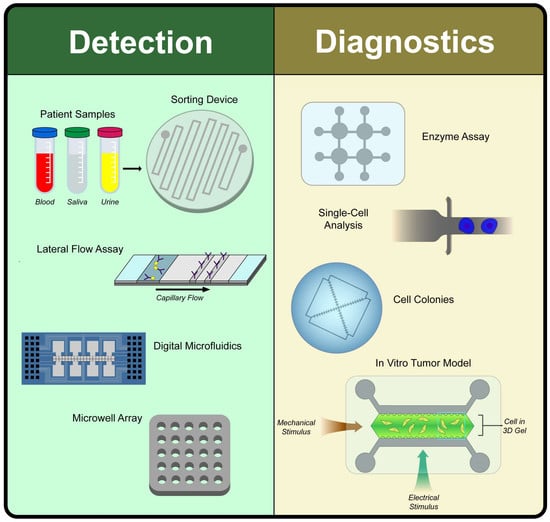
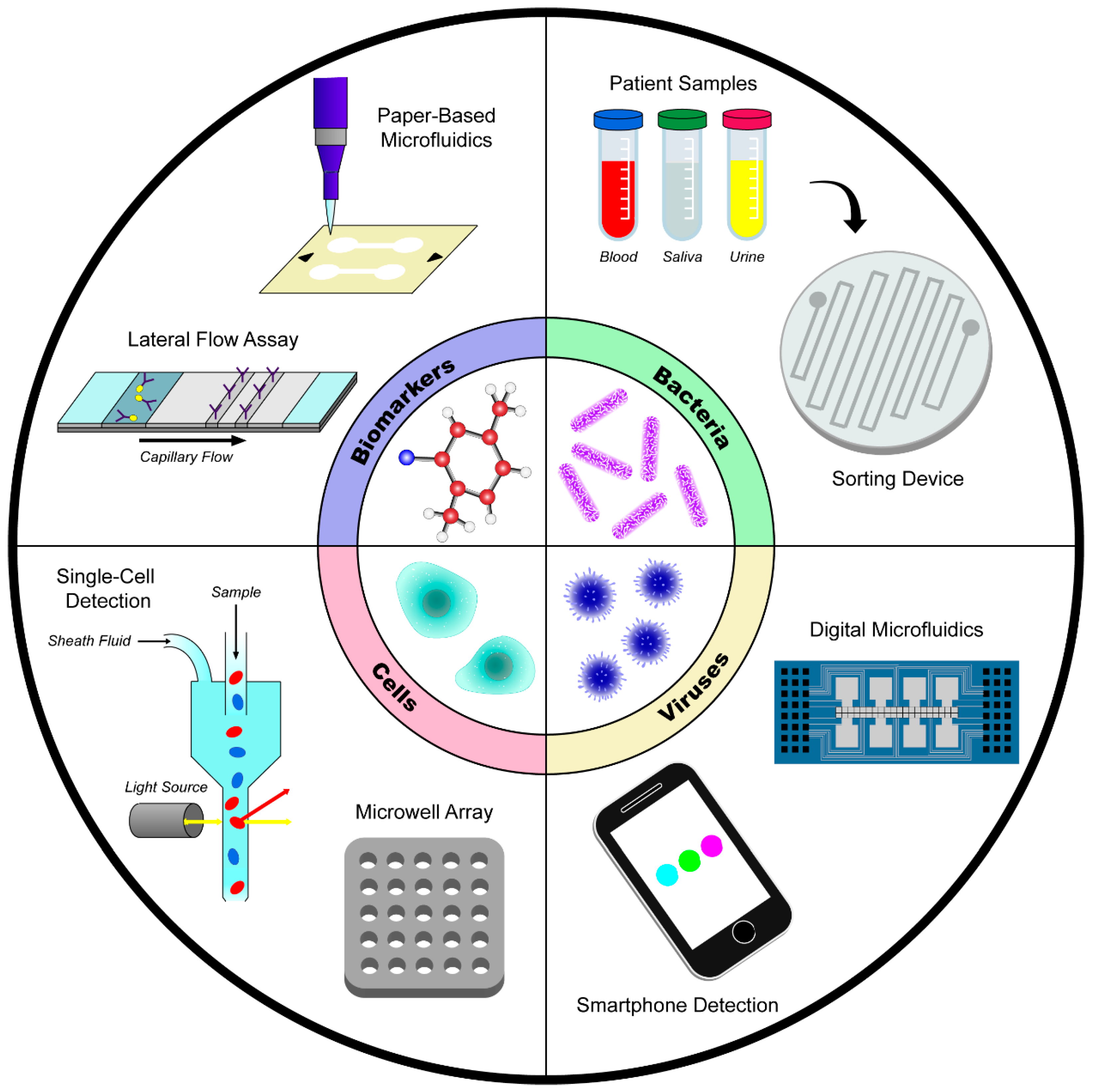
Microfluidic and Paper-Based Devices for Disease Detection and Diagnostic Research
Abstract
:1. Introduction
2. Disease Detection
2.1. Detection of Biomolecules and Biomarkers
2.1.1. Lateral Flow Strip Assays to Detect Biomolecules
2.1.2. Paper Microfluidic Devices to Detect Biomolecules and Biomarkers
2.1.3. Detection of Biologically Relevant Molecules using PDMS-Based Microfluidic Devices
2.1.4. Sensors, Chips, and Other Technologies to Detect Biomolecules
2.2. Detection of Human Cells
2.2.1. Methods to Detect Blood Cells
2.2.2. Methods to Detect Cancer Cells
2.3. Detection of Bacteria
2.3.1. Detection using Blood Samples
2.3.2. Detection using Saliva or Other Samples
2.4. Detection of Viruses
2.4.1. Methods to Detect Influenza
2.4.2. Methods to Detect Zika
2.4.3. Methods to Detect Sexually Transmitted Diseases
3. Disease Diagnostics
3.1. Cell Behavior
3.1.1. Single Cell Heterogeneity
3.1.2. 3D Cell Migration
3.1.3. Angiogenesis
3.1.4. Cell-to-Cell Communication
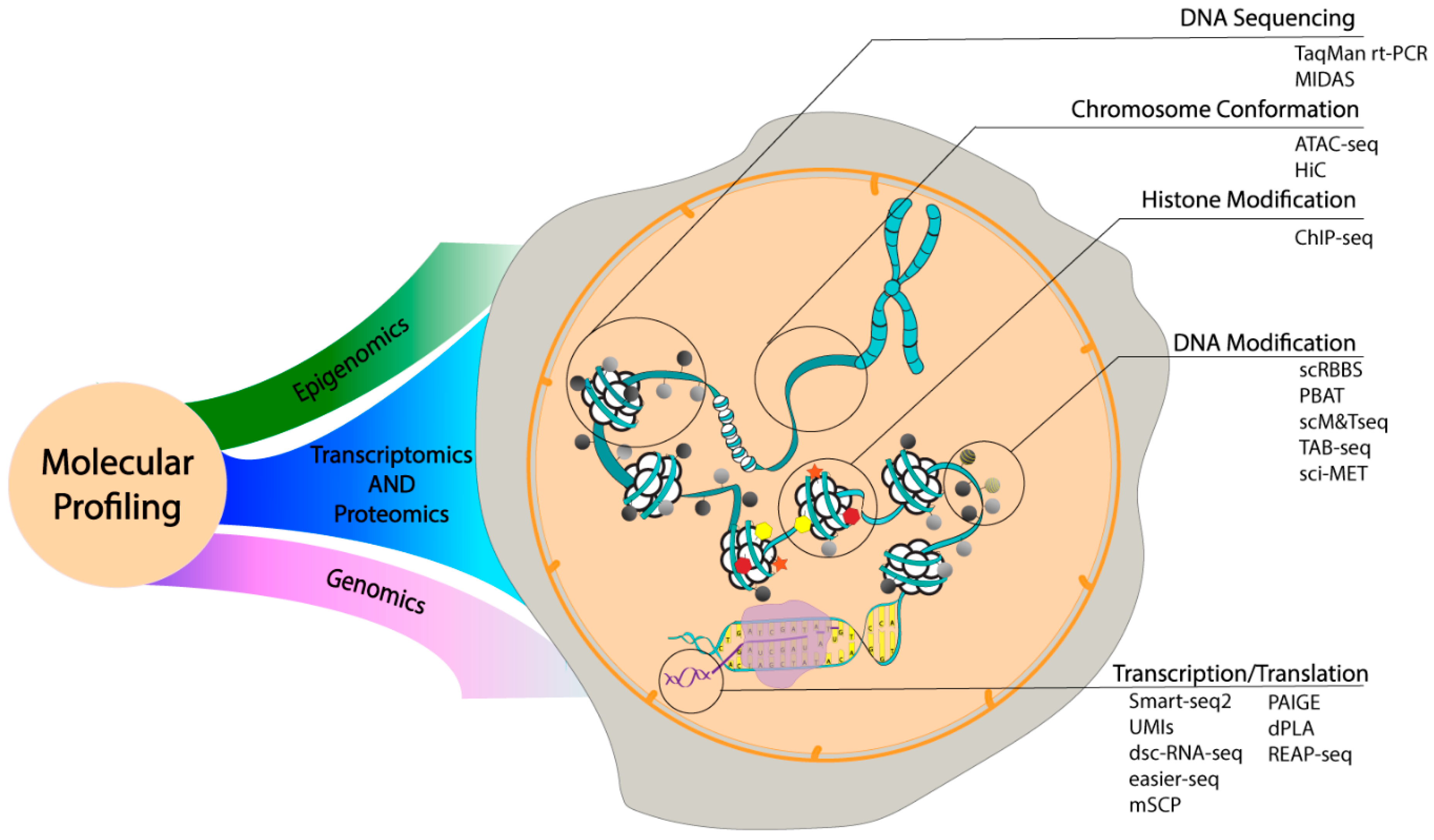
3.2. Analytical Developments in Molecular Profiling
3.2.1. Single Cell Epigenomics
3.2.2. Single Cell Genomics
3.2.3. Single Cell Transcriptomics
3.2.4. Single Cell Proteomics
3.3. Drug Screening
3.3.1. Automated High-Throughput Screening
3.3.2. Microfluidic Devices to Study Cytotoxic Effects and Pharmacokinetics
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| PSA | Prostate Specific Antigen |
| PDMS | Polydimethylsiloxane |
| CTCs | Circulating Tumor Cells |
| HIV | Human Immunodeficiency Virus |
| AIDS | Acquired Immune Deficiency Syndrome |
| LFSAs | Lateral Flow Strip Assays |
| TTX | Tetrodotoxin |
| ELISA | Enzyme Linked Immunosorbent Assay |
| PKU | Phenylketonuria |
| qRT-PCR | Quantitative Reverse Transcription Polymerase Chain Reaction |
| RBCs | Red Blood Cells |
| CRP | C-reactive protein |
| IL-2 | Interleukin-2 |
| HEPES | N-2-hydroxyethhylpiperazine-N-ethane-sulfonicacid |
| cTn I | Cardiac Troponine I |
| CK-MB | Creatine Kinase-MB |
| IFNγ | Interferon γ |
| WBCs | White Blood Cells |
| T2DM | Type 2 Diabetes Mellitus |
| HUVECs | Human Umbilical Vein Endothelial Cells |
| EpCAM | Epithelial Cell Adhesion Molecule |
| 2-NBDG | 2-Deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-d-glucose |
| ECL | Electrochemiluminescence |
| CD4 | Cluster of Differentiation 4 |
| EVs | Extracellular Vesicles |
| PtNPs | Platinum Nanoparticles |
| AuNPs | Gold Nanoparticles |
| Phe | Phenylalanine |
| PBS | Phosphate Buffered Saline |
| PCR | Polymerase Chain Reaction |
| PJI | Periprosthetic Joint Infection |
| LAMP | Loop Mediated Isothermal Amplification |
| CFU | Colony Forming Unit |
| AIV | Avian Influenza Virus |
| HAU | Hemagglutinin Unit |
| SERS | Surface-Enhanced Raman Scattering |
| DMF | Digital Microfluidics |
| BART | Bioluminescent Real Time Reporter |
| RT-LAMP | Reverse Transcription Loop Mediated Isothermal Amplification |
| STDs | Sexually Transmitted Diseases |
| tHDA | Thermophilic Helicase-Dependent Amplification |
| PFU | Plaque Forming Unit |
| EMA | Ethidium Monoazide |
| USD | United States Dollar |
| HTS | High-Throughput Screening |
| MALDI-MSI | Matrix-Assisted Laser Desorption Ionization-Mass Spectrometry Imaging |
| hiPSC | Human Induced Pluripotent Stem Cells |
| ECM | Extracellular Matrix |
| EMT | Epithelial to Mesenchymal Transition |
| TOF-SIMS | Time-of-Flight Secondary Ion Mass Spectrometry |
| PCA | Principal Component Analysis |
| CyTOF | Cytometry Time of Flight |
| EGF | Epidermal Growth Factor |
| CAFs | Cancer-Associated Fibroblasts |
| SDF-1α | Stromal Cell-Derived Factor 1α |
| VEGF | Vascular Epidermal Growth Factor |
| hLFs | Human Lung Fibroblasts |
| ECs | Endothelial Cells |
| PDGF | Platelet-Derived Growth Factor |
| ASCs | Adipose Stem Cells |
| MMP | Matrix Metalloproteinase |
| hMSC | Human Mesenchymal Stromal Cells |
| NK | Natural Killer |
| ICI | Immune Checkpoint Inhibitor |
| TILs | Tumor-Infiltrating Lymphocytes |
| RT | Reverse Transcription |
| DMRs | Differentially Methylated Regions |
| sc-GEM | Single-Cell Analysis of Genotype, Expression, and Methylation |
| ChIP | Chromatin Immunoprecipitation |
| NGS | Next Generation Sequencing |
| H3K4me3 | H3 Lysine 4 Trimethylation |
| H3K4me2 | H3 Lysine 4 Dimethylation |
| ES | Embryonic Stem |
| MEFs | Mouse Embryonic Fibroblasts |
| MOWChIP-seq | Microfluidic Oscillatory Washing-Based ChIP-seq |
| ATAC-seq | Assay for Transposase-Accessible Chromatin Sequencing |
| SISSOR | Single-Stranded Sequencing Using Microfluidic Reactors |
| SNPs | Single Nucleotide Polymorphisms |
| MPS | Massively Parallel Sequencing |
| MIDAS | Microwell Displacement Amplification System |
| gDNA | Genomic DNA |
| WTA | Whole Transcriptome Amplification |
| UMIs | Unique Molecular Identifiers |
| STAMPs | Single Cell Transcriptomes Attached to Microparticles |
| BHMs | Barcoded Hydrogel Microspheres |
| Hi-SCL | High-Throughput Single Cell Labeling |
| dscRNA-seq | Droplet Single-Cell RNA Sequencing |
| easier-seq | Emulsion-Based Amplification of Sequence Independent Evenly Transcribed RNA-seq |
| ddPC | Droplet Digital PCR |
| mSCP | Modular Three-Part Single-Cell Pipette |
| SCBC | Single Cell Barcode Chip |
| DEAL | DNA-Encoded Antibody Library |
| REAP | RNA Expression and Protein |
| FNAs | Fine Needle Aspirates |
| dPLA | Digital Proximity Ligation Assay |
| PAIGE | Protein Assay via Induced Gene Expression |
| PPI | Protein-Protein Interactions |
| FCCS | Fluorescence Cross-Correlation Spectroscopy |
| GFP | Green Fluorescence Protein |
| μPAD | Microfluidic Paper-Based Analytical Devices |
| LOD | Limit of Detection |
References
- Fleischmann, C.; Scherag, A.; Adhikari, N.; Hartog, C.; Tsaganos, T.; Schlattmann, P.; Angus, D.; Reinhart, K.; Trialists, I.F.A.C.; Trialists, I.F.A.C. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Am. J. Respir. Crit. Care Med. 2016, 193, 259–272. [Google Scholar] [CrossRef] [PubMed]
- World Malaria Report 2017; World Health Organization: Geneva, Switzerland, 2017; p. 41.
- Wu, J.; Dong, M.; Rigatto, C.; Liu, Y.; Lin, F. Lab-on-chip technology for chronic disease diagnosis. npj Digital Med. 2018. [Google Scholar] [CrossRef]
- Yuan, G.; Cai, L.; Elowitz, M.; Enver, T.; Fan, G.; Guo, G.; Irizarry, R.; Kharchenko, P.; Kim, J.; Orkin, S.; et al. Challenges and emerging directions in single-cell analysis. Genome Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Holmstrom, B.; Johansson, M.; Bergh, A.; Stenman, U.; Hallmans, G.; Stattin, P. Prostate specific antigen for early detection of prostate cancer: Longitudinal study. BMJ 2009. [Google Scholar] [CrossRef] [PubMed]
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. PMC 2014, 6, 25–64. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Xia, Y.; Tang, Y.; Zhang, W.; Yeh, Y.; Lu, H.; Zheng, S. A Nanostructured Microfluidic Immunoassay Platform for Highly Sensitive Infectious Pathogen Detection. Small 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chang, C.; Lee, G. Integrated microfluidic device using a single universal aptamer to detect multiple types of influenza viruses. Biosens. Bioelectron. 2016, 86, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhang, J.; Xu, F.; Wen, X.; Li, P.; Zhang, X.; Qiao, S.; Ge, S.; Xia, N.; Qian, S.; et al. A paper-based microfluidic Dot-ELISA system with smartphone for the detection of influenza A. Microfluid. Nanofluid. 2017. [Google Scholar] [CrossRef]
- Wang, Y.; Ruan, Q.; Lei, Z.; Ling, S.; Zhu, Z.; Zhou, L.; Yang, C. Highly Sensitive and Automated Surface Enhanced Raman Scattering-based Immunoassay for H5N1 Detection with Digital Microfluidics. Anal. Chem. 2018, 90, 5224–5231. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Pandian, V.; Mauk, M.; Bau, H.; Cherry, S.; Tisi, L.; Liu, C. Smartphone-Based Mobile Detection Platform for Molecular Diagnostics and Spatiotemporal Disease Mapping. Anal. Chem. 2018, 90, 4823–4831. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.; Takahashi, M.; Braff, D.; Lambert, G.; Lee, J.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.; Shin, Y.; Chung, S.; Hwang, K.; Yoon, D.; Lee, J. Simple and Highly Sensitive Molecular Diagnosis of Zika Virus by Lateral Flow Assays. Anal. Chem. 2016, 88, 12272–12278. [Google Scholar] [CrossRef] [PubMed]
- Surawathanawises, K.; Wiedorn, V.; Cheng, X. Micropatterned macroporous structures in microfluidic devices for viral separation from whole blood. Analyst 2017, 142, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Zhang, J.; Li, Y.; Zhang, C.; Wang, D.; Zhu, W.; Li, F.; Ge, S.; Xia, N.; Qian, S. A bead-based microfluidic system for joint detection in TORCH screening at point-of-care testing. Microsyst. Technol. 2018, 24, 2007–2015. [Google Scholar] [CrossRef]
- Altschuler, S.; Wu, L. Cellular Heterogeneity: Do Differences Make a Difference? Cell 2010, 141, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Reece, A.; Xia, B.; Jiang, Z.; Noren, B.; McBride, R.; Oakey, J. Microfluidic techniques for high throughput single cell analysis. Curr. Opin. Biotech. 2016, 40, 90–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sia, S.; Whitesides, G. Microfluidic devices fabricated in poly(dimethylsiloxane) for biological studies. Electrophoresis 2003, 24, 3563–3576. [Google Scholar] [CrossRef] [PubMed]
- Heath, J.; Ribas, A.; Mischel, P. Single-cell analysis tools for drug discovery and development. Nat. Rev. Drug Discov. 2016, 15, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, P.; Markowicz, M.; Mikiciuk-Olasik, E. Adaptation of High-Throughput Screening in Drug Discovery-Toxicological Screening Tests. Int. J. Mol. Sci. 2012, 13, 427–452. [Google Scholar] [CrossRef] [PubMed]
- Gawad, C.; Koh, W.; Quake, S. Single-cell genome sequencing: Current state of the science. Nat. Rev. Genet. 2016, 17, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Kim, J.; Song, H.; Sohn, K.Y.; Jeon, M.; Han, K.-H. Microlfuidic technologies for circulating tumor cell isolation. Analyst 2018, 143, 2936–2970. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.; Zu, Y. Detecting Circulating Tumor Cells: Current Challenges and New Trends. Theranostics 2013, 3, 377–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esmaeilsabzali, H.; Beischlag, T.; Cox, M.; Parameswaran, A.; Park, E. Detection and isolation of circulating tumor cells: Principles and methods. Biotechnol. Adv. 2013, 31, 1063–1084. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Zhang, Y.; Chen, W. Capturing Cancer: Emerging Microfluidic Technologies for the Capture and Characterization of Circulating Tumor Cells. Small 2015, 11, 3850–3872. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol. Adv. 2018, 36, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.; Abajobir, A.; Abbafati, C.; Abbas, K.; Abd-Allah, F.; Abera, S.; Aboyans, V.; Adetokunboh, O.; Arnlov, J.; Afshin, A.; et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef]
- Safavieh, M.; Coarsey, C.; Esiobu, N.; Memic, A.; Vyas, J.; Shafiee, H.; Asghar, W. Advances in Candida detection platforms for clinical and point-of-care applications. Crit. Rev. Biotechnol. 2017, 37, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.; Zhang, Q.; Hui, C.; Radford, K.; Nair, P.; Brennan, J. Development of a functional point-of-need diagnostic for myeloperoxidase detection to identify neutrophilic bronchitis. Analyst 2016, 141, 6438–6443. [Google Scholar] [CrossRef] [PubMed]
- Raston, N.; Nguyen, V.; Gu, M. A new lateral flow strip assay (LFSA) using a pair of aptamers for the detection of Vaspin. Biosens. Bioelectron. 2017, 93, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Guan, Z.; Song, Y.; Song, E.; Lu, Z.; Liu, D.; An, Y.; Zhu, Z.; Zhou, L.; Yang, C. Lateral flow assay with pressure meter readout for rapid point-of-care detection of disease-associated protein. Lab Chip 2018, 18, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Xu, F.; Xiao, M.; Fu, Q.; Cheng, Z.; Zhang, S.; Huang, C.; Tang, Y. A new lateral-flow immunochromatographic strip combined with quantum dot nanobeads and gold nanoflowers for rapid detection of tetrodotoxin. Analyst 2017, 142, 4393–4398. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Rodriguez, M.; Serrano-Pertierra, E.; Garcia, A.; Martin, S.; Mo, M.; Cernuda-Morollon, E.; Blanco-Lopez, M. Point-of-care detection of extracellular vesicles: Sensitivity optimization and multiple-target detection. Biosens. Bioelectron. 2017, 87, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Kong, T.; Flanigan, S.; Weinstein, M.; Kalwa, U.; Legner, C.; Pandey, S. A fast, reconfigurable flow switch for paper microfluidics based on selective wetting of folded paper actuator strips. Lab Chip 2017, 17, 3621–3633. [Google Scholar] [CrossRef] [PubMed]
- Messina, M.; Meli, C.; Conoci, S.; Petralia, S. A facile method for urinary phenylalanine measurement on paper-based lab-on-chip for PKU therapy monitoring. Analyst 2017, 142, 4629–4632. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Z.; Fang, C.; Lu, Y.; Yu, L.; Li, C. Integration of paper and micropipette tip to build a “sample-in, answer-out” point-of-care device. Microfluid. Nanofluid. 2017. [Google Scholar] [CrossRef]
- Yao, Y.; Li, H.; Wang, D.; Liu, C.; Zhang, C. An electrochemiluminescence cloth-based biosensor with smartphone-based imaging for detection of lactate in saliva. Analyst 2017, 142, 3715–3724. [Google Scholar] [CrossRef] [PubMed]
- Salim, B.; Athira, M.; Kandaswamy, A.; Vijayakumar, M.; Saravanan, T.; Sairam, T. Microfluidic device for novel breast cancer screening by blood test using miRNA beacon probe. Biomed. Microdevices 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.; Hsiung, C.; Chen, Y.; Huang, C.; Hsu, H.; Hsieh, P. Real-time CRP detection from whole blood using micropost-embedded microfluidic chip incorporated with label-free biosensor. Analyst 2018, 143, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Usuba, R.; Yokokawa, M.; Ackermann, T.; Llobera, A.; Fukunaga, K.; Murata, S.; Ohkohchi, N.; Suzuki, H. Photonic Lab-on-a-Chip for Rapid Cytokine Detection. ACS Sens. 2016, 1, 979–986. [Google Scholar] [CrossRef]
- Qiu, X.; Yang, S.; Wu, D.; Wang, D.; Qiao, S.; Ge, S.; Xia, N.; Yu, D.; Qian, S. Rapid enumeration of CD4+T lymphocytes using an integrated microfluidic system based on Chemiluminescence image detection at point-of-care testing. Biomed. Microdevices 2018, 20. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; He, M.; Mu, L.; Lin, Z.; Wang, Y.; Pang, W.; Reed, M.; Duan, X. Cellphone-Enabled Microwell-Based Microbead Aggregation Assay for Portable Biomarker Detection. ACS Sens. 2018, 3, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Liu, P.; Zhang, Y.; Yu, Z.; Chen, X.; Zhou, L.; Nie, B.; Zaczek, A.; Chen, J.; Liu, J. Sensitive Detection of Single-Cell Secreted H2O2 by Integrating a. Microfluidic Droplet Sensor and Au Nanoclusters. Anal. Chem. 2018, 90, 4478–4484. [Google Scholar] [CrossRef] [PubMed]
- Matta, D.; Tripathy, S.; Vanjari, S.; Sharma, C.; Singh, S. An ultrasensitive label free nanobiosensor platform for the detection of cardiac biomarkers. Biomed. Microdevices 2016, 18, 18. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Lee, M.; Kim, D. Detection of early stage prostate cancer by using a simple carbon nanotube@paper biosensor. Biosens. Bioelectron. 2018, 102, 345–350. [Google Scholar] [CrossRef] [PubMed]
- Castiello, F.; Tabrizian, M. Multiplex Surface Plasmon Resonance Imaging-Based Biosensor for Human Pancreatic Islets Hormones Quantification. Anal. Chem. 2018, 90, 3132–3139. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.; Du, Y.; Wu, T.; Chen, C.; Lee, D.; Chen, S.; Huang, T.; Wu, H.; Shaikh, M. Immunosensor for the ultrasensitive and quantitative detection of bladder cancer in point of care testing. Biosens. Bioelectron 2016, 84, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Mosher, C.; Lee, X.; Das, S.; Cargill, A.; Tang, X.; Chen, B.; McLamore, E.; Gomes, C.; Hostetter, J.; et al. Rapid and Label-Free Detection of Interferon Gamma via an Electrochemical Aptasensor Comprising a Ternary Surface Monolayer on a Gold Interdigitated Electrode Array. ACS Sensors 2017, 2, 210–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, W.; Goh, B.; Khor, S. Microfluidic paper-based analytical devices for potential use in quantitative and direct detection of disease biomarkers in clinical analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1060, 424–442. [Google Scholar] [CrossRef] [PubMed]
- Akyazi, T.; Basabe-Desmonts, L.; Benito-Lopez, F. Review on microfluidic paper-based analytical devices towards commercialisation. Anal. Chim. Acta 2018, 1001, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Hegener, M.; Li, H.; Han, D.; Steckl, A.; Pauletti, G. Point-of-care coagulation monitoring: First clinical experience using a paper-based lateral flow diagnostic device. Biomed. Microdevices 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.; Fernandes, S.; Rajaratnam, A.; DeChiara, N.; Mace, C. Measurement of the hematocrit using paper-based microfluidic devices. Lab Chip 2016, 16, 3689–3694. [Google Scholar] [CrossRef] [PubMed]
- Schie, I.; Ruger, J.; Mondol, A.; Ramoji, A.; Neugebauer, U.; Krafft, C.; Popp, J. High-Throughput Screening Raman Spectroscopy Platform for Label-Free Cellomics. Anal. Chem. 2018, 90, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Tay, H.; Dalan, R.; Li, K.; Boehm, B.; Hou, H. A Novel Microdevice for Rapid Neutrophil Purification and Phenotyping in Type 2 Diabetes Mellitus. Small 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, G.; Cui, G.; Zhou, X.; Liu, G. White blood cell counting on smartphone paper electrochemical sensor. Biosens. Bioelectron. 2017, 90, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Long, D.; Shang, Y.; Qiu, Y.; Zhou, B.; Yang, P. A single-cell analysis platform for electrochemiluminescent detection of platelets adhesion to endothelial cells based on Au@DL-ZnCQDs nanoprobes. Biosens. Bioelectron. 2018, 102, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wang, Z.; Li, Z.; Kim, J.; Deng, Y.; Li, Y.; Heath, J.; Wei, W.; Lu, S.; Shi, Q. High-throughput screening of rare metabolically active tumor cells in pleural effusion and peripheral blood of lung cancer patients. Proc. Natl. Acad. Sci. USA 2017, 114, 2544–2549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedrol, E.; Martinez, J.; Aguilo, M.; Garcia-Algar, M.; Nazarenus, M.; Guerrini, L.; Garcia-Rico, E.; Alvarez-Puebla, R.; Diaz, F.; Massons, J. Microfluidic device with dual-channel fluorescence acquisition for quantification/identification of cancer cells. Microfluid. Nanofluid. 2017. [Google Scholar] [CrossRef]
- Lee, T.; Bu, J.; Moon, J.; Kim, Y.; Kang, Y.; Cho, Y.; Kim, I. A multi-staining chip using hydrophobic valves for exfoliative cytology in cancer. J. Micromech. Microeng. 2017. [Google Scholar] [CrossRef]
- Roman, J.; Jarroux, N.; Patriarche, G.; Francais, O.; Pelta, J.; Le Pioufle, B.; Bacri, L. Functionalized Solid-State Nanopore Integrated in a Reusable Microfluidic Device for a Better Stability and Nanoparticle Detection. ACS Appl. Mater. Interfaces 2017, 9, 41634–41640. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, A.; Asghar, W.; Ahmed, S.; Lotan, Y.; Hsieh, J.; Kim, Y.; Iqbal, S. Electrophysiological analysis of biopsy samples using elasticity as an inherent cell marker for cancer detection. Anal. Methods 2014, 6, 7166–7174. [Google Scholar] [CrossRef]
- Ali, W.; Ilyas, A.; Bui, L.; Sayles, B.; Hur, Y.; Kim, Y.; Iqbal, S. Differentiating Metastatic and Non-metastatic Tumor Cells from Their Translocation Profile through Solid-State Micropores. Langmuir 2016, 32, 4924–4934. [Google Scholar] [CrossRef] [PubMed]
- Basiji, D.; Ortyn, W.; Liang, L.; Venkatachalam, V.; Morrissey, P. Cellular image analysis and imaging by flow cytometry. Clin. Lab. Med. 2007, 27, 653–670. [Google Scholar] [CrossRef] [PubMed]
- U.S. Centers for Disease Control and Prevention. Antimicrobial Resistance: Antibiotic Resistance Threats in the United States; U.S. Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013.
- Eid, C.; Santiago, J. Assay for Listeria monocytogenes cells in whole blood using isotachophoresis and recombinase polymerase amplification. Analyst 2017, 142, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, P.; Evander, M.; Petersson, K.; Mellhammar, L.; Lehmusvuori, A.; Karhunen, U.; Soikkeli, M.; Seppa, T.; Tuunainen, E.; Spangar, A.; et al. Integrated Acoustic Separation, Enrichment, and Microchip Polymerase Chain Reaction Detection of Bacteria from Blood for Rapid Sepsis Diagnostics. Anal. Chem. 2016, 88, 9403–9411. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.; Song, D.; Shrestha, S.; Miao, J.; Cui, L.; Guan, W. A field-deployable mobile molecular diagnostic system for malaria at the point of need. Lab Chip 2016, 16, 4341–4349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horst, A.; Rosenbohm, J.; Kolluri, N.; Hardick, J.; Gaydos, C.; Cabodi, M.; Klapperich, C.; Linnes, J. A paperfluidic platform to detect Neisseria gonorrhoeae in clinical samples. Biomed. Microdevices 2018, 20, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chang, W.; Wang, C.; You, H.; Wu, J.; Liu, T.; Lee, M.; Lee, G. An integrated microfluidic system for live bacteria detection from human joint fluid samples by using ethidium monoazide and loop-mediated isothermal amplification. Microfluid. Nanofluid. 2017. [Google Scholar] [CrossRef]
- Hoyos-Nogues, M.; Brosel-Oliu, S.; Abramova, N.; Munoz, F.; Bratov, A.; Mas-Moruno, C.; Gil, F. Impedimetric antimicrobial peptide-based sensor for the early detection of periodontopathogenic bacteria. Biosens. Bioelectron. 2016, 86, 377–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, X.; Lan, Y.; Wang, B.; Zhang, Y.; Liu, B.; Yang, P.; Zhang, W.; Qiao, L. Microfluidic Air Sampler for Highly Efficient Bacterial Aerosol Collection and Identification. Anal. Chem. 2016, 88, 11504–11512. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Jing, W.; Sun, X.; Liu, Q.; Yang, C.; Liu, S.; Qin, K.; Sui, G. High-Throughput Microfluidic Device for LAMP Analysis of Airborne Bacteria. ACS Sens. 2016, 1, 958–962. [Google Scholar] [CrossRef]
- U.S. Centers for Disease Control and Prevention. Disease Burden of Influenza. Available online: https://www.cdc.gov/flu/about/disease/burden.htm (accessed on 15 July 2018).
- Prakash, R.; Pabbaraju, K.; Wong, S.; Wong, A.; Tellier, R.; Kaler, K. Multiplex, Quantitative, Reverse Transcription PCR Detection of Influenza Viruses Using Droplet Microfluidic Technology. Micromachines 2015, 6, 63–79. [Google Scholar] [CrossRef]
- U.S. Centers for Disease Control and Prevention. 2016 Case Counts in the US. Available online: https://www.cdc.gov/zika/reporting/2016-case-counts.html (accessed on 15 July 2018).
- Yang, Y.; Wang, K.; Gu, X.; Leong, K.W. Biophysical Regulation of Cell Behavior-Cross Talk between Substrate Stiffness and Nanotopography. Engineering 2017, 3, 36–54. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Shin, J.Y.; Leijten, J.; Jeon, O.; Camci-Unal, G.; Dikina, A.D.; Brinegar, K.; Ghaemmaghami, A.M.; Alsberg, E.; Khademhosseini, A. High-throughput approaches for screening and analysis of cell behaviors. Biomaterials 2018, 153, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ma, Y.; Zhao, M.; Hu, Z.; Wei, Z. Device To Study the Cell Invasion Behavior and Phenotypic Profile at Single Cell Level. Anal. Chem. 2018, 90, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, Y.; Weng, L.T.; Leung, M.; Xing, X.; Fan, Z.; Wu, H. Fast Single-Cell Patterning for Study of Drug-Induced Phenotypic Alterations of HeLa Cells Using Time-of-Flight Secondary Ion Mass Spectrometry. Anal. Chem. 2016, 88, 12196–12203. [Google Scholar] [CrossRef] [PubMed]
- Anchang, B.; Davis, K.L.; Fienberg, H.G.; Williamson, B.D.; Bendall, S.C.; Karacosta, L.G.; Tibshirani, R.; Nolan, G.P.; Plevritis, S.K. DRUG-NEM: Optimizing drug combinations using single-cell perturbation response to account for intratumoral heterogeneity. Proc. Natl. Acad. Sci. USA 2018, 115, E4294–E4303. [Google Scholar] [CrossRef] [PubMed]
- Mokbel, M.; Mokbel, D.; Mietke, A.; Träber, N.; Girardo, S.; Otto, O.; Guck, J.; Aland, S. Numerical Simulation of Real-Time Deformability Cytometry To Extract Cell Mechanical Properties. ACS Biomater. Sci. Eng. 2017, 3, 2962–2973. [Google Scholar] [CrossRef]
- Deng, Y.; Davis, S.; Yang, F.; Paulsen, K.; Kumar, M.; DeVaux, R.; Wang, X.; Conklin, D.; Oberai, A.; Herschkowitz, J.; et al. Inertial Microfluidic Cell Stretcher (iMCS): Fully Automated, High-Throughput, and Near Real-Time Cell Mechanotyping. Small 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, D.; Zhou, Y.; Khoo, B.; Han, J.; Ai, Y. Characterizing Deformability and Electrical Impedance of Cancer Cells in a Microfluidic Device. Anal. Chem. 2018, 90, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Hou, J.; Chen, S.; Chow, Y.; Wang, R.; Ma, D.; Zhu, R.; Sun, D. Microfluidic single-cell array platform enabling week-scale clonal expansion under chemical/electrical stimuli. Biomicrofluidics 2017, 11. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.J.; Mu, L.; Xue, Q.; Wu, Y.; Wu, P.H.; Li, J.; Vortmeyer, A.O.; Miller-Jensen, K.; Wirtz, D.; et al. High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal. Chem. 2013, 85, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Li, L.; Liu, M.; Yan, S.; Miao, C.; Li, R.; Luo, Y.; Liu, T.; Lin, B.; Ji, Y.; et al. Paper-Based 3D Scaffold for Multiplexed Single Cell Secretomic Analysis. Anal. Chem. 2018, 90, 5825–5832. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Luni, C.; Elvassore, N. Microfluidics for secretome analysis under enhanced endogenous signaling. Biochem. Biophys. Res. Commun. 2018, 497, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Huang, M.; Wang, X.K.; Zhu, Y.; Li, J.S.; Wong, C.C.L.; Fang, Q. Nanoliter-Scale Oil-Air-Droplet Chip-Based Single Cell Proteomic Analysis. Anal. Chem. 2018, 90, 5430–5438. [Google Scholar] [CrossRef] [PubMed]
- Truong, D.; Puleo, J.; Llave, A.; Mouneimne, G.; Kamm, R.D.; Nikkhah, M. Breast Cancer Cell Invasion into a Three Dimensional Tumor-Stroma Microenvironment. Sci. Rep. 2016, 6, 34094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, A.D.; Searson, P.C. Live-cell imaging of invasion and intravasation in an artificial microvessel platform. Cancer Res. 2014, 74, 4937–4945. [Google Scholar] [CrossRef] [PubMed]
- Erdogan, B.; Ao, M.; White, L.M.; Means, A.L.; Brewer, B.M.; Yang, L.; Washington, M.K.; Shi, C.; Franco, O.E.; Weaver, A.M.; et al. Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J. Cell Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chanasakulniyom, M.; Glidle, A.; Cooper, J. Cell proliferation and migration inside single cell arrays. Lab Chip 2015, 15, 208–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hockemeyer, K.; Janetopoulos, C.; Terekhov, A.; Hofmeister, W.; Vilgelm, A.; Costa, L.; Wikswo, J.P.; Richmond, A. Engineered three-dimensional microfluidic device for interrogating cell-cell interactions in the tumor microenvironment. Biomicrofluidics 2014, 8, 044105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.J.; Hannanta-anan, P.; Chau, M.; Kim, Y.S.; Swartz, M.A.; Wu, M. Cooperative roles of SDF-1α and EGF gradients on tumor cell migration revealed by a robust 3D microfluidic model. PLoS ONE 2013, 8, e68422. [Google Scholar] [CrossRef] [PubMed]
- Blaha, L.; Zhang, C.; Cabodi, M.; Wong, J. A microfluidic platform for modeling metastatic cancer cell matrix invasion. Biofabrication 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Bray, L.J.; Werner, C. Evaluation of Three-Dimensional in Vitro Models to Study Tumor Angiogenesis. ACS Biomater. Sci. Eng. 2017, 4, 337–346. [Google Scholar] [CrossRef]
- Du, X.; Li, W.; Du, G.; Cho, H.; Yu, M.; Fang, Q.; Lee, L.; Fang, J. Droplet Array-Based 3D Coculture System for High-Throughput Tumor Angiogenesis Assay. Anal. Chem. 2018, 90, 3253–3261. [Google Scholar] [CrossRef] [PubMed]
- Nashimoto, Y.; Hayashi, T.; Kunita, I.; Nakamasu, A.; Torisawa, Y.S.; Nakayama, M.; Takigawa-Imamura, H.; Kotera, H.; Nishiyama, K.; Miura, T.; et al. Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device. Integr. Biol. 2017, 9, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Sun, Y.; Yu, X.; Shao, Y.; Zhang, P.; Dai, G.; Fu, J. Angiogenesis in Liquid Tumors: An In Vitro Assay for Leukemic-Cell-Induced Bone Marrow Angiogenesis. Adv. Healthc. Mater. 2016, 5, 1014–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Lin, X.; Lin, L.; Feng, Q.; Kitamori, T.; Lin, J.; Sun, J. Integrated Microfluidic Platform with Multiple Functions To Probe Tumor-Endothelial Cell Interaction. Anal. Chem. 2017, 89, 10037–10044. [Google Scholar] [CrossRef] [PubMed]
- Borsig, L.; Wolf, M.; Roblek, M.; Lorentzen, A.; Heikenwalder, M. Inflammatory chemokines and metastasis-tracing the accessory. Oncogene 2014, 33, 3217–3224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paino, F.; La Noce, M.; Di Nucci, D.; Nicoletti, G.F.; Salzillo, R.; De Rosa, A.; Ferraro, G.A.; Papaccio, G.; Desiderio, V.; Tirino, V. Human adipose stem cell differentiation is highly affected by cancer cells both in vitro and in vivo: Implication for autologous fat grafting. Cell Death Dis. 2017, 8, e2568. [Google Scholar] [CrossRef] [PubMed]
- Rothbauer, M.; Zirath, H.; Ertl, P. Recent advances in microfluidic technologies for cell-to-cell interaction studies. Lab Chip 2018, 18, 249–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sai, J.; Rogers, M.; Hockemeyer, K.; Wikswo, J.P.; Richmond, A. Study of Chemotaxis and Cell-Cell Interactions in Cancer with Microfluidic Devices. Methods Enzymol. 2016, 570, 19–45. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.; Sobolik, T.; Schaffer, D.K.; Samson, P.C.; Johnson, A.C.; Owens, P.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A.; Wikswo, J.P.; et al. Engineered microfluidic bioreactor for examining the three-dimensional breast tumor microenvironment. Biomicrofluidics 2018, 12, 034102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estrada, M.; Rebelo, S.; Davies, E.; Pinto, M.; Pereira, H.; Santo, V.; Smalley, M.; Barry, S.; Gualda, E.; Alves, P.; et al. Modelling the tumour microenvironment in long-term microencapsulated 3D co-cultures recapitulates phenotypic features of disease progression. Biomaterials 2016, 78, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.J.R.; Hall, J.K.; Kiyotake, E.A.; Christensen, T.; Balasubramaniam, V.; Anseth, K.S. Epithelial-mesenchymal crosstalk influences cellular behavior in a 3D alveolus-fibroblast model system. Biomaterials 2018, 155, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, A.; Aldrian, C.; Bronsert, P.; Thomann, Y.; Nanko, N.; Melin, N.; Rucker, G.; Follo, M.; Grosu, A.; Niedermann, G.; et al. A deep conical agarose microwell array for adhesion independent three-dimensional cell culture and dynamic volume measurement. Lab Chip 2018, 18, 179–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Dugan, C.; Kennedy, R. Microfluidic Chip with Integrated Electrophoretic Immunoassay for Investigating Cell-Cell Interactions. Anal. Chem. 2018, 90, 5171–5178. [Google Scholar] [CrossRef] [PubMed]
- Ke, L.; Kuo, Z.; Chen, Y.; Yen, T.; Dong, M.; Tseng, H.; Liu, C. Cancer immunotherapy mu-environment LabChip: Taking advantage of optoelectronic tweezers. Lab Chip 2018, 18, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Su, H.; Li, P.; Chiang, P.; Huang, C.; Chen, C.; Hsieh, M. Prognostic impact of tumor infiltrating lymphocytes on patients with metastatic urothelial carcinoma receiving platinum based chemotherapy. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.; Doty, D.; Zielstorff, M.; Kariv, I.; Moy, L.Y.; Gimbel, A.; Chevillet, J.R.; Lowry, N.; Santos, J.; Mott, V.; et al. A multiplexed microfluidic system for evaluation of dynamics of immune-tumor interactions. Lab Chip 2018, 18, 1844–1858. [Google Scholar] [CrossRef] [PubMed]
- Liotta, L.; Petricoin, E. Molecular profiling of human cancer. Nat. Rev. Genet. 2000, 1, 48. [Google Scholar] [CrossRef] [PubMed]
- Lecault, V.; White, A.K.; Singhal, A.; Hansen, C.L. Microfluidic single cell analysis: From promise to practice. Curr. Opin. Chem. Biol. 2012, 16, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Armbrecht, L.; Dittrich, P.S. Recent advances in the analysis of single cells. Anal. Chem. 2016, 89, 2–21. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, Y.; Shivalila, C.S.; Soldner, F.; Markoulaki, S.; Jaenisch, R. Tracing dynamic changes of DNA methylation at single-cell resolution. Cell 2015, 163, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321. [Google Scholar] [CrossRef] [PubMed]
- Corces, M.R.; Buenrostro, J.D.; Wu, B.; Greenside, P.G.; Chan, S.M.; Koenig, J.L.; Snyder, M.P.; Pritchard, J.K.; Kundaje, A.; Greenleaf, W.J. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 2016, 48, 1193. [Google Scholar] [CrossRef] [PubMed]
- Cusanovich, D.A.; Daza, R.; Adey, A.; Pliner, H.A.; Christiansen, L.; Gunderson, K.L.; Steemers, F.J.; Trapnell, C.; Shendure, J. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015, 348, 910–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagano, T.; Lubling, Y.; Várnai, C.; Dudley, C.; Leung, W.; Baran, Y.; Cohen, N.M.; Wingett, S.; Fraser, P.; Tanay, A. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 2017, 547, 61. [Google Scholar] [CrossRef] [PubMed]
- Lorthongpanich, C.; Cheow, L.F.; Balu, S.; Quake, S.R.; Knowles, B.B.; Burkholder, W.F.; Solter, D.; Messerschmidt, D.M. Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 2013, 341, 1110–1112. [Google Scholar] [CrossRef] [PubMed]
- Cheow, L.F.; Courtois, E.T.; Tan, Y.; Viswanathan, R.; Xing, Q.; Tan, R.Z.; Tan, D.S.; Robson, P.; Loh, Y.-H.; Quake, S.R. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat. Methods 2016, 13, 833. [Google Scholar] [CrossRef] [PubMed]
- Rotem, A.; Ram, O.; Shoresh, N.; Sperling, R.A.; Goren, A.; Weitz, D.A.; Bernstein, B.E. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat. Biotechnol. 2015, 33, 1165. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Chen, C.; He, B.; Tan, K.; Lu, C. A microfluidic device for epigenomic profiling using 100 cells. Nat. Methods 2015, 12, 959. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.D.; Wu, B.; Litzenburger, U.M.; Ruff, D.; Gonzales, M.L.; Snyder, M.P.; Chang, H.Y.; Greenleaf, W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015, 523, 486. [Google Scholar] [CrossRef] [PubMed]
- Davidson, P.M.; Sliz, J.; Isermann, P.; Denais, C.; Lammerding, J. Design of a microfluidic device to quantify dynamic intra-nuclear deformation during cell migration through confining environments. Integr. Biol. 2015, 7, 1534–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, S.H.; Jang, S.-C.; Lee, B.; Jeong, H.-H.; Jeong, S.-G.; Lee, S.S.; Kim, K.P.; Lee, C.-S. Monitoring of chromosome dynamics of single yeast cells in a microfluidic platform with aperture cell traps. Lab Chip 2016, 16, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Baslan, T.; Kendall, J.; Rodgers, L.; Cox, H.; Riggs, M.; Stepansky, A.; Troge, J.; Ravi, K.; Esposito, D.; Lakshmi, B. Genome-wide copy number analysis of single cells. Nat. Protoc. 2012, 7, 1024. [Google Scholar] [CrossRef] [PubMed]
- Bogaerts, P.; Cuzon, G.; Evrard, S.; Hoebeke, M.; Naas, T.; Glupczynski, Y. Evaluation of a DNA microarray for rapid detection of the most prevalent extended-spectrum β-lactamases, plasmid-mediated cephalosporinases and carbapenemases in Enterobacteriaceae, Pseudomonas and Acinetobacter. Int. J. Antimicrob. Agents 2016, 48, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [PubMed]
- Chapman, A.R.; He, Z.; Lu, S.; Yong, J.; Tan, L.; Tang, F.; Xie, X.S. Single cell transcriptome amplification with MALBAC. PLoS ONE 2015, 10, e0120889. [Google Scholar] [CrossRef] [PubMed]
- Zahn, H.; Steif, A.; Laks, E.; Eirew, P.; VanInsberghe, M.; Shah, S.P.; Aparicio, S.; Hansen, C.L. Scalable whole-genome single-cell library preparation without preamplification. Nat. Methods 2017, 14, 167. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.K.; Edge, P.; Lee, H.S.; Bansal, V.; Bafna, V.; Huang, X.; Zhang, K. Ultraaccurate genome sequencing and haplotyping of single human cells. Proc. Natl. Acad. Sci. USA 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Qiu, L.; Xu, Y.; Li, G.; Mu, Y. Single cell digital polymerase chain reaction on self-priming compartmentalization chip. Biomicrofluidics 2017, 11, 014109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, D.; Zhang, H.; Guan, Z.; Song, Y.; Liu, R.; Zhu, Z.; Yang, C. Microwell Array Method for Rapid Generation of Uniform Agarose Droplets and Beads for Single Molecule Analysis. Anal. Chem. 2018, 90, 2570–2577. [Google Scholar] [CrossRef] [PubMed]
- Eastburn, D.J.; Sciambi, A.; Abate, A.R. Ultrahigh-throughput Mammalian single-cell reverse-transcriptase polymerase chain reaction in microfluidic drops. Anal. Chem. 2013, 85, 8016–8021. [Google Scholar] [CrossRef] [PubMed]
- Gole, J.; Gore, A.; Richards, A.; Chiu, Y.-J.; Fung, H.-L.; Bushman, D.; Chiang, H.-I.; Chun, J.; Lo, Y.-H.; Zhang, K. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat. Biotechnol. 2013, 31, 1126. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.A.; Jiang, P.; Zheng, Y.W.L.; Liao, G.J.W.; Sun, H.; Wong, J.; Siu, S.S.N.; Chan, W.C.; Chan, S.L.; Chan, A.T.C.; et al. Cancer Genome Scanning in Plasma: Detection of Tumor-Associated Copy Number Aberrations, Single-Nucleotide Variants, and Tumoral Heterogeneity by Massively Parallel Sequencing. Clin. Chem. 2013, 59, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Reinholt, S.J.; Craighead, H.G. Microfluidic Device for Aptamer-Based Cancer Cell Capture and Genetic Mutation Detection. Anal. Chem. 2018, 90, 2601–2608. [Google Scholar] [CrossRef] [PubMed]
- Manjunatha, S.D.; Erik, K.L.; Bethany, H.K.; Pratim, C.P.; Britton, O.P.; Anushree, C.; Prashant, N. High-Throughput Block Optical DNA Sequence Identification. Small 2018, 14, 1703165. [Google Scholar] [CrossRef]
- Bustin, S.A.; Mueller, R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. 2005, 109, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Picelli, S.; Faridani, O.R.; Björklund, Å.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.; Zeisel, A.; Joost, S.; La Manno, G.; Zajac, P.; Kasper, M.; Lönnerberg, P.; Linnarsson, S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 2013, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef] [PubMed]
- Rotem, A.; Ram, O.; Shoresh, N.; Sperling, R.A.; Schnall-Levin, M.; Zhang, H.; Basu, A.; Bernstein, B.E.; Weitz, D.A. High-throughput single-cell labeling (Hi-SCL) for RNA-Seq using drop-based microfluidics. PLoS ONE 2015, 10, e0116328. [Google Scholar] [CrossRef] [PubMed]
- Adamson, B.; Norman, T.M.; Jost, M.; Cho, M.Y.; Nuñez, J.K.; Chen, Y.; Villalta, J.E.; Gilbert, L.A.; Horlbeck, M.A.; Hein, M.Y.; et al. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 2016, 167, 1867.e21–1882.e21. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Subramaniam, M.; Targ, S.; Nguyen, M.; Maliskova, L.; McCarthy, E.; Wan, E.; Wong, S.; Byrnes, L.; Lanata, C.M.; et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 2017, 36, 89. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Chen, H.; Liu, L.; Huang, Y. Single Cell Total RNA Sequencing through Isothermal Amplification in Picoliter-Droplet Emulsion. Anal. Chem. 2016, 88, 10795–10799. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Wang, J. Assay of Genome-Wide Transcriptome and Secreted Proteins on the Same Single Immune Cells by Microfluidics and RNA Sequencing. Anal. Chem. 2016, 88, 10309–10315. [Google Scholar] [CrossRef] [PubMed]
- Shalek, A.K.; Satija, R.; Shuga, J.; Trombetta, J.J.; Gennert, D.; Lu, D.; Chen, P.; Gertner, R.S.; Gaublomme, J.T.; Yosef, N.; et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 2014, 510, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.C.; Clark, I.C.; Shahi, P.; Abate, A.R. Single-cell RT-PCR in microfluidic droplets with integrated chemical lysis. Anal. Chem. 2018, 90, 1273–1279. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Sun, Y.; Liu, C.; Duan, X.; Tang, W.; Li, Z. Precise quantitation of microRNA in a single cell with droplet digital PCR based on ligation reaction. Anal. Chem. 2016, 88, 11384–11389. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Gao, M.; Chong, Z.; Li, Y.; Han, X.; Chen, R.; Qin, L. Single-Cell Isolation by Modular Single-Cell Pipette for RNA-Sequencing. Lab Chip 2016, 16, 4742–4748. [Google Scholar] [CrossRef] [PubMed]
- Irish, J.M.; Kotecha, N.; Nolan, G.P. Mapping normal and cancer cell signalling networks: Towards single-cell proteomics. Nat. Rev. Cancer 2006, 6, 146. [Google Scholar] [CrossRef] [PubMed]
- Dovichi, N.J.; Hu, S.; Michels, D.; Mao, D.; Dambrowitz, A. Proteomics for Biological Discovery: Single Cell Proteomics; John Wiley & Sons: Oxford, UK, 2006; pp. 225–245. [Google Scholar]
- Ma, C.; Fan, R.; Ahmad, H.; Shi, Q.; Comin-Anduix, B.; Chodon, T.; Koya, R.C.; Liu, C.-C.; Kwong, G.A.; Radu, C.G. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T. cells. Nat. Med. 2011, 17, 738. [Google Scholar] [CrossRef] [PubMed]
- Peterson, V.M.; Zhang, K.X.; Kumar, N.; Wong, J.; Li, L.; Wilson, D.C.; Moore, R.; McClanahan, T.K.; Sadekova, S.; Klappenbach, J.A. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017, 35, 936. [Google Scholar] [CrossRef] [PubMed]
- Ullal, A.V.; Peterson, V.; Agasti, S.S.; Tuang, S.; Juric, D.; Castro, C.M.; Weissleder, R. Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci. Transl. Med. 2014, 6, 219ra9. [Google Scholar] [CrossRef] [PubMed]
- Albayrak, C.; Jordi, C.A.; Zechner, C.; Lin, J.; Bichsel, C.A.; Khammash, M.; Tay, S. Digital quantification of proteins and mRNA in single mammalian cells. Mol. Cell 2016, 61, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Abbaspourrad, A.; Zhang, H.; Tao, Y.; Cui, N.; Asahara, H.; Zhou, Y.; Yue, D.; Koehler, S.A.; Ung, L.W.; Heyman, J. Label-free single-cell protein quantification using a drop-based mix-and-read system. Sci. Rep. 2015, 5, 12756. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Yu, J.; Li, F.; Deng, L.; Wu, F.; Huang, X.; Bergstrand, J.; Widengren, J.; Dong, C.; Ren, J. In Situ Monitoring of p53 Protein and MDM2 Protein Interaction in Single Living Cells Using Single-Molecule Fluorescence Spectroscopy. Anal. Chem. 2018, 90, 6144–6151. [Google Scholar] [CrossRef] [PubMed]
- Jing, T.; Lai, Z.; Wu, L.; Han, J.; Lim, C.T.; Chen, C.-H. Single cell analysis of leukocyte protease activity using integrated continuous-flow microfluidics. Anal. Chem. 2016, 88, 11750–11757. [Google Scholar] [CrossRef] [PubMed]
- Herper, M. The Cost of Creating A New Drug Now $5 Billion, Pushing Big Pharma To Change. Forbes. com. 11 August 2013. [Google Scholar]
- Jabs, J.; Zickgraf, F.; Park, J.; Wagner, S.; Jiang, X.; Jechow, K.; Kleinheinz, K.; Toprak, U.; Schneider, M.; Meister, M.; et al. Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations. Mol. Syst. Biol. 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Boehnke, K.; Iversen, P.; Schumacher, D.; Lallena, M.; Haro, R.; Amat, J.; Haybaeck, J.; Liebs, S.; Lange, M.; Schafer, R.; et al. Assay Establishment and Validation of a High-Throughput Screening Platform for Three-Dimensional Patient-Derived Colon Cancer Organoid Cultures. J. Biomol. Screen. 2016, 21, 931–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Leslie, K.; Guest, D.; Yeshcheulova, O.; Roy, I.; Piva, M.; Moriceau, G.; Zangle, T.; Lo, R.; Teitell, M.; et al. High-Speed Live-Cell Interferometry: A New Method for Quantifying Tumor Drug Resistance and Heterogeneity. Anal. Chem. 2018, 90, 3299–3306. [Google Scholar] [CrossRef] [PubMed]
- Damiati, S.; Kompella, U.; Damiati, S.; Kodzius, R. Microfluidic Devices for Drug Delivery Systems and Drug Screening. Genes 2018, 9, 103. [Google Scholar] [CrossRef] [PubMed]
- Skardal, A.; Shupe, T.; Atala, A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 2016, 21, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ma, Y.; Lou, Q.; Zhu, H.; Yang, B.; Fang, Q. Three-Dimensional Cell Culture and Drug Testing in a Microfluidic Sidewall-Attached Droplet Array. Anal. Chem. 2017, 89, 10153–10157. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, L.; Liu, Z.; Ding, M.; Luo, G.; Liang, Q. A microfluidic chip of multiple-channel array with various oxygen tensions for drug screening. Microfluid. Nanofluid. 2016, 20. [Google Scholar] [CrossRef]
- Sun, J.; Liu, W.; Li, Y.; Gholamipour-Shirazi, A.; Abdulla, A.; Ding, X. An on-chip cell culturing and combinatorial drug screening system. Microfluid. Nanofluid. 2017, 21. [Google Scholar] [CrossRef]
- Stevens, M.; Maire, C.; Chou, N.; Murakami, M.; Knoff, D.; Kikuchi, Y.; Kimmerling, R.; Liu, H.; Haidar, S.; Calistri, N.; et al. Drug sensitivity of single cancer cells is predicted by changes in mass accumulation rate. Nat. Biotechnol. 2016, 34, 1161–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- LaBonia, G.; Ludwig, K.; Mousseau, C.; Hummon, A. iTRAQ Quantitative Proteomic Profiling and MALDI-MSI of Colon Cancer Spheroids Treated with Combination Chemotherapies in a 3D Printed Fluidic Device. Anal. Chem. 2018, 90, 1423–1430. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Wu, L.; Zhang, Y.; Zong, S.; Wang, Z.; Cui, Y. Pharmacokinetics-on-a-Chip Using Label-Free SERS Technique for Programmable Dual-Drug Analysis. ACS Sens. 2017, 2, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, W.; Choi, G.; Yang, X.; Miao, J.; Cui, L.; Guan, W. Arbitrarily Accessible 3D Microfluidic Device for Combinatorial High-Throughput Drug Screening. Sensors 2016, 16, 1616. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Kim, Y.; Son, J.; Son, M.; Lee, T. On-Chip Peptide Mass Spectrometry Imaging for Protein Kinase Inhibitor Screening. Anal. Chem. 2017, 89, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Popova, A.; Schillo, S.; Demir, K.; Ueda, E.; Nesterov-Mueller, A.; Levkin, P. Droplet-Array (DA) Sandwich Chip: A Versatile Platform for High-Throughput Cell Screening Based on Superhydrophobic-Superhydrophilic Micropatterning. Adv. Mater. 2015, 27, 5217–5222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitebread, S.; Hamon, J.; Bojanic, D.; Urban, L. In vitro safety pharmacology profiling: An essential tool for successful drug development. Drug Discov. Today 2005, 10, 1421–1433. [Google Scholar] [CrossRef]
- Espulgar, W.; Aoki, W.; Ikeuchi, T.; Mita, D.; Saito, M.; Lee, J.; Tamiya, E. Centrifugal microfluidic platform for single-cell level cardiomyocyte-based drug profiling and screening. Lab Chip 2015, 15, 3572–3580. [Google Scholar] [CrossRef] [PubMed]
- Mathur, A.; Loskill, P.; Shao, K.; Huebsch, N.; Hong, S.; Marcus, S.; Marks, N.; Mandegar, M.; Conklin, B.; Lee, L.; et al. Human iPSC-based Cardiac Microphysiological System For Drug Screening Applications. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]


| Biomolecule(s) Detected | Type of Device | Method of Detection | Results Readout | Limit of Detection | Total Time (min) | Sample Type | Quantitative | Ref. |
|---|---|---|---|---|---|---|---|---|
| Myoglobin | LFSA | Antibodies with PtNPs, H2O2 reaction forming O2 gas | Increase in pressure | 2.9 ng/mL | 20 | Dilute serum | yes | [31] |
| Tetrodotoxin | LFSA | Gold nanoflower conjugated antibodies, quantum dots | Quantum dot fluorescence | 0.2 ng/mL | 8 | TTX spiked PBS | yes | [32] |
| Vaspin | LFSA | Complimentary aptamers and AuNPs | Colorimetric intensity | 0.137 nM in buffer 0.105 nM in serum | 5 | Vaspin spiked buffer and serum | yes | [30] |
| Extracellular vesicles | LFSA | Colloidal gold, carbon black, magnetic nanoparticle conjugated antibodies | Colorimetric intensity | 3.4 × 106 EVs/μL | 15 | Human plasma | yes | [33] |
| Myeloperoxidase | LFSA | AuNP conjugated antibodies | Colorimetric intensity | 250 ng/mL | 15 | Human sputum | yes | [29] |
| Glucose, nitrites, and protein | μPAD | Chemical reactions with biomolecules and paper actuator | Colorimetric | n.a. | 12 | Artificial saliva | no | [34] |
| Phenylalanine | μPAD | Phe reaction forming NH3 and pH change | Colorimetric intensity | 20 μM | 20 | Urine | yes | [35] |
| Glucose, pH, and protein | μPAD | Enzymes and chromogenic agents | Colorimetric | 2 mM 0.6 mg/mL | 5 | Artificial Urine | yes | [36] |
| Lactate | μPAD | Electrochemilumin-escence reaction | ECL intensity | 0.035 mM | n.a. | Saliva | yes | [37] |
| miRNA 21 | PDMS | Molecular beacon probe | Fluorescence | n.a. | 30 | Blood | yes | [38] |
| CRP | PDMS | Capture antibodies | Spectrometry shift | 3.2 ng/mL | 60 | Blood | yes | [39] |
| IL-2 | PDMS | Capture antibodies | Fluorescence | 50 pg/mL | 30 | Blood | yes | [40] |
| CD4 | PDMS | 2 mm beads and chemiluminescence assay | Chemiluminescence | 75 cells/μL | 45 | Blood | yes | [41] |
| PSA | PDMS | Poly styrene beads with antibodies | Droplet counting | 3.67 pM | 45 | Spiked HEPES | yes | [42] |
| H2O2 | PDMS | Horseradish peroxidase-Au nanoclusters and droplets | Fluorescence | 200 amol | 90 | Cell cultures | yes | [43] |
| Myoglobin, cTn I, CK-MB | Chip | Carbon nanotubes and antibodies | Conductance | 6 fg/mL 50 fg/mL 20 fg/mL | <1 | Spiked PBS | yes | [44] |
| PSA | Chip | Carbon nanotubes and antibodies | Resistance | 1.18 ng/mL | 120 | PSA solution | yes | [45] |
| Insulin, glucagon, and somatostatin | Chip | Antibodies | Surface plasmon resonance | 1 nM 4 nM 246 nM | ~20 | Spiked solution | yes | [46] |
| Galectin-1 | Chip | Alumina nanoparticles, antibodies | Impedance | 7.8 μg/mL | 30 | T24 cell lysates | yes | [47] |
| IFN-γ | Chip | RNA aptamer on gold electrode array | Impedance | 11.56 pM | <35 | Spiked solutions | yes | [48] |
| Type of Device | Advantages | Disadvantages | Ref. |
|---|---|---|---|
| LFSA | Easy to use at home or in clinic, inexpensive, quick results | Some currently have poor limit of detection, most in research setting only, majority not quantitative | [29,30,31,32,33] |
| μPAD | Easy to use, cheapest type of device, quick results, easy storage and disposal, can be quantitative, require small sample | Some currently have poor limit of detection, must in research setting only, some required non-ambient conditions | [34,35,36,37] |
| PDMS | Highly sensitive, easily controllable, relatively inexpensive, requires small amount of sample, high throughput | Requires special training and equipment for use, almost no use in clinics currently | [38,39,40,41,42,43] |
| Chip | Easy to use, quick results, requires small amount of sample, sensitive limits of detection, easy to manufacture | Many require special equipment, can be expensive depending on test | [44,45,46,47,48] |
| Cell(s) Detected | Type of Device | Method of Detection | Results Readout | Limit of Detection | Total Time (min) | Sample Type | Quantitative | Ref. |
|---|---|---|---|---|---|---|---|---|
| RBCs | LFSA | RBC migration distance to determine coagulation | RBC migration distance | n.a. | 4 | Whole blood | no | [51] |
| RBCs | μPAD | Directed flow of cells to determine hematocrit | Blood travel distance | n.a. | 30 | Whole blood | yes | [52] |
| WBCs | HTS-RS | Combined automated imaging microscopy with Raman spectroscopy | Raman spectra | n.a. | 20 | Extracted WBCs | yes | [53] |
| WBCs | Chip | Small electrodes patterned onto a thin layer of gold | Voltage | 195 cells/µL | 20 | WBCs in 1 mM ferricyanide/ferrocyanide | yes | [55] |
| WBCs | PDMS | Chemotaxis and NETosis for neutrophil sorting and phenotyping | Fluorescence | n.a. | 120 | Whole blood | yes | [54] |
| Platelet | ECL | Adhesion molecule E-selectin as marker site on damaged HUVEC | ECL intensity | 1 platelet | 12 | Platelet-rich plasma | yes | [56] |
| Cancer Cell | Microwell | Fluorescent glucose analog (2-NBDG) to detect high glucose uptake | Fluorescence | n.a. | 10 | PE sample | yes | [57] |
| Cancer Cell | PDMS | Fluorescence-tagged antibodies | Colori-metric | 106 cells/mL | 1.5 | Serum sample | yes | [58] |
| Cancer Cell | PDMS | Six different antibodies for staining | Staining | n.a. | 140 | Serum sample | no | [59] |
| Pathogen(s) Detected | Type of Device | Method of Detection | Results Readout | LOD | Total Time (min) | Sample Type | Quant. | Ref. |
|---|---|---|---|---|---|---|---|---|
| Listeria monocytogenes | PDMS | Isotachophoresis purification and recombinase polymerase amplification | Fluorescence | 5000 cells/mL | <50 | Spiked Blood | yes | [65] |
| Pseudomonas putida and Escherichia coli | PDMS | Acoustic RBC separation and PCR | Fluorescence | 1000 cells/mL | n.a. | Blood | no | [66] |
| Plasmodium falciparum | Chip | LAMP | Fluorescence | 0.6 cells/μL | <40 | Blood | yes | [67] |
| Neisseria gonorrhoeae | μPAD | tHDA | Colorimetric | 10 cells | 60 | Genital swabs | no | [68] |
| Acinetobacter baumannii, CNS, Escherichia coli, Staphylococcus aureus, and MRSA | PDMS | LAMP and ethidium monoazide (EMA) | Fluorescence | ~1 CFU | ~60 | Spiked solution | no | [69] |
| Streptococcus sanguinis | Chip | Immobilized antimicrobial peptides on electrodes | Impedance | 10 CFU/mL | ~60 | Artificial saliva | yes | [70] |
| Vibrio parahaemolyticus | PDMS | Cell trapping | Mass spectrometry | 15 CFU | 20 | Spiked air | yes | [71] |
| Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, Citrobacter koseri, and Klebsiella pneumonia | PDMS | LAMP | Fluorescence | 24 cells | <60 | Airborne bacterial lysates | yes | [72] |
| H5N2 avian influenza virus | Chip | ZnO nanorods functionalized with antibodies | Fluorescence | 3.6 × 103 EID50/mL | 25 | Dilute sample | yes | [7] |
| H1N1, H3N2, and influenza B | PDMS | Universal aptamer conjugated to magnetic beads | Fluorescence | 3.2 HAU | 20 | Purified RNA | no | [8] |
| H1N1 and influenza A | Chip | Nitrocellulose membrane functionalized with antibodies for ELISA | Colorimetric | 32 × 10−4 HA | 20 | Lysed sample | yes | [9] |
| H5N1 avian influenza virus | DMF | SERS-based immunoassay | Absorbance | 74 pg/mL | 50 | Human serum | yes | [10] |
| Zika virus and HIV | Phone | Bioluminescent assay with BART-LAMP | Luminescence | 5 PFU | 45 | Blood, saliva, urine | yes | [11] |
| Zika virus | μPAD | Toehold sensor linked to RNA amplification | Colorimetric | 3 fM | 30 | RNA in serum | yes | [12] |
| Zika virus | LFSA | Incorporation of RT-LAMP | Colorimetric | One copy of RNA | 35 | Blood | yes | [13] |
| HIV | PDMS | Traps from porous silica beads and polystyrene | Fluorescence | n.a. | 60 | Blood plasma | yes | [14] |
| TOX, RUB, CMV, HSV-1, and HSV-2 herpes | Chip | Chemiluminescence immunoassay | Luminescence | 32-fold dilution | 30 | Serum sample | yes | [15] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campbell, J.M.; Balhoff, J.B.; Landwehr, G.M.; Rahman, S.M.; Vaithiyanathan, M.; Melvin, A.T. Microfluidic and Paper-Based Devices for Disease Detection and Diagnostic Research. Int. J. Mol. Sci. 2018, 19, 2731. https://doi.org/10.3390/ijms19092731
Campbell JM, Balhoff JB, Landwehr GM, Rahman SM, Vaithiyanathan M, Melvin AT. Microfluidic and Paper-Based Devices for Disease Detection and Diagnostic Research. International Journal of Molecular Sciences. 2018; 19(9):2731. https://doi.org/10.3390/ijms19092731
Chicago/Turabian StyleCampbell, Joshua M., Joseph B. Balhoff, Grant M. Landwehr, Sharif M. Rahman, Manibarathi Vaithiyanathan, and Adam T. Melvin. 2018. "Microfluidic and Paper-Based Devices for Disease Detection and Diagnostic Research" International Journal of Molecular Sciences 19, no. 9: 2731. https://doi.org/10.3390/ijms19092731