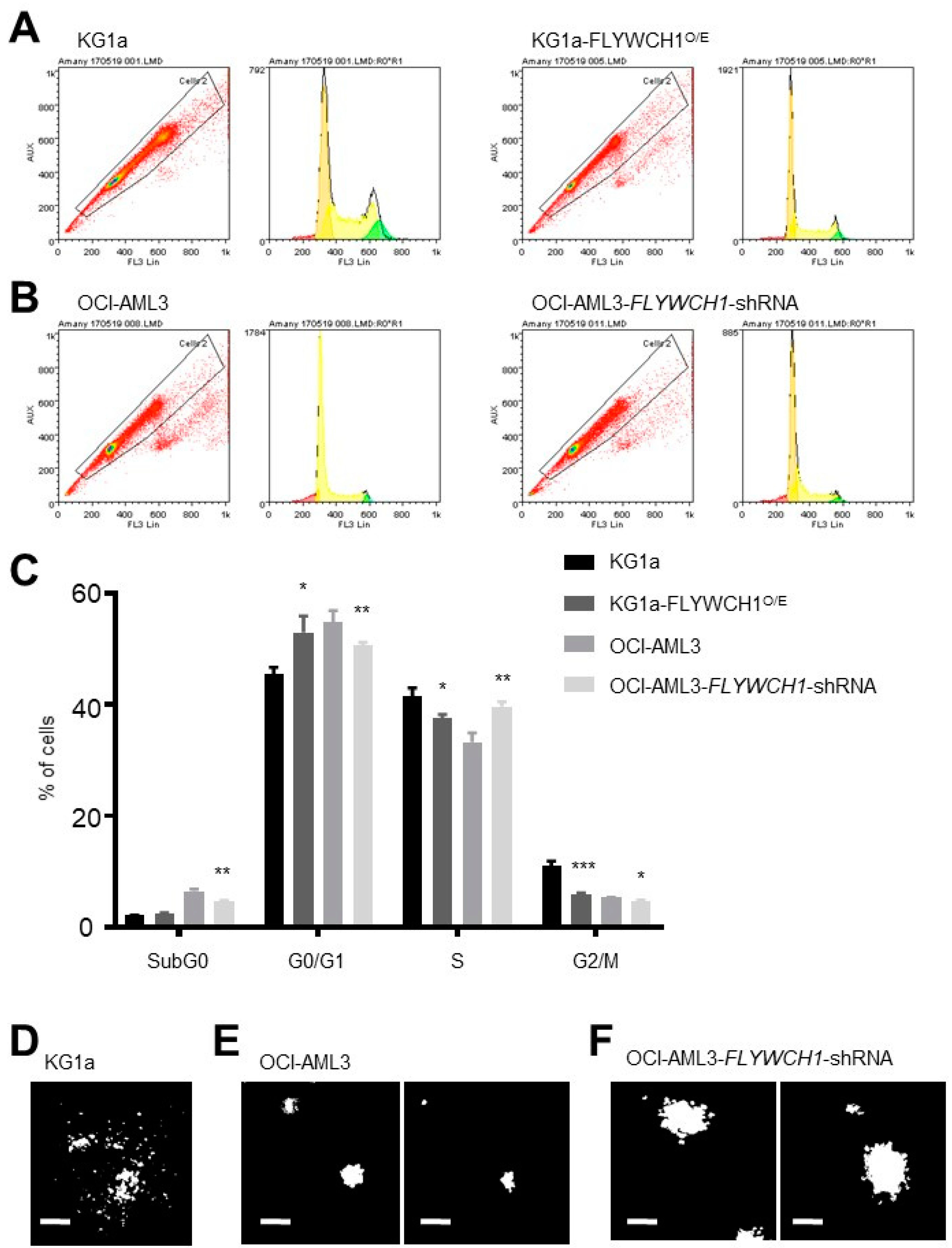
2.4. Significant Differences between sub-G0 and G0/G1 Phases of Low and High FLYWCH1 Expression Cell Lines
2.5. Low FLYWCH1-Expressing KG1a Cells Formed Scattered Colonies in Comparison with Typical Colony Formation in the High FLYWCH1-Expressing OCI-AML3 Cells
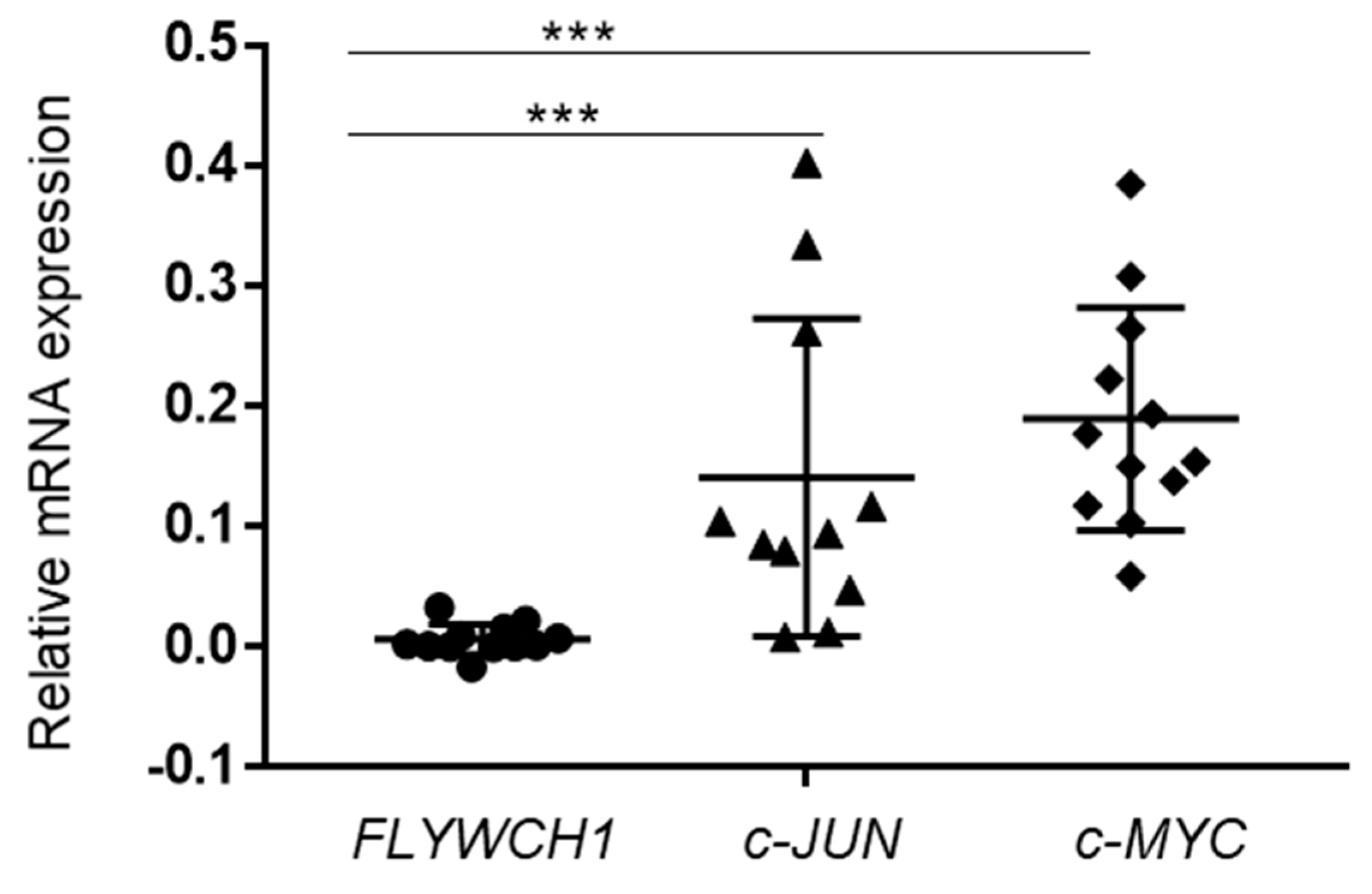
2.6. The c-JUN and c-MYC Expressed at Higher Levels in Comparison to FLYWCH1 in Primary AML Samples
3. Discussion
4. Material and Methods
4.1. Cell Culture
4.2. RNA Isolation, Reverse Transcription (RT), and Quantitative Real-Time PCR (qRT-PCR)
4.3. Western Blotting
4.4. Immunofluorescence Assay
4.5. Plasmids
4.6. Lentivirus Production and Cell Transduction
4.7. Cell Cycle Analysis
4.8. Colony Formation Assay
4.9. Patient Primary Samples
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AML | Acute myeloid leukaemia |
| CFU | Colony forming unit |
| CML | Chronic myeloid leukaemia |
| CSCs | Cancer stem cells |
| FACS | Fluorescence activated cell sorting |
| FLYWCH1 | FLYWCH-type zinc finger 1 |
| GADPH | Glyceraldehyde 3-phosphate dehydrogenase |
| HSC | Hematopoietic stem cell |
| LSCs | Leukaemia stem cells |
| mRNA | Messenger RNA |
| qPCR | Quantitative real-time polymerase chain reaction |
| PI | Propidium iodide |
| RIPA | Radio-immunoprecipitation assay |
| RNA | Ribonucleic acid |
| RRS | Ras-Recruitment System |
| scsRNA | Scrambled control shRNA |
| shRNA | small hairpin RNA |
| T-ALL | T-cell acute lymphoblastic leukaemia |
| ZNF | Zinc Finger Protein |
References
- Ferrara, F.; Schiffer, C.A. Acute myeloid leukaemia in adults. Lancet 2013, 381, 484–495. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Xu, J.; Ward, E. CA: A cancer journal for clinicians. Cancer Stat. 2010, 60, 277–300. [Google Scholar]
- Visser, O.; Trama, A.; Maynadié, M.; Stiller, C.; Marcos-Gragera, R.; De Angelis, R.; Mallone, S.; Tereanu, C.; Allemani, C.; Ricardi, U.; et al. Incidence, survival and prevalence of myeloid malignancies in Europe. Eur. J. Cancer 2012, 48, 3257–3266. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Odenike, O.; Rowley, J.D. Leukaemogenesis: More than mutant genes. Nat. Rev. Cancer 2010, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Amit, S.; Hatzubai, A.; Birman, Y.; Andersen, J.S.; Ben-Shushan, E.; Mann, M.; Ben-Neriah, Y.; Alkalay, I. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: A molecular switch for the Wnt pathway. Genes Dev. 2002, 16, 1066–1076. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef]
- Reya, T.; Duncan, A.W.; Ailles, L.; Domen, J.; Scherer, D.C.; Willert, K.; Hintz, L.; Nusse, R.; Weissman, I.L. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 2003, 423, 409. [Google Scholar] [CrossRef]
- Scheller, M.; Huelsken, J.; Rosenbauer, F.; Taketo, M.M.; Birchmeier, W.; Tenen, D.G.; Leutz, A. Hematopoietic stem cell and multilineage defects generated by constitutive β-catenin activation. Nat. Immunol. 2006, 7, 1037. [Google Scholar] [CrossRef]
- Kirstetter, P.; Anderson, K.; Porse, B.T.; Jacobsen, S.E.; Nerlov, C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nat. Immunol. 2006, 7, 1048–1056. [Google Scholar] [CrossRef]
- Wang, Y.; Krivtsov, A.V.; Sinha, A.U.; North, T.E.; Goessling, W.; Feng, Z.; Zon, L.I.; Armstrong, S.A. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science 2010, 327, 1650–1653. [Google Scholar] [CrossRef]
- Ysebaert, L.; Chicanne, G.; Demur, C.; De Toni, F.; Prade-Houdellier, N.; Ruidavets, J.B.; Mansat-De Mas, V.; Rigal-Huguet, F.; Laurent, G.; Payrastre, B.; et al. Expression of beta-catenin by acute myeloid leukemia cells predicts enhanced clonogenic capacities and poor prognosis. Leukemia 2006, 20, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Okuhashi, Y.; Itoh, M.; Nara, N.; Tohda, S. Effects of combination of notch inhibitor plus hedgehog inhibitor or Wnt inhibitor on growth of leukemia cells. Anticancer Res. 2011, 31, 893–896. [Google Scholar] [PubMed]
- Gandillet, A.; Park, S.; Lassailly, F.; Griessinger, E.; Vargaftig, J.; Filby, A.; Lister, T.A.; Bonnet, D. Heterogeneous sensitivity of human acute myeloid leukemia to beta-catenin down-modulation. Leukemia 2011, 25, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Siapati, E.K.; Papadaki, M.; Kozaou, Z.; Rouka, E.; Michali, E.; Savvidou, I.; Gogos, D.; Kyriakou, D.; Anagnostopoulos, N.I.; Vassilopoulos, G. Proliferation and bone marrow engraftment of AML blasts is dependent on beta-catenin signalling. Br. J. Haematol. 2011, 152, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.J.; Buske, C. Knocking the Wnt out of the sails of leukemia stem cell development. Cell Stem Cell 2007, 1, 597–598. [Google Scholar] [CrossRef] [PubMed]
- Olson, L.E.; Tollkuhn, J.; Scafoglio, C.; Krones, A.; Zhang, J.; Ohgi, K.A.; Wu, W.; Taketo, M.M.; Kemler, R.; Grosschedl, R. Homeodomain-mediated beta-catenin-dependent switching events dictate cell-lineage determination. Cell 2006, 125, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.F.; Ng, D.Y.; Jayakumaran, G.; Wood, G.A.; Koide, H.; Doble, B.W. beta-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent mechanism. Cell Stem Cell 2011, 8, 214–227. [Google Scholar] [CrossRef]
- Barker, N.; Hurlstone, A.; Musisi, H.; Miles, A.; Bienz, M.; Clevers, H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. Embo J. 2001, 20, 4935–4943. [Google Scholar] [CrossRef] [Green Version]
- Nateri, A.S.; Spencer-Dene, B.; Behrens, A. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature 2005, 437, 281–285. [Google Scholar] [CrossRef]
- Nateri, A.S.; Riera-Sans, L.; Da Costa, C.; Behrens, A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 2004, 303, 1374–1378. [Google Scholar] [CrossRef]
- Li, N.; Babaei-Jadidi, R.; Lorenzi, F.; Spencer-Dene, B.; Clarke, P.; Domingo, E.; Tulchinsky, E.; Vries, R.G.J.; Kerr, D.; Pan, Y.; et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis 2019, 8, 019–0125. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, B.A.; Almozyan, S.; Babaei-Jadidi, R.; Onyido, E.K.; Saadeddin, A.; Kashfi, S.H.; Spencer-Dene, B.; Ilyas, M.; Lourdusamy, A.; Behrens, A.; Nateri, A.S. FLYWCH1, a Novel Suppressor of Nuclear beta-Catenin, Regulates Migration and Morphology in Colorectal Cancer. Mol. Cancer Res. 2018, 16, 1977–1990. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.M.; Chen, X.; Zhang, W.; Liu, C. KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol. Cell Biol. 2010, 30, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Kang, H.S.; Jetten, A.M. The Kruppel-like zinc finger protein Glis2 functions as a negative modulator of the Wnt/beta-catenin signaling pathway. FEBS Lett. 2007, 581, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Razin, S.V.; Borunova, V.V.; Maksimenko, O.G.; Kantidze, O.L. Cys2His2 zinc finger protein family: Classification, functions, and major members. Biochemistry 2012, 77, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Jen, J.; Wang, Y.C. Zinc finger proteins in cancer progression. J. Biomed. Sci. 2016, 23, 53. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.S.; Sun, X.X.; Qin, J.; Smolik, S.M.; Lu, H. Identification and characterization of a novel Drosophila melanogaster glutathione S-transferase-containing FLYWCH zinc finger protein. Gene 2004, 342, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Krauss, V.; Dorn, R. Evolution of the trans-splicing Drosophila locus mod(mdg4) in several species of Diptera and Lepidoptera. Gene 2004, 331, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Beaster-Jones, L.; Okkema, P.G. DNA binding and in vivo function of C.elegans PEB-1 require a conserved FLYWCH motif. J. Mol. Biol. 2004, 339, 695–706. [Google Scholar] [CrossRef]
- Ow, M.C.; Martinez, N.J.; Olsen, P.H.; Silverman, H.S.; Barrasa, M.I.; Conradt, B.; Walhout, A.J.; Ambros, V. The FLYWCH transcription factors FLH-1, FLH-2, and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes Dev. 2008, 22, 2520–2534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, H.; Li, Q.; O’Neal, J.; Kreisel, F.; Le Beau, M.M.; Tomasson, M.H. c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood 2005, 106, 2452–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, K.R.; Ohta, K.; Nakamae, H.; Hino, M.; Yamane, T.; Takubo, T.; Tatsumi, N. Differential effects of fibroblast growth factor-4, epidermal growth factor and transforming growth factor-beta1 on functional development of stromal layers in acute myeloid leukemia. Leuk. Res. 2002, 26, 933–938. [Google Scholar] [CrossRef]
- Piragyte, I.; Clapes, T.; Polyzou, A.; Klein Geltink, R.I.; Lefkopoulos, S.; Yin, N.; Cauchy, P.; Curtis, J.D.; Klaeylé, L.; Langa, X.; et al. A metabolic interplay coordinated by HLX regulates myeloid differentiation and AML through partly overlapping pathways. Nat. Commun. 2018, 9, 3090–3107. [Google Scholar] [CrossRef] [PubMed]
- Kühnl, A.; Valk, P.J.; Sanders, M.A.; Ivey, A.; Hills, R.K.; Mills, K.I.; Gale, R.E.; Kaiser, M.F.; Dillon, R.; Joannides, M.; et al. Downregulation of the Wnt inhibitor CXXC5 predicts a better prognosis in acute myeloid leukemia. Blood 2015, 125, 2985–2994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Azim, H.; Zhu, Y.; Hollis, R.; Wang, X.; Ge, S.; Hao, Q.L.; Smbatyan, G.; Kohn, D.B.; Rosol, M.; Crooks, G.M. Expansion of multipotent and lymphoid-committed human progenitors through intracellular dimerization of Mpl. Blood 2008, 111, 4064–4074. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikesch, J.; Steffen, B.; Berdel, W.E.; Serve, H.; Müller-Tidow, C. The emerging role of Wnt signaling in the pathogenesis of acute myeloid leukemia. Leukemia 2007, 21, 1638. [Google Scholar] [CrossRef]
- Kode, A.; Manavalan, J.S.; Mosialou, I.; Bhagat, G.; Rathinam, C.V.; Luo, N.; Khiabanian, H.; Lee, A.; Murty, V.V.; Friedman, R.; et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature 2014, 506, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Xiao, G.; Hu, J. Regulation of Wnt/β-catenin signaling by posttranslational modifications. Cell Biosci. 2014, 4, 13. [Google Scholar]
- Clevers, H.; Nusse, R. Wnt/beta-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Saland, E.; Boutzen, H.; Castellano, R.; Pouyet, L.; Griessinger, E.; Larrue, C.; de Toni, F.; Scotland, S.; David, M.; Danet-Desnoyers, G.; et al. A robust and rapid xenograft model to assess efficacy of chemotherapeutic agents for human acute myeloid leukemia. Blood Cancer J. 2015, 5, e297. [Google Scholar] [CrossRef] [PubMed]
- OH, N.; Erbilgin, Y.; Firtina, S.; Celkan, T.; Karakas, Z.; Aydogan, G.; Turkkan, E.; Yildirmak, Y.; Timur, C.; Zengin, E.; et al. Deregulated WNT signaling in childhood T-cell acute lymphoblastic leukemia. Blood Cancer J. 2014, 4, e192. [Google Scholar]
- Famili, F.; Brugman, M.H.; Taskesen, E.; Naber, B.E.A.; Fodde, R.; Staal, F.J.T. High levels of canonical Wnt signaling lead to loss of stemness and increased differentiation in hematopoietic stem cells. Stem Cell Rep. 2016, 6, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Coulombel, L. Identification of hematopoietic stem/progenitor cells: Strength and drawbacks of functional assays. Oncogene 2004, 23, 7210. [Google Scholar] [CrossRef] [PubMed]
- Hogge, D.; Lansdorp, P.M.; Reid, D.; Gerhard, B.; Eaves, C.J. Enhanced detection, maintenance, and differentiation of primitive human hematopoietic cells in cultures containing murine fibroblasts engineered to produce human steel factor, interleukin-3, and granulocyte colony-stimulating factor. Blood 1996, 88, 3765–3773. [Google Scholar] [PubMed]
- Grandori, C.; Cowley, S.M.; James, L.P.; Eisenman, R.N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000, 16, 653–699. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.H.; Loboda, A.P.; Showe, M.K.; Showe, L.C.; McMahon, S.B. Analysis of genomic targets reveals complex functions of MYC. Nat. Rev. Cancer 2004, 4, 562. [Google Scholar] [CrossRef] [PubMed]
- Karn, J.; Watson, J.V.; Lowe, A.D.; Green, S.M.; Vedeckis, W. Regulation of cell cycle duration by c-myc levels. Oncogene 1989, 4, 773–787. [Google Scholar]
- Shichiri, M.; Hanson, K.D.; Sedivy, J.M. Effects of c-myc Expression on Proliferation, Quiescence, and the G~ 0 to G~ 1 Transition in Nontransformed Cells. Cell Growth Differ. 1993, 4, 93–104. [Google Scholar]
- Jiang, W.; Kahn, S.M.; Zhou, P.; Zhang, Y.J.; Cacace, A.M.; Infante, A.S.; Doi, S.; Santella, R.M.; Weinstein, I.B. Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene 1993, 8, 3447–3457. [Google Scholar] [PubMed]
- Quelle, D.; Ashmun, R.A.; Shurtleff, S.A.; Kato, J.Y.; Bar-Sagi, D.; Roussel, M.F.; Sherr, C.J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993, 7, 1559–1571. [Google Scholar] [CrossRef] [PubMed]
- Scandura, J.M.; Boccuni, P.; Massagué, J.; Nimer, S.D. Transforming growth factor beta-induced cell cycle arrest of human hematopoietic cells requires p57KIP2 up-regulation. Proc. Natl. Acad. Sci. USA 2004, 101, 15231–15236. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almars, A.; Chondrou, P.S.; Onyido, E.K.; Almozyan, S.; Seedhouse, C.; Babaei-Jadidi, R.; Nateri, A.S. Increased FLYWCH1 Expression is Negatively Correlated with Wnt/β-catenin Target Gene Expression in Acute Myeloid Leukemia Cells. Int. J. Mol. Sci. 2019, 20, 2739. https://doi.org/10.3390/ijms20112739
Almars A, Chondrou PS, Onyido EK, Almozyan S, Seedhouse C, Babaei-Jadidi R, Nateri AS. Increased FLYWCH1 Expression is Negatively Correlated with Wnt/β-catenin Target Gene Expression in Acute Myeloid Leukemia Cells. International Journal of Molecular Sciences. 2019; 20(11):2739. https://doi.org/10.3390/ijms20112739
Chicago/Turabian StyleAlmars, Amany, Panagiota S. Chondrou, Emenike K. Onyido, Sheema Almozyan, Claire Seedhouse, Roya Babaei-Jadidi, and Abdolrahman S. Nateri. 2019. "Increased FLYWCH1 Expression is Negatively Correlated with Wnt/β-catenin Target Gene Expression in Acute Myeloid Leukemia Cells" International Journal of Molecular Sciences 20, no. 11: 2739. https://doi.org/10.3390/ijms20112739




