Mutations in the Rice OsCHR4 Gene, Encoding a CHD3 Family Chromatin Remodeler, Induce Narrow and Rolled Leaves with Increased Cuticular Wax
Abstract
:1. Introduction
2. Results
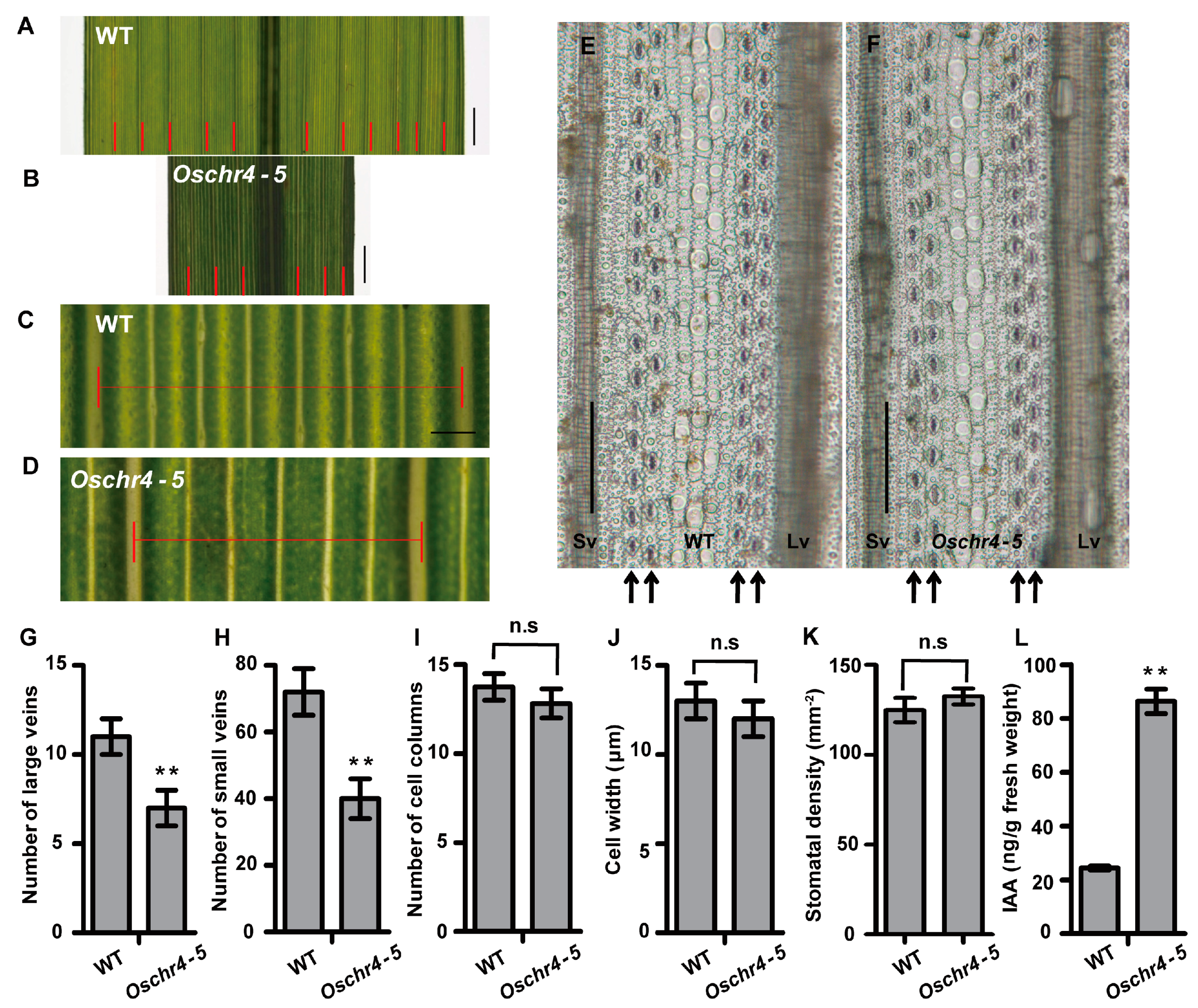
2.1. Pleiotropic Phenotype of the Oschr4-5 Mutant
2.2. Anatomical and Physiological Basis of Reduced Leaf Width
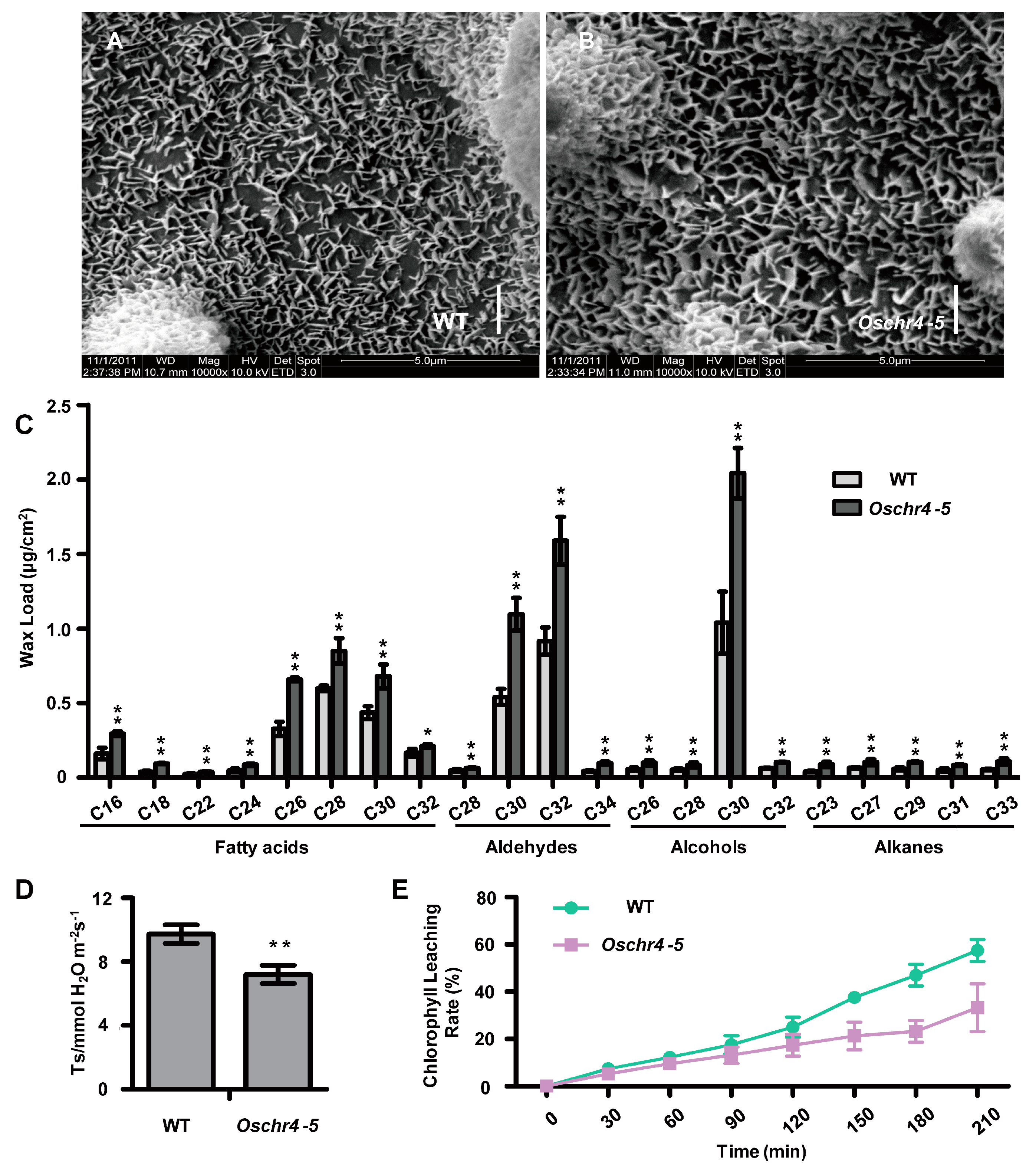
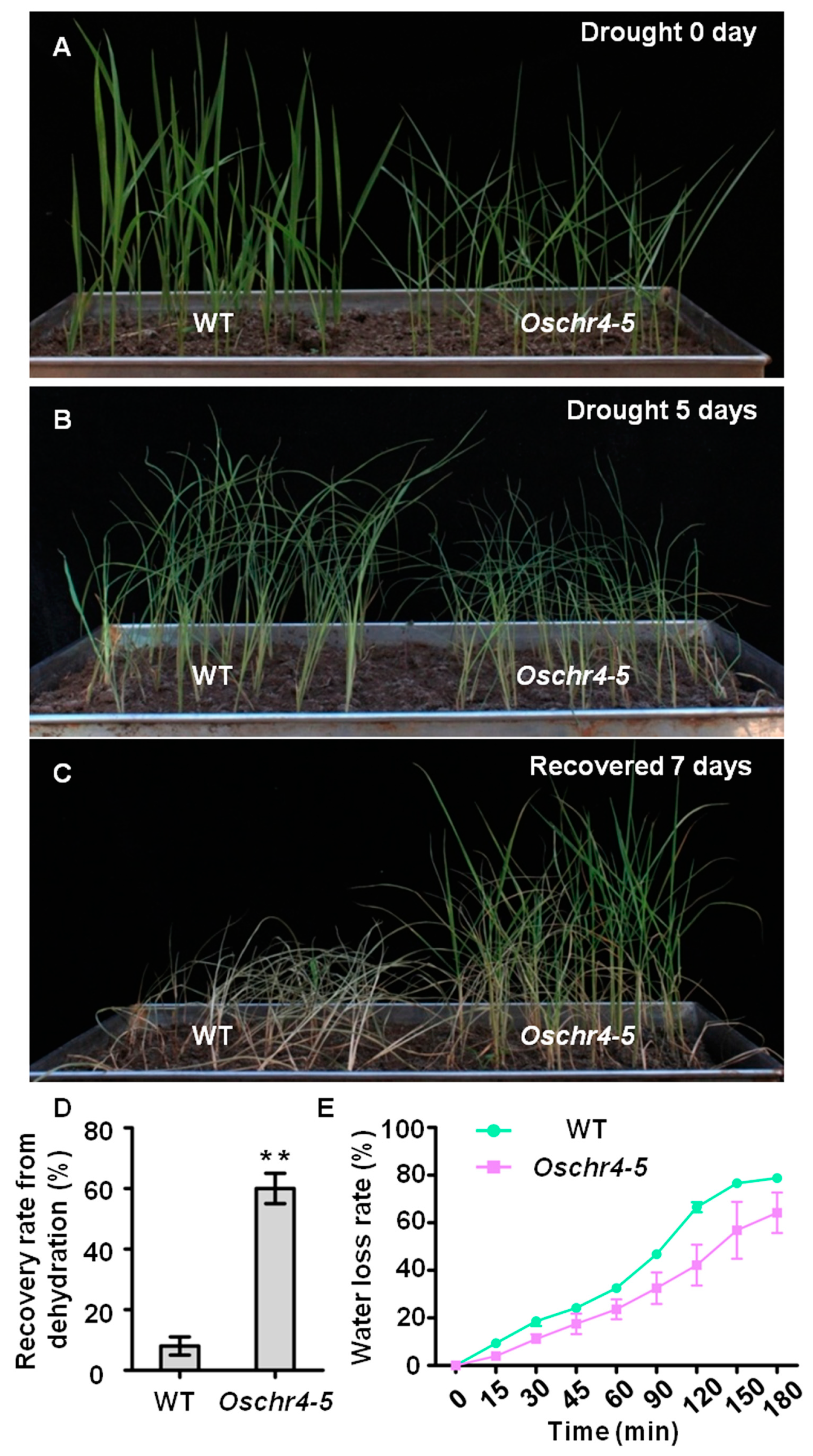
2.3. Increased Cuticular Wax on the Surface of Leaves with Enhanced Drought Tolerance in Oschr4-5
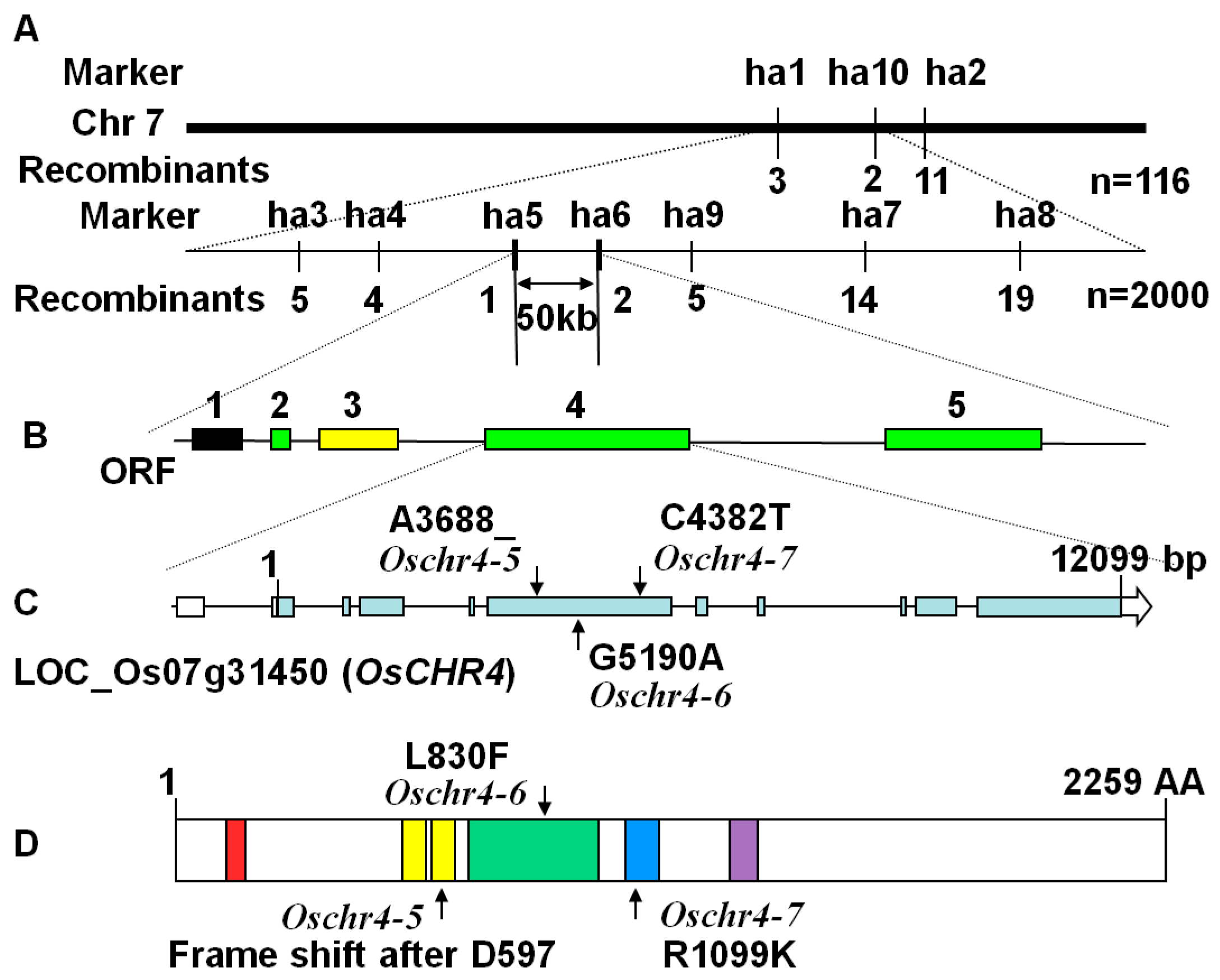
2.4. Characterization of the Molecular Lesions in the Oschr4 Mutant
2.5. Alignment and Phylogenetic Analysis
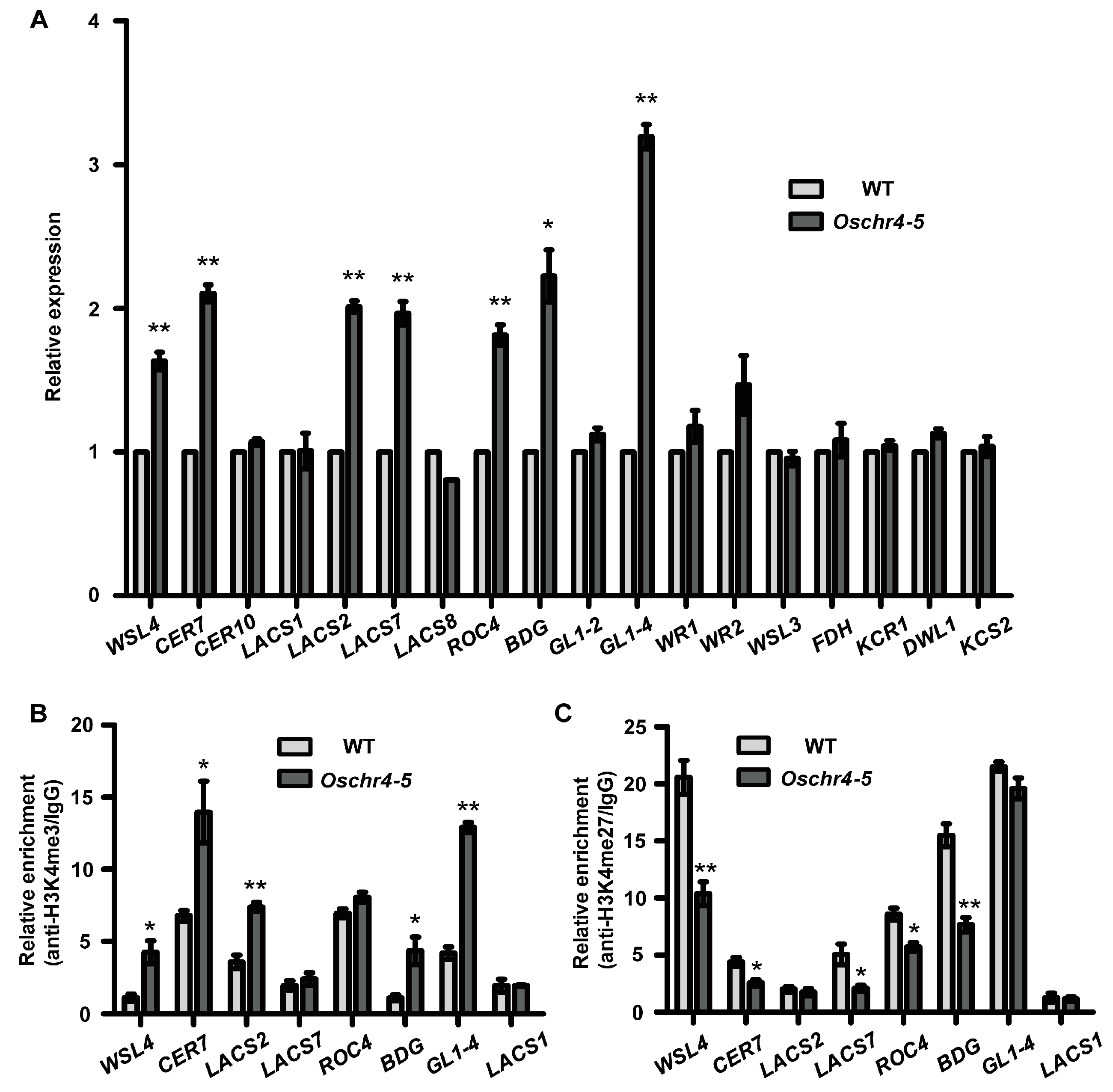
2.6. Expression of Several Wax and Auxin Biosynthesis Related Genes Were Upregulated in Oschr4-5
2.7. Histone Modifications of the Wax and Auxin Biosynthesis-Related Genes Were Affected in Oschr4-5
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Condition
4.2. Microscopic Observation
4.3. Measurement of Chlorophyll and Indole Acetic Acid (IAA)
4.4. Measurement of Cuticular Waxes
4.5. Water Loss and Chlorophyll-Leaching Assays
4.6. Mapping and Sequencing
4.7. Real-Time Polymerase Chain Reaction (RT-PCR)
4.8. Sequence and Phylogenetic Analyses
4.9. Chromatin Immunoprecipitation (ChIP) Analysis
4.10. Accession Number
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CHD3 | Chromodomain Helicase DNA-binding (CHD) protein 3 |
| ChIP | Chromatin immunoprecipitation |
| CHR4 | CHD-related protein 4 |
| NAL1 | Narrow leaf 1 |
References
- Lang, Y.; Zhang, Z.; Gu, X.; Yang, J.; Zhu, Q. Physiological and ecological effect of crimpy leaf character in rice (Oryza sativa L.). II. Photosynthetic character, dry mass production and yield forming. Acta Agron. Sin. 2004, 30, 883–887. [Google Scholar]
- Jiang, D.; Fang, J.; Lou, L.; Zhao, J.; Yuan, S.; Yin, L.; Sun, W.; Peng, L.; Guo, B.; Li, X. Characterization of a null allelic mutant of the rice NAL1 gene reveals its role in regulating cell division. PLoS ONE 2015, 10, e0118169. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Qian, Q.; Bu, Q.; Li, S.; Chen, Q.; Sun, J.; Liang, W.; Zhou, Y.; Chu, C.; Li, X.; et al. Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008, 147, 1947–1959. [Google Scholar] [CrossRef] [PubMed]
- Takashi, S.; Noriko, K.; Takeshi, N.; Kozue, O.; Yutaka, S.; Kohei, I.; Yasuo, N.; Tomokazu, K.; Yoshiaki, N.; Motoyuki, A. A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 2009, 60, 227–241. [Google Scholar]
- Woo, Y.M.; Park, H.J.; Su’udi, M.; Yang, J.I.; Park, J.J.; Back, K.; Park, Y.M.; An, G. Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 2007, 65, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Matsuda, Y.; Ozawa, K.; Nishimura, T.; Koshiba, T.; Fraaije, M.W.; Sekiguchi, H. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 2008, 279, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Morinaka, Y.; Inukai, Y.; Kitano, H.; Fujioka, S. Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 2013, 73, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, C.; Hu, G.; Xing, L.; Qian, W.; Si, H.; Sun, Z.; Wang, X.; Fu, Y. Characterization and Fine Mapping of a Novel Rice Narrow Leaf Mutant nal9. J. Integr. Plant Biol. 2013, 55, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Luo, L.; Chen, L.; Tao, X.; Huang, M.; Wang, H.; Chen, Z.; Xiao, W. Chromosome mapping, molecular cloning and expression analysis of a novel gene response for leaf width in rice. Biochem. Biophys. Res. Commun. 2016, 480, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yan, C.; Zeng, X.; Yang, Y.; Fang, Y.; Tian, C.; Sun, Y.; Cheng, Z.; Gu, M. ROLLED LEAF 9, encoding a GARP protein, regulates the leaf abaxial cell fate in rice. Plant Mol. Biol. 2008, 68, 239–250. [Google Scholar]
- Zhang, G.; Xu, Q.; Zhu, X.; Qian, Q.; Xue, H. SHALLOT-LIKE1 Is a KANADI Transcription Factor That Modulates Rice Leaf Rolling by Regulating Leaf Abaxial Cell Development. Plant Cell 2009, 21, 719–735. [Google Scholar] [CrossRef] [Green Version]
- Ken-Ichiro, H.; Mari, O.; Emi, H.; Masashi, A.; Tsutomu, I.; Hikaru, S.; Jun-Ichi, I.; Yasuo, N. The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev. Biol. 2009, 334, 345–354. [Google Scholar]
- Li, L.; Shi, Z.; Li, L.; Shen, G.; Wang, X.; An, L.; Zhang, J. Overexpression of ACL 1 (abaxially curled leaf 1) Increased Bulliform Cells and Induced Abaxial Curling of Leaf Blades in Rice. Mol. Plant. 2010, 3, 807–817. [Google Scholar] [CrossRef]
- Shi, Z.; Wang, J.; Wan, X.; Shen, G.; Wang, X.; Zhang, J. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 2007, 226, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Zhang, G.; Qian, Q.; Xue, H. Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 2012, 159, 1488–1500. [Google Scholar] [CrossRef]
- Wu, C.; Fu, Y.; Hu, G.; Si, H.; Cheng, S.; Liu, W. Isolation and characterization of a rice mutant with narrow and rolled leaves. Planta 2010, 232, 313–324. [Google Scholar] [CrossRef]
- Hu, J.; Zhu, L.; Zeng, D.; Gao, Z.; Guo, L.; Fang, Y.; Zhang, G.; Dong, G.; Yan, M.; Liu, J. Identification and characterization of NARROW ANDROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 2010, 73, 283–292. [Google Scholar] [CrossRef]
- Zhao, S.; Zhao, L.; Liu, F.; Wu, Y.; Zhu, Z.; Sun, C.; Tan, L. NARROW AND ROLLED LEAF 2 regulates leaf shape, male fertility, and seed size in rice. J. Integr. Plant Biol. 2016, 58, 983–996. [Google Scholar] [CrossRef]
- Chen, W.; Sheng, Z.; Cai, Y.; Li, Q.; Wei, X.; Xie, L.; Jiao, G.; Shao, G.; Tang, S.; Wang, J.; et al. Rice Morphogenesis and chlorophyll accumulation is regulated by the protein encoded by NRL3 and its interaction with NAL9. Front. Plant Sci. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Xia, Y.; Nikolau, B.J.; Schnable, P.S. Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell 1996, 8, 1291–1304. [Google Scholar] [CrossRef]
- Islam, M.A.; Hao, D.; Jing, N.; Ye, H.; Xiong, L. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 2009, 70, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.; Tang, D.; Huang, J.; Li, M.; Wu, X.; Lu, L.; Wang, J.; Yu, X.; Chen, J.; Gu, M. Rice OsGL 1-1 is involved in leaf cuticular wax and cuticle membrane. Mol. Plant. 2011, 4, 985–995. [Google Scholar] [CrossRef]
- Zhou, L.; Ni, E.; Yang, J.; Zhou, H.; Liang, H.; Li, J.; Jiang, D.; Wang, Z.; Liu, Z.; Zhuang, C. Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS ONE 2013, 8, e65139. [Google Scholar] [CrossRef]
- Mao, B.; Cheng, Z.; Lei, C.; Xu, F.; Gao, S.; Ren, Y.; Wang, J.; Zhang, X.; Wang, J.; Wu, F. Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 2012, 235, 39–52. [Google Scholar] [CrossRef]
- Jung, K.; Han, M.; Lee, D.; Lee, Y.; Lukas, S.; Rochus, F.; Andrea, F.; Alexander, Y.; Heinz, S.; Kim, Y.; et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 2006, 18, 3015–3032. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wan, L.; Zhang, L.; Zhang, Z.; Zhang, H.; Quan, R.; Zhou, S.; Huang, R. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 2012, 78, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jenks, M.A.; Liu, J.; Liu, A.; Zhang, X.; Xiang, J.; Zou, J.; Peng, Y.; Chen, X. Overexpression of Transcription Factor OsWR2 Regulates Wax and Cutin Biosynthesis in Rice and Enhances its Tolerance to Water Deficit. Plant Mol. Biol. Rep. 2014, 32, 719–731. [Google Scholar] [CrossRef]
- Wang, Z.; Tian, X.; Zhao, Q.; Liu, Z.; Li, X.; Ren, Y.; Tang, J.; Fang, J.; Xu, Q.; Bu, Q. The E3 Ligase DROUGHT HYPERSENSITIVE Negatively Regulates Cuticular Wax Biosynthesis by Promoting the Degradation of Transcription Factor ROC4 in Rice. Plant Cell 2018, 30, 228–244. [Google Scholar] [CrossRef]
- Bernard, A.; Joubès, J. Arabidopsis cuticular waxes: Advances in synthesis, export and regulation. Prog. Lipid Res. 2013, 52, 110–129. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Cairns, B.R. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Hall, J.A.; Georgel, P.T. CHD proteins: A diverse family with strong ties. Biochem. Cell Biol. 2007, 85, 463–476. [Google Scholar] [CrossRef]
- Stanley, D.R.; Henderson, J.T.; Jerome, R.E.; Edenberg, H.J.; Jeanne, R.S.; Joe, O. Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 2010, 35, 33–43. [Google Scholar]
- Hay, A.; Kaur, H.; Phillips, A.; Hedden, P.; Hake, S.; Tsiantis, M. The Gibberellin Pathway Mediates KNOTTED1-Type Homeobox Function in Plants with Different Body Plans. Curr. Biol. 2002, 12, 1557–1565. [Google Scholar] [CrossRef] [Green Version]
- Franziska, T.; Francois, R.; Sara, F.; Marie-Laure, M.M.; Elodie, G.; Nicolas, B.; Séverine, G.; Martienssen, R.A.; George, C.; Vincent, C. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007, 3, 855–866. [Google Scholar]
- Hu, Y.; Lai, Y.; Zhu, D. Transcription regulation by CHD proteins to control plant development. Front. Plant Sci. 2014, 5, 223. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Liu, D.; Zhong, X.; Zhang, C.; Zhang, Q.; Zhou, D. CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. USA 2012, 109, 5773–5778. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Xu, J.; Chen, Y.; Mao, C.; Zhang, S.; Bai, Y.; Jiang, D.; Wu, P. Molecular cloning and characterization of OsCHR4, a rice chromatin-remodeling factor required for early chloroplast development in adaxial mesophyll. Planta 2012, 236, 1165–1176. [Google Scholar] [CrossRef]
- Ma, X.; Ma, J.; Zhai, H.; Xin, P.; Chu, J.; Qiao, Y.; Han, L. CHR729 Is a CHD3 Protein That Controls Seedling Development in Rice. PLoS ONE 2015, 10, e0138934. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Gan, T.; Liu, L.; Long, W.; Wang, Y.; Niu, M.; Li, X.; Zheng, M.; Jiang, L.; et al. CRL6, a member of the CHD protein family, is required for crown root development in rice. Plant Physiol. Biochem. 2016, 105, 185–194. [Google Scholar] [CrossRef]
- Cho, S.; Lee, C.; Gi, E.; Yim, Y.; Koh, H.; Kang, K.; Paek, N. The Rice Rolled Fine Striped (RFS) CHD3/Mi-2 Chromatin Remodeling Factor Epigenetically Regulates Genes Involved in Oxidative Stress Responses During Leaf Development. Front. Plant Sci. 2018, 9, 364. [Google Scholar] [CrossRef] [Green Version]
- Enrico, S.; Michalis, B.; Miltos, T. Control of leaf and vein development by auxin. Cold Spring Harbor Perspect. Biol. 2010, 2, a001511. [Google Scholar]
- Klee, H.J.; Horsch, R.B.; Hinchee, M.A.; Hein, M.B.; Hoffmann, N.L. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Gene Dev. 1987, 1, 86–96. [Google Scholar] [CrossRef]
- Rafael, T.S.; Mattijs, B.; Karin, L.; GöRan, S.; Mol, J.N.M.; Erik, S.; Ronald, K. FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev. 2002, 16, 753. [Google Scholar]
- Zhu, X.; Xiong, L. Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc. Natl. Acad. Sci. USA 2013, 110, 17790–17795. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Guan, Y.; Zhang, D.; Dong, X.; Tian, L.; Qu, L. A β-ketoacyl-CoA synthase is Involved in Rice Leaf Cuticular Wax Synthesis and requires a CER2-LIKE Protein as a Cofactor. Plant Physiol. 2017, 173, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef]
- Abu-Zaitoon, Y.M.; Bennett, K.; Normanly, J.; Nonhebel, H.M. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol. Plant. 2012, 146, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, X.; He, K.; Ma, Y.; Su, N.; He, H.; Stolc, V.; Tongprasit, W.; Jin, W.; Jiang, J.; et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 2008, 20, 259–276. [Google Scholar] [CrossRef]
- Wang, X.; Elling, A.; Li, X.; Li, N.; Peng, Z.; He, G.; Sun, H.; Qi, Y.; Liu, X.; Deng, X. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 2009, 21, 1053–1069. [Google Scholar] [CrossRef]
- Wang, F.; Tang, Y.; Miao, R.; Xu, F.; Lin, T.; He, G.; Sang, X. Identification and gene mapping of a narrow and upper-albino leaf mutant in rice (Oryza sativa L.). Chin. Sci. Bull. 2012, 57, 3798–3803. [Google Scholar] [CrossRef]
- Ernst, A.; Villar, C.B.R.; Sara, F.; Reyes, J.C.; Lars, H.; Claudia, K.H. CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 2009, 5, e1000605. [Google Scholar]
- Ogas, J.; Cheng, J.; Sung, Z.; Somerville, C. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 1997, 277, 91–94. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.T.; Li, H.-C.; Rider, S.D.; Mordhorst, A.P.; Romero-Severson, J.; Cheng, J.-C.; Robey, J.; Sung, Z.R.; de Vries, S.C.; Ogas, J. PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 2004, 134, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; King, C.; Henderson, J.T.; Stanley Dean, R.; Yinglin, B.; Heng, Z.; Matthew, F.; Jacob, G.; Joe, O. PICKLE acts during germination to repress expression of embryonic traits. Plant J. 2005, 44, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Ogas, J.; Kaufmann, S.; Henderson, J.; Somerville, C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 13839–13844. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, L.; Xiang, J.; Gao, G.; Xu, F.; Liu, A.; Zhang, X.; Peng, Y.; Chen, X.; Wan, X. OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS ONE 2015, 10, e116676. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Zhu, S.; Zhao, Z.; Liu, L.; Wang, X.; Zhang, Z.; Zhang, X.; Wang, J.; Wang, J.; Guo, X.; et al. Wax Crystal-Sparse Leaf 4, encoding a b-ketoacyl-coenzyme A synthase 6, is involved in rice cuticular wax accumulation. Plant Cell Rep. 2017, 36, 1655–1666. [Google Scholar] [CrossRef]
- Lco, S.; Song, T.; Kosma, D.K.; Parsons, E.P.; Rowland, O.; Jenks, M.A. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2010, 59, 553–564. [Google Scholar]
- Zhao, Y.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; José, A.; Ecker, J.R.; Jennifer, N.; Joanne, C.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002, 16, 3100–3112. [Google Scholar] [CrossRef]
- Kakei, Y.; Nakamura, A.; Yamamoto, M.; Ishida, Y.; Yamazaki, C.; Sato, A.; Narukawa-Nara, M.; Soeno, K.; Shimada, Y. Biochemical and chemical biology study of rice OsTAR1 revealed that tryptophan aminotransferase is involved in auxin biosynthesis; identification of a potent OsTAR1 inhibitor, pyruvamine2031. Plant Cell Physiol. 2017, 58, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Kiyoshi, M.; Chen, Q.; Hiroyuki, K.; Yuji, K.; Sunil, O.; Jennifer, D.B.; David, B.; Zhao, Y. The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 2013, 288, 1448–1457. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of Auxin-Regulated Gene Expression in Plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Jörg, F.; Dmitri, D.; Andreas, H.; Ingo, S. Chromosomal histone modification patterns—From conservation to diversity. Trends Plant Sci. 2006, 11, 199–208. [Google Scholar]
- Ernst, A.; Villar, C.B.R.; Riccardo, D.M.; Sabrina, S.; Claudia, K. The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 2011, 23, 1047–1060. [Google Scholar]
- Gou, J.; Strauss, S.H.; Tsai, C.J.; Fang, K.; Chen, Y.; Jiang, X.; Busov, V.B. Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones. Plant Cell 2010, 22, 623–639. [Google Scholar] [CrossRef] [PubMed]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Gendrel, A.V.; Lippman, Z.; Martienssen, R.; Colot, V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2005, 2, 213–218. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, T.; Wang, D.; Fang, J.; Zhao, J.; Yuan, S.; Xiao, L.; Li, X. Mutations in the Rice OsCHR4 Gene, Encoding a CHD3 Family Chromatin Remodeler, Induce Narrow and Rolled Leaves with Increased Cuticular Wax. Int. J. Mol. Sci. 2019, 20, 2567. https://doi.org/10.3390/ijms20102567
Guo T, Wang D, Fang J, Zhao J, Yuan S, Xiao L, Li X. Mutations in the Rice OsCHR4 Gene, Encoding a CHD3 Family Chromatin Remodeler, Induce Narrow and Rolled Leaves with Increased Cuticular Wax. International Journal of Molecular Sciences. 2019; 20(10):2567. https://doi.org/10.3390/ijms20102567
Chicago/Turabian StyleGuo, Tingting, Daofeng Wang, Jingjing Fang, Jinfeng Zhao, Shoujiang Yuan, Langtao Xiao, and Xueyong Li. 2019. "Mutations in the Rice OsCHR4 Gene, Encoding a CHD3 Family Chromatin Remodeler, Induce Narrow and Rolled Leaves with Increased Cuticular Wax" International Journal of Molecular Sciences 20, no. 10: 2567. https://doi.org/10.3390/ijms20102567




