A Cell Junctional Protein Network Associated with Connexin-26
Abstract
:1. Introduction
2. Results
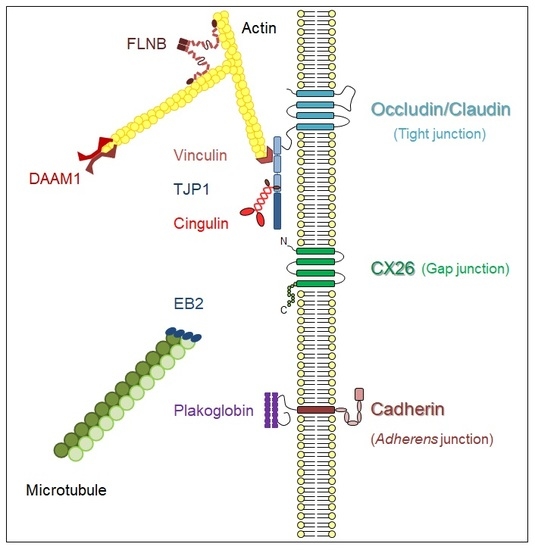
2.1. A Membrane-Cytoskeleton Protein Network Associated with CX26
2.2. Known CX Binding Partners in the CX26 Molecular Complex
2.3. Expanding the Protein Interaction Network with Cx26
3. Discussion
4. Materials and Methods
4.1. Animals and Tissue
4.2. DNA Clones
4.3. Antibodies
4.4. Bacteria Expression of Fusion Protein
4.5. Affinity Capture Assay
4.6. Mass Spectrometry Analyses
4.7. NCBI Protein Reference Sequence Accession Numbers
4.8. In Silico Analyses
4.9. Yeast Two-Hybrid Assay
4.10. Immunoprecipitation and Western Blotting
4.11. Indirect Immunofluorescence
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhu, Y.; Zong, L.; Mei, L.; Zhao, H.-B. Connexin26 Gap Junction Mediates MiRNA Intercellular Genetic Communication in the Cochlea and Is Required for Inner Ear Development. Sci. Rep. 2015, 5, 15647. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.L.; Ceriani, F.; Houston, O.; Polishchuk, R.; Polishchuk, E.; Crispino, G.; Zorzi, V.; Mammano, F.; Marcotti, W. Connexin-Mediated Signaling in Nonsensory Cells Is Crucial for the Development of Sensory Inner Hair Cells in the Mouse Cochlea. J. Neurosci. 2017, 37, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Beltramello, M.; Piazza, V.; Bukauskas, F.F.; Pozzan, T.; Mammano, F. Impaired Permeability to Ins(1,4,5)P3 in a Mutant Connexin Underlies Recessive Hereditary Deafness. Nat. Cell Biol. 2005, 7, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, W.; Ahmad, S.; Sipp, J.A.; Chen, P.; Lin, X. Gap Junction-Mediated Intercellular Biochemical Coupling in Cochlear Supporting Cells Is Required for Normal Cochlear Functions. Proc. Natl. Acad. Sci. USA 2005, 102, 15201–15206. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 12 April 2018).
- Kelsell, D.P.; Dunlop, J.; Stevens, H.P.; Lench, N.J.; Liang, J.N.; Parry, G.; Mueller, R.F.; Leigh, I.M. Connexin 26 Mutations in Hereditary Non-Syndromic Sensorineural Deafness. Nature 1997, 387, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, P.; Estivill, X.; Volpini, V.; Totaro, A.; Castellvi-Bel, S.; Govea, N.; Mila, M.; Della, M.M.; Ventruto, V.; De, M.B.; et al. Linkage of DFNB1 to Non-Syndromic Neurosensory Autosomal-Recessive Deafness in Mediterranean Families. Eur. J. Hum. Genet. 1997, 5, 83–88. [Google Scholar] [PubMed]
- Angeli, S.; Lin, X.; Liu, X.Z. Genetics of Hearing and Deafness. Anat. Rec. 2012, 295, 1812–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, R.J.; Jones, M.-K.N. Nonsyndromic Hearing Loss and Deafness, DFNB1. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Shearer, A.E.; Hildebrand, M.S.; Smith, R.J. Hereditary Hearing Loss and Deafness Overview. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Donahue, H.J.; Qu, R.W.; Genetos, D.C. Joint Diseases: From Connexins to Gap Junctions. Nat. Rev. Rheumatol. 2018, 14, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Derangeon, M.; Spray, D.C.; Bourmeyster, N.; Sarrouilhe, D.; Hervé, J.-C. Reciprocal Influence of Connexins and Apical Junction Proteins on Their Expressions and Functions. Biochim. Biophys. Acta 2009, 1788, 768–778. [Google Scholar] [CrossRef] [PubMed]
- Dbouk, H.A.; Mroue, R.M.; El-Sabban, M.E.; Talhouk, R.S. Connexins: A Myriad of Functions Extending beyond Assembly of Gap Junction Channels. Cell Commun. Signal. 2009, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.W. The Gap Junction Proteome and Its Relationship to Disease. Trends Cell Biol. 2010, 20, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Hervé, J.-C.; Derangeon, M.; Sarrouilhe, D.; Giepmans, B.N.G.; Bourmeyster, N. Gap Junctional Channels Are Parts of Multiprotein Complexes. Biochim. Biophys. Acta 2012, 1818, 1844–1865. [Google Scholar] [CrossRef] [PubMed]
- Stains, J.P.; Civitelli, R. Connexins in the Skeleton. Semin. Cell Dev. Biol. 2016, 50, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Leithe, E.; Mesnil, M.; Aasen, T. The Connexin 43 C-Terminus: A Tail of Many Tales. Biochim. Biophys. Acta 2018, 1860, 48–64. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Nakagawa, S.; Suga, M.; Yamashita, E.; Oshima, A.; Fujiyoshi, Y.; Tsukihara, T. Structure of the Connexin 26 Gap Junction Channel at 3.5 Å Resolution. Nature 2009, 458, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Nusrat, A.; Chen, J.A.; Foley, C.S.; Liang, T.W.; Tom, J.; Cromwell, M.; Quan, C.; Mrsny, R.J. The Coiled-Coil Domain of Occludin Can Act to Organize Structural and Functional Elements of the Epithelial Tight Junction. J. Biol. Chem. 2000, 275, 29816–29822. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.F.; Glenn, K.A.; Zhang, Y.; Wen, H.; Knutson, T.; Gouvion, C.M.; Robinson, B.K.; Zhou, Z.; Yang, B.; Smith, R.J.H.; et al. Selective Cochlear Degeneration in Mice Lacking the F-Box Protein, Fbx2, a Glycoprotein-Specific Ubiquitin Ligase Subunit. J. Neurosci. 2007, 27, 5163–5171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, D.J.F.; Cohen-Salmon, M.; Charollais, A.; Caille, D.; Lampe, P.D.; Chavrier, P.; Meda, P.; Petit, C. Consortin, a Trans-Golgi Network Cargo Receptor for the Plasma Membrane Targeting and Recycling of Connexins. Hum. Mol. Genet. 2010, 19, 262–275. [Google Scholar] [CrossRef] [PubMed]
- STRING: Functional Protein Association Networks. Available online: https://string-db.org/cgi/input.pl (accessed on 15 July 2018).
- Masson, D.; Kreis, T.E. Binding of E-MAP-115 to Microtubules Is Regulated by Cell Cycle-Dependent Phosphorylation. J. Cell Biol. 1995, 131, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.F.; Mandai, K.; Sakisaka, T.; Okabe, N.; Yamamoto, Y.; Yokoyama, S.; Mizoguchi, A.; Shiozaki, H.; Monden, M.; Takai, Y. Ankycorbin: A Novel Actin Cytoskeleton-associated Protein. Genes Cells 2001, 5, 1001–1008. [Google Scholar] [CrossRef]
- Ajima, R.; Kajiya, K.; Inoue, T.; Tani, M.; Shiraishi-Yamaguchi, Y.; Maeda, M.; Segawa, T.; Furuichi, T.; Sutoh, K.; Yokota, J. HOMER2 Binds MYO18B and Enhances Its Activity to Suppress Anchorage Independent Growth. Biochem. Biophys. Res. Commun. 2007, 356, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi-Yamaguchi, Y.; Sato, Y.; Sakai, R.; Mizutani, A.; Knöpfel, T.; Mori, N.; Mikoshiba, K.; Furuichi, T. Interaction of Cupidin/Homer2 with Two Actin Cytoskeletal Regulators, Cdc42 Small GTPase and Drebrin, in Dendritic Spines. BMC Neurosci. 2009, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Metzger, T.; Gache, V.; Xu, M.; Cadot, B.; Folker, E.S.; Richardson, B.E.; Gomes, E.R.; Baylies, M.K. MAP and Kinesin-Dependent Nuclear Positioning Is Required for Skeletal Muscle Function. Nature 2012, 484, 120–124. [Google Scholar] [CrossRef] [PubMed]
- Flam, B.R.; Hartmann, P.J.; Harrell-Booth, M.; Solomonson, L.P.; Eichler, D.C. Caveolar Localization of Arginine Regeneration Enzymes, Argininosuccinate Synthase, and Lyase, with Endothelial Nitric Oxide Synthase. Nitric Oxide 2001, 5, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Giepmans, B.N.G.; Moolenaar, W.H. The Gap Junction Protein Connexin43 Interacts with the Second PDZ Domain of the Zona Occludens-1 Protein. Curr. Biol. 1998, 8, 931–934. [Google Scholar] [CrossRef]
- Toyofuku, T.; Yabuki, M.; Otsu, K.; Kuzuya, T.; Hori, M.; Tada, M. Direct Association of the Gap Junction Protein Connexin-43 with ZO-1 in Cardiac Myocytes. J. Biol. Chem. 1998, 273, 12725–12731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, T.; Sawada, N.; Chiba, H.; Kokai, Y.; Yamamoto, M.; Urban, M.; Lee, G.-H.; Hertzberg, E.L.; Mochizuki, Y.; Spray, D.C. Induction of Tight Junctions in Human Connexin 32 (HCx32)-Transfected Mouse Hepatocytes: Connexin 32 Interacts with Occludin. Biochem. Biophys. Res. Commun. 1999, 266, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.M.; Fay, A.J.; Puthenveedu, M.A.; von Zastrow, M.; Jan, Y.-N.; Jan, L.Y. Microtubule Plus-End-Tracking Proteins Target Gap Junctions Directly from the Cell Interior to Adherens Junctions. Cell 2007, 128, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Fowler, S.L.; Akins, M.; Zhou, H.; Figeys, D.; Bennett, S.A.L. The Liver Connexin32 Interactome Is a Novel Plasma Membrane-Mitochondrial Signaling Nexus. J. Proteome Res. 2013, 12, 2597–2610. [Google Scholar] [CrossRef] [PubMed]
- Zemljic-Harpf, A.E.; Godoy, J.C.; Platoshyn, O.; Asfaw, E.K.; Busija, A.R.; Domenighetti, A.A.; Ross, R.S. Vinculin Directly Binds Zonula Occludens-1 and Is Essential for Stabilizing Connexin-43-Containing Gap Junctions in Cardiac Myocytes. J. Cell Sci. 2014, 127, 1104–1116. [Google Scholar] [CrossRef] [PubMed]
- Kausalya, P.J.; Reichert, M.; Hunziker, W. Connexin45 Directly Binds to ZO-1 and Localizes to the Tight Junction Region in Epithelial MDCK Cells. FEBS Lett. 2001, 505, 92–96. [Google Scholar] [CrossRef]
- Nielsen, P.A.; Baruch, A.; Shestopalov, V.I.; Giepmans, B.N.G.; Dunia, I.; Benedetti, E.L.; Kumar, N.M. Lens Connexins Α3Cx46 and Α8Cx50 Interact with Zonula Occludens Protein-1 (ZO-1). Mol. Biol. Cell 2003, 14, 2470–2481. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Olson, C.; Lu, S.; Kamasawa, N.; Yasumura, T.; Rash, J.E.; Nagy, J.I. Neuronal Connexin36 Association with Zonula Occludens-1 Protein (ZO-1) in Mouse Brain and Interaction with the First PDZ Domain of ZO-1. Eur. J. Neurosci. 2004, 19, 2132–2146. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Solan, J.L.; Taffet, S.M.; Javier, R.; Lampe, P.D. Connexin 43 Interacts with Zona Occludens-1 and -2 Proteins in a Cell Cycle Stage-Specific Manner. J. Biol. Chem. 2005, 280, 30416–30421. [Google Scholar] [CrossRef] [PubMed]
- Penes, M.C.; Li, X.; Nagy, J.I. Expression of Zonula Occludens-1 (ZO-1) and the Transcription Factor ZO-1-associated Nucleic Acid-binding Protein (ZONAB)–MsY3 in Glial Cells and Colocalization at Oligodendrocyte and Astrocyte Gap Junctions in Mouse Brain. Eur. J. Neurosci. 2005, 22, 404–418. [Google Scholar] [CrossRef] [PubMed]
- Flores, C.E.; Li, X.; Bennett, M.V.L.; Nagy, J.I.; Pereda, A.E. Interaction between Connexin35 and Zonula Occludens-1 and Its Potential Role in the Regulation of Electrical Synapses. Proc. Natl. Acad. Sci. USA 2008, 105, 12545–12550. [Google Scholar] [CrossRef] [PubMed]
- Cordenonsi, M.; D’Atri, F.; Hammar, E.; Parry, D.A.; Kendrick-Jones, J.; Shore, D.; Citi, S. Cingulin Contains Globular and Coiled-Coil Domains and Interacts with ZO-1, ZO-2, ZO-3, and Myosin. J. Cell Biol. 1999, 147, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Yum, S.W.; Zhang, J.; Valiunas, V.; Kanaporis, G.; Brink, P.R.; White, T.W.; Scherer, S.S. Human Connexin26 and Connexin30 Form Functional Heteromeric and Heterotypic Channels. Am. J. Physiol. Cell Physiol. 2007, 293, 1032–1048. [Google Scholar] [CrossRef] [PubMed]
- Karademir, L.B.; Aoyama, H.; Yue, B.; Chen, H.; Bai, D. Engineered Cx26 Variants Established Functional Heterotypic Cx26/Cx43 and Cx26/Cx40 Gap Junction Channels. Biochem. J. 2016, 473, 1391–1403. [Google Scholar] [CrossRef] [PubMed]
- Schubert, A.-L.; Schubert, W.; Spray, D.C.; Lisanti, M.P. Connexin Family Members Target to Lipid Raft Domains and Interact with Caveolin-1. Biochemistry 2002, 41, 5754–5764. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Chen, S.; Shao, Q.; Chen, J.; Bijian, K.; Laird, D.W.; Alaoui-Jamali, M.A. Dynamin 2 Interacts with Connexin 26 to Regulate Its Degradation and Function in Gap Junction Formation. Int. J. Biochem. Cell Biol. 2014, 55, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Henzl, M.T.; Thalmann, I.; Larson, J.D.; Ignatova, E.G.; Thalmann, R. The Cochlear F-Box Protein OCP1 Associates with OCP2 and Connexin 26. Hear. Res. 2004, 191, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Sorgen, P.L.; Trease, A.J.; Spagnol, G.; Delmar, M.; Nielsen, M.S. Protein–Protein Interactions with Connexin 43: Regulation and Function. Int. J. Mol. Sci. 2018, 19, 1428. [Google Scholar] [CrossRef] [PubMed]
- Thévenin, A.F.; Kowal, T.J.; Fong, J.T.; Kells, R.M.; Fisher, C.G.; Falk, M.M. Proteins and Mechanisms Regulating Gap-Junction Assembly, Internalization, and Degradation. Physiology 2013, 28, 93–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.G.; Peracchia, C. Chemical Gating of Heteromeric and Heterotypic Gap Junction Channels. J. Membr. Biol. 1998, 162, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.A.; Wong, W.-C.; Eisenhaber, B.; Warwicker, J.; Eisenhaber, F. Charged Residues next to Transmembrane Regions Revisited: “Positive-inside Rule” Is Complemented by the “Negative inside Depletion/Outside Enrichment Rule”. BMC Biol. 2017, 15, 66. [Google Scholar] [CrossRef] [PubMed]
- Kojima, T.; Kokai, Y.; Chiba, H.; Yamamoto, M.; Mochizuki, Y.; Sawada, N. Cx32 but Not Cx26 Is Associated with Tight Junctions in Primary Cultures of Rat Hepatocytes. Exp. Cell Res. 2001, 263, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Komarova, Y.; Lansbergen, G.; Galjart, N.; Grosveld, F.; Borisy, G.G.; Akhmanova, A. EB1 and EB3 Control CLIP Dissociation from the Ends of Growing Microtubules. Mol. Biol. Cell 2005, 16, 5334–5345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, J.; Xie, M.; Gou, X.; Lee, P.; Schneider, M.D.; Wu, X. Microtubules Regulate Focal Adhesion Dynamics through MAP4K4. Dev. Cell 2014, 31, 572–585. [Google Scholar] [CrossRef] [PubMed]
- Chkourko, H.S.; Guerrero-Serna, G.; Lin, X.; Darwish, N.; Pohlmann, J.R.; Cook, K.E.; Martens, J.R.; Rothenberg, E.; Musa, H.; Delmar, M. Remodeling of Mechanical Junctions and of Microtubule-Associated Proteins Accompany Cardiac Connexin43 Lateralization. Heart Rhythm 2012, 9, 1133–1140.e6. [Google Scholar] [CrossRef] [PubMed]
- Agullo-Pascual, E.; Lin, X.; Leo-Macias, A.; Zhang, M.; Liang, F.-X.; Li, Z.; Pfenniger, A.; Lübkemeier, I.; Keegan, S.; Fenyö, D.; et al. Super-Resolution Imaging Reveals That Loss of the C-Terminus of Connexin43 Limits Microtubule plus-End Capture and NaV1.5 Localization at the Intercalated Disc. Cardiovasc. Res. 2014, 104, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, W.H.; Gingras, A.R.; Critchley, D.R.; Emsley, J. Integrin Connections to the Cytoskeleton through Talin and Vinculin. Biochem. Soc. Trans. 2008, 36, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Francis, R.; Wei, C.J.; Linask, K.L.; Lo, C.W. Connexin 43-Mediated Modulation of Polarized Cell Movement and the Directional Migration of Cardiac Neural Crest Cells. Dev. Camb. Engl. 2006, 133, 3629–3639. [Google Scholar] [CrossRef] [PubMed]
- Haines, R.J.; Pendleton, L.C.; Eichler, D.C. Argininosuccinate Synthase: At the Center of Arginine Metabolism. Int. J. Biochem. Mol. Biol. 2011, 2, 8–23. [Google Scholar] [PubMed]
- Rodriguez-Sinovas, A.; Boengler, K.; Cabestrero, A.; Gres, P.; Morente, M.; Ruiz-Meana, M.; Konietzka, I.; Miró, E.; Totzeck, A.; Heusch, G.; et al. Translocation of Connexin 43 to the Inner Mitochondrial Membrane of Cardiomyocytes through the Heat Shock Protein 90-Dependent TOM Pathway and Its Importance for Cardioprotection. Circ. Res. 2006, 99, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, P.M.; Ryu, S.-Y.; Pruzansky, D.P.; Kuriakose, M.; Gilmore, A.; Kinnally, K.W. Mitochondrial Apoptosis Is Amplified through Gap Junctions. Biochem. Biophys. Res. Commun. 2009, 390, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Sajan, S.A.; Warchol, M.E.; Lovett, M. Toward a Systems Biology of Mouse Inner Ear Organogenesis: Gene Expression Pathways, Patterns and Network Analysis. Genetics 2007, 177, 631–653. [Google Scholar] [CrossRef] [PubMed]
- Home-UniGene-NCBI. Available online: https://www.ncbi.nlm.nih.gov/unigene (accessed on 24 April 2018).
- Shen, J.; Scheffer, D.I.; Kwan, K.Y.; Corey, D.P. SHIELD: An Integrative Gene Expression Database for Inner Ear Research. Database 2015, 2015, bav071. [Google Scholar] [CrossRef] [PubMed]
- MGI-Mouse Genome Informatics. The International Database Resource for the Laboratory Mouse. Available online: http://www.informatics.jax.org/ (accessed on 24 April 2018).
- International Mouse Phenotyping Consortium. Available online: http://www.mousephenotype.org/ (accessed on 24 April 2018).
- Azaiez, H.; Decker, A.R.; Booth, K.T.; Simpson, A.C.; Shearer, A.E.; Huygen, P.L.M.; Bu, F.; Hildebrand, M.S.; Ranum, P.T.; Shibata, S.B.; et al. HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice. PLOS Genet. 2015, 11, e1005137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuse, M.; Fujita, K.; Hiiragi, T.; Fujimoto, K.; Tsukita, S. Claudin-1 and -2: Novel Integral Membrane Proteins Localizing at Tight Junctions with No Sequence Similarity to Occludin. J. Cell Biol. 1998, 141, 1539–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riazuddin, S.; Ahmed, Z.M.; Fanning, A.S.; Lagziel, A.; Kitajiri, S.; Ramzan, K.; Khan, S.N.; Chattaraj, P.; Friedman, P.L.; Anderson, J.M.; et al. Tricellulin Is a Tight-Junction Protein Necessary for Hearing. Am. J. Hum. Genet. 2006, 79, 1040–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dror, A.A.; Avraham, K.B. Hearing Loss: Mechanisms Revealed by Genetics and Cell Biology. Annu. Rev. Genet. 2009, 43, 411–437. [Google Scholar] [CrossRef] [PubMed]
- Wangemann, P. K+ Cycling and Its Regulation in the Cochlea and the Vestibular Labyrinth. Audiol. Neurotol. 2002, 7, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef] [PubMed]
- Havlis, J.; Thomas, H.; Sebela, M.; Shevchenko, A. Fast-Response Proteomics by Accelerated in-Gel Digestion of Proteins. Anal. Chem. 2003, 75, 1300–1306. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical Statistical Model to Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef] [PubMed]
- The European Bioinformatics Institute < EMBL-EBI. Available online: https://www.ebi.ac.uk/ (accessed on 15 July 2018).
- ExPASy: SIB Bioinformatics Resource Portal—Home. Available online: https://www.expasy.org/ (accessed on 15 July 2018).
- Hereditary Hearing Loss. Hereditary Hearing Loss Homepage. Available online: http://hereditaryhearingloss.org/ (accessed on 15 July 2018).
- Genomics and Bioinformatics @ Davidson College. Available online: http://gcat.davidson.edu/ (accessed on 15 July 2018).
- PSORT WWW Server. Available online: https://psort.hgc.jp/ (accessed on 15 July 2018).
- The Connexin-Deafness Homepage. Available online: http://davinci.crg.es/deafness/ (accessed on 15 July 2018).
- Uniprot < EMBL-EBI. Available online: https://www.ebi.ac.uk/uniprot (accessed on 15 July 2018).
- BioGRID. Database of Protein, Chemical, and Genetic Interactions. Available online: https://thebiogrid.org/ (accessed on 15 July 2018).
- WebGestalt GSAT. Available online: http://www.webgestalt.org/option.php (accessed on 15 July 2018).
- Clustal Omega < Multiple Sequence Alignment < EMBL-EBI. Available online: https://www.ebi.ac.uk/Tools/msa/clustalo/ (accessed on 15 July 2018).







| Human Gene Acronym | Human Gene | Protein Name, Aliases, and Acronyms |
|---|---|---|
| CGN | Cingulin | Cingulin |
| DAAM1 | Disheveled-associated activator of morphogenesis 1 | DAAM1 |
| FLNB | Filamin B | Filamin-B, Filamin-3, β-filamin |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH |
| HOMER2 | Homer scaffold protein 2 | Homer-2, cupidin |
| JUP | Junction plakoglobin | Plakoglobin, γ-catenin, desmoplakin-3 |
| MAP7 | Microtubule-associated protein 7 | MAP7, EMAP-115, ensconsin |
| MAPRE2 | Microtubule-associated RP/EB family member 2 | EB2 |
| PTK2B | Protein tyrosine kinase 2β | Focal adhesion kinase 2, FAK2, PYK2, PTK2B |
| RAI14 | Retinoic acid-induced 14 | RAI14, ankycorbin, NORPEG |
| TJP1 | Tight Junction Protein 1 | Zonula occludens 1, ZO-1, TJP1 |
| VCL | Vinculin | Vinculin, VCL |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batissoco, A.C.; Salazar-Silva, R.; Oiticica, J.; Bento, R.F.; Mingroni-Netto, R.C.; Haddad, L.A. A Cell Junctional Protein Network Associated with Connexin-26. Int. J. Mol. Sci. 2018, 19, 2535. https://doi.org/10.3390/ijms19092535
Batissoco AC, Salazar-Silva R, Oiticica J, Bento RF, Mingroni-Netto RC, Haddad LA. A Cell Junctional Protein Network Associated with Connexin-26. International Journal of Molecular Sciences. 2018; 19(9):2535. https://doi.org/10.3390/ijms19092535
Chicago/Turabian StyleBatissoco, Ana C., Rodrigo Salazar-Silva, Jeanne Oiticica, Ricardo F. Bento, Regina C. Mingroni-Netto, and Luciana A. Haddad. 2018. "A Cell Junctional Protein Network Associated with Connexin-26" International Journal of Molecular Sciences 19, no. 9: 2535. https://doi.org/10.3390/ijms19092535