Host-Induced Gene Silencing: A Powerful Strategy to Control Diseases of Wheat and Barley
Abstract
:1. Introduction
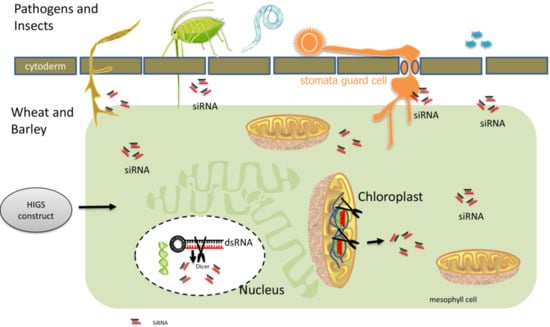
2. General Mechanism of HIGS
3. HIGS Used for Controlling Diseases of Wheat and Barely
3.1. HIGS against Insect Pests
3.2. HIGS against Nematodes
3.3. HIGS against Viruses
3.4. HIGS against Fungi
4. Current Challenges of HIGS
5. Conclusions and Future Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Blanco, A.; Cenci, A.; Simeone, R.; Gadaleta, A.; Pignone, D.; Galasso, I. The cytogenetics and molecular characteristics of a translocated chromosome 1AS.1AL-1DL with a Glu-D1 locus in durum wheat. Cell. Mol. Biol. Lett. 2002, 7, 559–567. [Google Scholar] [PubMed]
- Xie, W.; Xiong, W.; Pan, J.; Ali, T.; Cui, Q.; Guan, D.; Meng, J.; Mueller, N.D.; Lin, E.; Davis, S.J. Decreases in global beer supply due to extreme drought and heat. Nat. Plants 2018, 4, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Newton, A.C. Climate change, plant diseases and food security: An overview. Plant Pathol. 2011, 60, 2–14. [Google Scholar] [CrossRef]
- Fisher, M.C.; Henk, D.A.; Briggs, C.J.; Brownstein, J.S.; Madoff, L.C.; McCraw, S.L.; Gurr, S.J. Emerging fungal threats to animal, plant and ecosystem health. Nature 2012, 484, 186–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godfray, H.C.J.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Nisbett, N.; Pretty, J.; Robinson, S.; Toulmin, C.; Whiteley, R. The future of the global food system. Philos. Trans. R. Soc. Lond. 2010, 365, 2769–2777. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.G.; Peng, Y.L.; Chen, W.Q.; Zhang, Z.Y. First detection of virulence in Puccinia striiformis f. sp. tritici in China to resistance genes Yr24 (=Yr26) present in wheat cultivar Chuanmai 42. Plant Dis. 2010, 94, 1163. [Google Scholar] [CrossRef]
- Keller, B.; Wicker, T.; Krattinger, S.G. Advances in wheat and pathogen genomics: Implications for disease control. Annu. Rev. Phytopathol. 2018, 56, 67–87. [Google Scholar] [CrossRef]
- Csorba, T.; Pantaleo, V.; Burgyan, J. RNA silencing: An antiviral mechanism. Adv. Virus Res. 2009, 75, 35–71. [Google Scholar]
- Harvey, J.J.W.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstädt, S.; Carr, J.P.; Baulcombe, D.C. An antiviral defense role of AGO2 in plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef]
- Hu, Q.; Niu, Y.; Zhang, K.; Liu, Y.; Zhou, X. Virus-derived transgenes expressing hairpin RNA give immunity to Tobacco mosaic virus and Cucumber mosaic virus. Virol. J. 2011, 8, 41. [Google Scholar] [CrossRef]
- Guozhong, H.; Rex, A.; Davis, E.L.; Baum, T.J.; Hussey, R.S. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. USA 2006, 103, 14302–14306. [Google Scholar] [Green Version]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 1990, 2, 279–289. [Google Scholar] [CrossRef]
- Jorgensen, R. Plants, RNAi, and the Nobel Prize. Science 2006, 314, 1242–1243. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Qi, T.; Yang, Q.; He, F.; Tan, C.; Ma, W.; Voegele, R.T.; Kang, Z.; Guo, J. Host-induced gene silencing of the MAPKK gene PsFUZ7 confers stable resistance to wheat stripe rust. Plant Physiol. 2017, 175, 1853–1863. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Stein, N.; Choulet, F.; Distelfeld, A.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, 6403. [Google Scholar]
- Wicker, T.; Schulman, A.H.; Tanskanen, J.; Spannagl, M.; Twardziok, S.; Mascher, M.; Springer, N.M.; Li, Q.; Waugh, R.; Li, C.; et al. The repetitive landscape of the 5100 Mbp barley genome. Mob. DNA 2017, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Holzberg, S.; Brosio, P.; Gross, C.; Pogue, G.P. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002, 30, 315–327. [Google Scholar] [CrossRef]
- Saurabh, S.; Vidyarthi, A.S.; Prasad, D. RNA interference: Concept to reality in crop improvement. Planta 2014, 239, 543–564. [Google Scholar] [CrossRef]
- Llave, C. Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci. 2010, 15, 701–707. [Google Scholar] [CrossRef]
- Tang, G.; Reinhart, B.J.; Bartel, D.P.; Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 2003, 17, 49–63. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Hannon, G.J. Uncovering RNAi mechanisms in plants: Biochemistry enters the foray. FEBS Lett. 2005, 579, 5899–5903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, D.; Uauy, C.; Blechl, A.; Dubcovsky, J. RNA interference for wheat functional gene analysis. Transgenic Res. 2007, 16, 689–701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miki, D. RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 2005, 138, 1903–1913. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.J.; Li, H.; Li, Y.W.; Zhu, Y.F.; Guo, J.K.; Liu, Y.J.; Chen, Y.; Jia, X. Generation of transgenic wheat lines with altered expression levels of 1Dx5 high-molecular weight glutenin subunit by RNA interference. J. Cereal Sci. 2008, 47, 153–161. [Google Scholar] [CrossRef]
- McGinnis, K.; Murphy, N.; Carlson, A.R.; Akula, A.; Akula, C.; Basinger, H.; Carlson, M.; Hermanson, P.; Kovacevic, N.; McGill, M.A.; et al. Assessing the efficiency of RNA interference for maize functional genomics. Plant Physiol. 2007, 143, 1441–1451. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Duan, X.; Lv, Y.; Zhang, X.; Nie, Z.; Xie, C.; Ni, Z.; Liang, R. Silencing of an aphid carboxylesterase gene by use of plant-mediated RNAi impairs Sitobion avenae tolerance of Phoxim insecticides. Transgenic Res. 2014, 23, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Abdellatef, E.; Will, T.; Koch, A.; Imani, J.; Vilcinskas, A.; Kogel, K.H. Silencing the expression of the salivary sheath protein causes transgenerational feeding suppression in the aphid Sitobion avenae. Plant Biotechnol. J. 2015, 13, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Lilley, C.J.; Bakhetia, M.; Charlton, W.L.; Urwin, P.E. Recent progress in the development of RNA interference for plant parasitic nematodes. Mol. Plant Pathol. 2007, 8, 701–711. [Google Scholar] [CrossRef]
- Tan, J.C.H.; Jones, M.G.K.; Fosu-Nyarko, J. Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasitol. 2013, 133, 166–178. [Google Scholar] [CrossRef]
- Fahim, M.; Millar, A.A.; Wood, C.C.; Larkin, P.J. Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol. J. 2012, 10, 150–163. [Google Scholar] [CrossRef]
- Kis, A.; Tholt, G.; Ivanics, M.; Várallyay, É.; Jenes, B.; Havelda, Z. Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol. Plant Pathol. 2016, 17, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Cruz, L.F.; Rupp, J.L.S.; Trick, H.N.; Fellers, J.P. Stable resistance to Wheat streak mosaic virus in wheat mediated by RNAi. In Vitro Cell. Dev. Plant 2014, 50, 665–672. [Google Scholar] [CrossRef]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef] [PubMed]
- Pliego, C.; Nowara, D.; Bonciani, G.; Gheorghe, D.M.; Xu, R. Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol. Plant Microbe Interact. 2013, 26, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Zhu, X.; Tan, C.; Liu, P.; Guo, J.; Kang, Z.; Guo, J. Host-induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol. J. 2017, 16, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Panwar, V.; Jordan, M.; McCallum, B.; Bakkeren, G. Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat. Plant Biotechnol. J. 2018, 16, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K.H. Host-induced gene silencing of cytochrome P450 lanosterol C14 -demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329. [Google Scholar] [CrossRef]
- Cheng, W.; Song, X.; Li, H.; Cao, L.; Sun, K.; Qiu, X.; Xu, Y.; Yang, P.; Huang, T.; Zhang, J.; et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015, 13, 1335–1345. [Google Scholar] [CrossRef]
- Chen, W.; Kastner, C.; Nowara, D.; Oliveira-Garcia, E.; Rutten, T.; Zhao, Y.; Deising, H.B.; Kumlehn, J.; Schweizer, P. Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 2016, 67, 4979–4991. [Google Scholar] [CrossRef]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Wang, D.; Liu, Q.; Li, X.; Sun, Y.; Wang, H.; Xia, L. Double-stranded RNA in the biological control of grain aphid (Sitobion avenae F.). Funct. Integr. Genom. 2015, 15, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhou, Y.; Wang, H.; Jones, H.D.; Gao, Q.; Wang, D.; Ma, Y.; Xia, L. Identifying potential RNAi targets in grain aphid (Sitobion avenae F.) based on transcriptome profiling of its alimentary canal after feeding on wheat plants. BMC Genom. 2013, 14, 560. [Google Scholar] [CrossRef]
- Richter, K.; Buchner, J. Hsp90: Chaperoning signal transduction. J. Cell. Physiol. 2010, 188, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Will, T.; Vilcinskas, A. Aphid-Proof Plants: Biotechnology-based approaches for aphid control. Adv. Biochem. Eng. Biotechnol. 2013, 179–203. [Google Scholar] [CrossRef]
- Arakane, Y.; Specht, C.A.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W. Chitin synthases are required for survival, fecundity and egg hatch in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. 2008, 38, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Aranda, M.; Marquessouza, H.; Bayer, T.; Tautz, D. The role of the segmentation gene hairy in Tribolium. Dev. Genes Evol. 2008, 218, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Chen, H.; Sun, Y.; Yu, X.; Xia, L. RNA interference of the ecdysone receptor genes EcR and USP in grain aphid (Sitobion avenae F.) affects its survival and fecundity upon feeding on wheat plants. Int. J. Mol. Sci. 2016, 17, 2098. [Google Scholar] [CrossRef]
- Eileen, K.; Henrike, S.; Andreas, V.; Boran, A. MMPs regulate both development and immunity in the tribolium model insect. PLoS ONE 2009, 4, e4751. [Google Scholar]
- Yu, X.D.; Liu, Z.C.; Huang, S.L.; Chen, Z.Q.; Sun, Y.W.; Duan, P.F.; Ma, Y.Z.; Xia, L.Q. RNAi-mediated plant protection against aphids. Pest Manag. Sci. 2016, 72, 1090–1098. [Google Scholar] [CrossRef]
- Christina, K.; Mohammad-Reza, H.; Alisdair, R.F.; Ute, R.; Tomasz, C.; Brigitte, H.; Wolf, B.F. The sucrose transporter StSUT1 localizes to sieve elements in potato tuber phloem and influences tuber physiology and development. Plant Physiol. 2003, 131, 102–113. [Google Scholar]
- Fraley, R.T.; Rogers, S.G.; Horsch, R.B.; Sanders, P.R.; Flick, J.S.; Adams, S.P.; Bittner, M.L.; Brand, L.A.; Fink, C.L.; Fry, J.S. Expression of bacterial genes in plant cells. Proc. Natl. Acad. Sci. USA 1983, 80, 4803–4807. [Google Scholar] [CrossRef] [PubMed]
- Carolan, J.C.; Fitzroy, C.I.J.; Ashton, P.D.; Douglas, A.E.; Wilkinson, T.L. The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics 2010, 9, 2457–2467. [Google Scholar] [CrossRef] [PubMed]
- Mashela, P.W.; Ndhlala, A.R.; Pofu, K.M.; Dube, Z.P. Phytochemicals of nematode-resistant transgenic plants. Transgenes. Second. Metab. 2017, 553–568. [Google Scholar] [CrossRef]
- Bernard, G.; Egnin, M.; Bonsi, C. The impact of plant-parasitic nematodes on sgriculture and methods of control. Intechopen 2017, e68958. [Google Scholar] [CrossRef]
- Joseph, S.; Gheysen, G.; Subramaniam, K. RNA interference in Pratylenchus coffeae: Knock down of Pc-pat-10 and Pc-unc-87 impedes migration. Mol. Biochem. Parasitol. 2012, 186, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Yadav, B.C.; Veluthambi, K.; Subramaniam, K.; Yadav, B.C.; Veluthambi, K.; Subramaniam, K. Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol. Biochem. Parasitol. 2006, 148, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Ghag, S.B. Host induced gene silencing, an emerging science to engineer crop resistance against harmful plant pathogens. Physiol. Mol. Plant Pathol. 2017, 100. [Google Scholar] [CrossRef]
- Sivamani, E.; Brey, C.W.; Dyer, W.E.; Talbert, L.E.; Qu, R. Resistance to wheat streak mosaic virus in transgenic wheat expressing the viral replicase (NIb) gene. Mol. Breed. 2000, 6, 469–477. [Google Scholar] [CrossRef]
- Sivamani, E.; Brey, C.W.; Talbert, L.E.; Young, M.A.; Dyer, W.E.; Kaniewski, W.K.; Qu, R. Resistance to wheat streak mosaic virus in transgenic wheat engineered with the viral coat protein gene. Transgenic Res. 2002, 11, 31–41. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.; Berger, P.H. Transgenic silencing in wheat transformed with the WSMV-CP gene. Biotechnology 2005, 4, 62–68. [Google Scholar]
- Shoup Rupp, J.L. RNAi-mediated, stable resistance to Triticum mosaic virus in wheat. Crop. Sci. 2016, 56, 1602–1610. [Google Scholar] [CrossRef]
- Brown, J.K.M. Durable resistance of crops to disease: A darwinian perspective. Annu. Rev. Phytopathol. 2015, 53, 513–539. [Google Scholar] [CrossRef] [PubMed]
- Larkin, P.J.; Banks, P.M.; Lagudah, E.S.; Appels, R.; Xiao, C.; Zhiyong, X.; Ohm, H.W.; McIntosh, R.A. Disomic Thinopyrum intermedium addition lines in wheat with Barley yellow dwarf virus resistance and with rust resistances. Genome 1995, 38, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Roelfs, A.P.; McCallum, B.; McVey, D.V.; Groth, J.V. Comparison of virulence and isozyme phenotypes of Pgt-QCCJ and great plains races of Puccinia graminis f. sp. tritici. Phytopathology 1997, 87, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Jurgenson, J.E.; Hulbert, S.H. Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 2011, 24, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Panwar, V.; McCallum, B.; Bakkeren, G. Endogenous silencing of Puccinia triticina pathogenicity genes throughin planta-expressed sequences leads to the suppression of rust diseases on wheat. Plant J. 2013, 73, 521–532. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, X.; Yao, J.; Voegele, R.T.; Zhang, Y.; Wang, W.; Huang, L.; Kang, Z. Characterization of protein kinase PsSRPKL, a novel pathogenicity factor in the wheat stripe rust fungus. Environ. Microbiol. 2015, 17, 2601–2617. [Google Scholar] [CrossRef]
- Cheng, Y.; Yao, J.; Zhang, Y.; Li, S.; Kang, Z. Characterization of a Ran gene from Puccinia striiformis f. sp. tritici involved in fungal growth and anti-cell death. Sci. Rep. 2016, 6, 35248. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, W.; Yao, J.; Huang, L.; Voegele, R.T.; Wang, X.; Kang, Z. Two distinct Ras genes from Puccinia striiformis exhibit differential roles in rust pathogenicity and cell death. Environ. Microbiol. 2016, 18, 3910–3922. [Google Scholar] [CrossRef]
- Zhu, X.; Jiao, M.; Guo, J.; Liu, P.; Tan, C.; Yang, Q.; Zhang, Y.; Thomas Voegele, R.; Kang, Z.; Guo, J. A novel MADS-box transcription factor PstMCM1-1 is responsible for full virulence of Puccinia striiformis f. sp. tritici. Environ. Microbiol. 2018, 20, 1452–1463. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, W.; Chu, X.; Sun, Q.; Tan, C.; Yang, Q.; Jiao, M.; Guo, J.; Kang, Z. The transcription factor PstSTE12 is required for virulence of Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 2017, 19, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Guo, J.; He, F.; Zhang, Y.; Tan, C.; Yang, Q.; Huang, C.; Kang, Z.; Guo, J. Silencing PsKPP4, a MAP kinase kinase kinase gene, reduces pathogenicity of the stripe rust fungus. Mol. Plant Pathol. 2018, 19, 2590–2602. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, J.; Ji, S.; Chen, Z.; Xu, J.; Tang, C.; Chen, S.; Kang, Z.; Wang, X. Candidate effector Pst_8713 impairs the plant immunity and contributes to virulence of Puccinia striiformis f. sp. tritici. Front. Plant Sci. 2018, 9, 1294. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wu, K.; Yao, J.; Li, S.; Wang, X.; Huang, L.; Kang, Z. PSTha5a23, a candidate effector from the obligate biotrophic pathogen Puccinia striiformis f. sp. tritici, is involved in plant defense suppression and rust pathogenicity. Environ. Microbiol. 2017, 19, 1717–1729. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Pedersen, C.; Schultz Larsen, T.; Aguilar, G.B.; Amp, M.O.; Ana, K. The stripe rust fungal effector PEC6 suppresses pattern-triggered immunity in a host species-independent manner and interacts with adenosine kinases. New Phytol. 2016, 7, 14034. [Google Scholar] [CrossRef]
- Liu, J.; Guan, T.; Zheng, P.; Chen, L.; Yang, Y.; Huai, B.; Li, D.; Chang, Q.; Huang, L.; Kang, Z. An extracellular Zn-only superoxide dismutase from Puccinia striiformis confers enhanced resistance to host-derived oxidative stress. Environ. Microbiol. 2016, 18, 4118–4135. [Google Scholar] [CrossRef]
- Machado, A.K.; Brown, N.A.; Urban, M.; Kanyuka, K.; Hammond-Kosack, K.E. RNAi as an emerging approach to control Fusarium head blight disease and mycotoxin contamination in cereals. Pest Manag. Sci. 2018, 74, 790–799. [Google Scholar] [CrossRef]
- Wegulo, S.N.; Baenziger, P.S.; Hernandez Nopsa, J.; Bockus, W.W.; Hallen-Adams, H. Management of Fusarium head blight of wheat and barley. Crop. Prot. 2015, 73, 100–107. [Google Scholar] [CrossRef]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, H.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef]
- Khatri, M.; Rajam, M.V. Targeting polyamines of Aspergillus nidulans by siRNA specific to fungal ornithine decarboxylase gene. Med. Mycol. 2007, 45, 211–220. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.M.; Thomma, B.P.; Huang, H.D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Dongxia, H.; Xi, C.; Donghai, L.; Lingyun, Z.; Yujing, Z.; Jing, L.; Zhen, B.; Xiangying, L.; Xing, C. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar]
- Yin, C.; Hulbert, S. Host Induced Gene Silencing (HIGS), a promising strategy for developing disease resistant crops. In Omics Int. 2015, 4, 1–2. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Heckel, D.G.; Bock, R. Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 2017, 35, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Koch, A.; Kogel, K.H. New wind in the sails: Improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol. J. 2015, 12, 821–831. [Google Scholar] [CrossRef]
- Li, W.; Koutmou, K.S.; Leahy, D.J.; Li, M. Systemic RNA Interference Deficiency-1 (SID-1) extracellular domain selectively binds long double-stranded RNA and is required for RNA transport by SID-1. J. Biol. Chem. 2015, 290, 18904–18913. [Google Scholar] [CrossRef] [Green Version]
- Arne, W.; Ming, W.; Feng-Mao, L.; Hongwei, Z.; Zhihong, Z.; Isgouhi, K.; Hsien-Da, H.; Hailing, J. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar]
- Quintana, J.F.; Babayan, S.A.; Buck, A.H. Small RNAs and extracellular vesicles in filarial nematodes: From nematode development to diagnostics. Parasite Immunol. 2016, 39. [Google Scholar] [CrossRef]
- Katsir, L.; Bahar, O. Bacterial outer membrane vesicles at the plant-pathogen interface. PLoS Pathog. 2017, 13, e1006306. [Google Scholar] [CrossRef]
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. Plant Sci. 2018, 9, 1245. [Google Scholar] [CrossRef] [PubMed]

| Species | Host | Target Gene | Remark | Major Phenotype | Ref. | |
|---|---|---|---|---|---|---|
| Insects | Sitobion avenae | Wheat | CbE E4 | Carboxylesterase gene | Impaired tolerance of phoxim insecticides | [26] |
| Sitobion avenae | Barley | shp | Structural sheath protein | Reduced fecundity and inhibited feeding behavior | [27] | |
| Nematodes | Meloidogyne incognita | Wheat | HSP90, ICL, and Mi-cpl-1 | Heat-shock protein 90, isocitrate lyase, and Mi-cpl-1 | Reduced reproduction | [28] |
| Pratylenchus spp. | Wheat | pat-10, unc-87 | Troponin C (Pat-10) and Calponin (unc-87) | Reduced reproduction | [29] | |
| Viruses | Wheat streak mosaic virus (WSMV) | Wheat | pre-miR395 | Artificial microRNA (amiRNA) | Stable resistance | [30] |
| Wheat dwarf virus (WDV) | Barley | amiR1, amiR6, and amiR8 | amiRNAs | Highly efficient resistance | [31] | |
| WSMV | Wheat | coat protein gene | Coat protein | Consistent resistance | [32] | |
| Fungi | Blumeria graminis | Wheat, Barley | Avra10 | Virulence effector | Reduced virulence | [33] |
| Blumeria graminis | Wheat, Barley | BEC1011, BEC1054, BEC1038, BEC1016, BEC1005, BEC1019, BEC1040, and BEC1018 | Effectors | Reduced virulence | [34] | |
| Puccinia striiformis f. sp. tritici | Wheat | PsCPK1 | PKA catalytic subunit | Stable resistance | [35] | |
| Puccinia striiformis f. sp. tritici | Wheat | PsFuz7 | MAP kinase kinase | Stable resistance | [14] | |
| Puccinia triticina | Wheat | PtMAPK1 and PtCYC1 | MAP kinase, cyclophilin | Enhanced resistance | [36] | |
| Fusarium species | Barley | CYP51 | Cytochrome P450 lanosterol C14-demethylase | Enhanced resistance | [37] | |
| Fusarium graminearum | Wheat | Chs3b | Chitin synthase 3b | Stable resistance | [38] | |
| Fusarium culmorum | Wheat | FcGls1 | β-1, 3-glucan synthase | Enhanced resistance | [39] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, T.; Guo, J.; Peng, H.; Liu, P.; Kang, Z.; Guo, J. Host-Induced Gene Silencing: A Powerful Strategy to Control Diseases of Wheat and Barley. Int. J. Mol. Sci. 2019, 20, 206. https://doi.org/10.3390/ijms20010206
Qi T, Guo J, Peng H, Liu P, Kang Z, Guo J. Host-Induced Gene Silencing: A Powerful Strategy to Control Diseases of Wheat and Barley. International Journal of Molecular Sciences. 2019; 20(1):206. https://doi.org/10.3390/ijms20010206
Chicago/Turabian StyleQi, Tuo, Jia Guo, Huan Peng, Peng Liu, Zhensheng Kang, and Jun Guo. 2019. "Host-Induced Gene Silencing: A Powerful Strategy to Control Diseases of Wheat and Barley" International Journal of Molecular Sciences 20, no. 1: 206. https://doi.org/10.3390/ijms20010206
APA StyleQi, T., Guo, J., Peng, H., Liu, P., Kang, Z., & Guo, J. (2019). Host-Induced Gene Silencing: A Powerful Strategy to Control Diseases of Wheat and Barley. International Journal of Molecular Sciences, 20(1), 206. https://doi.org/10.3390/ijms20010206