Opposite Polarity Monospore Genome De Novo Sequencing and Comparative Analysis Reveal the Possible Heterothallic Life Cycle of Morchella importuna
Abstract
:1. Introduction
2. Results
2.1. Sequencing Strategy and Data Statistics
2.2. De Novo Genomic Assembly
2.3. Protein Prediction and Functional Annotation
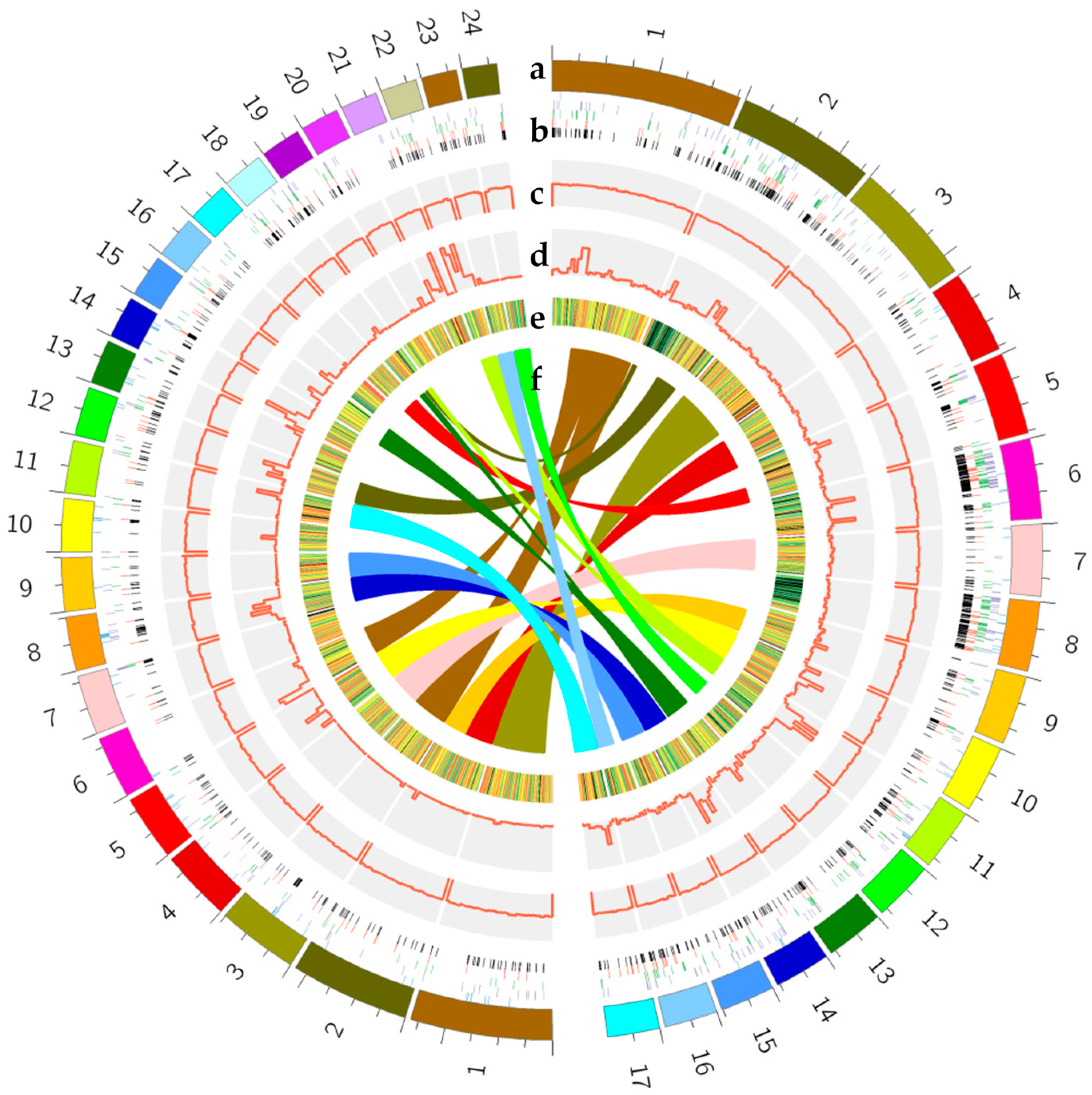
2.4. Genomic Structure and Characteristic Analysis
2.5. Genome Comparative Analysis of the Two Opposite-Polarity Monospores of M. Importuna
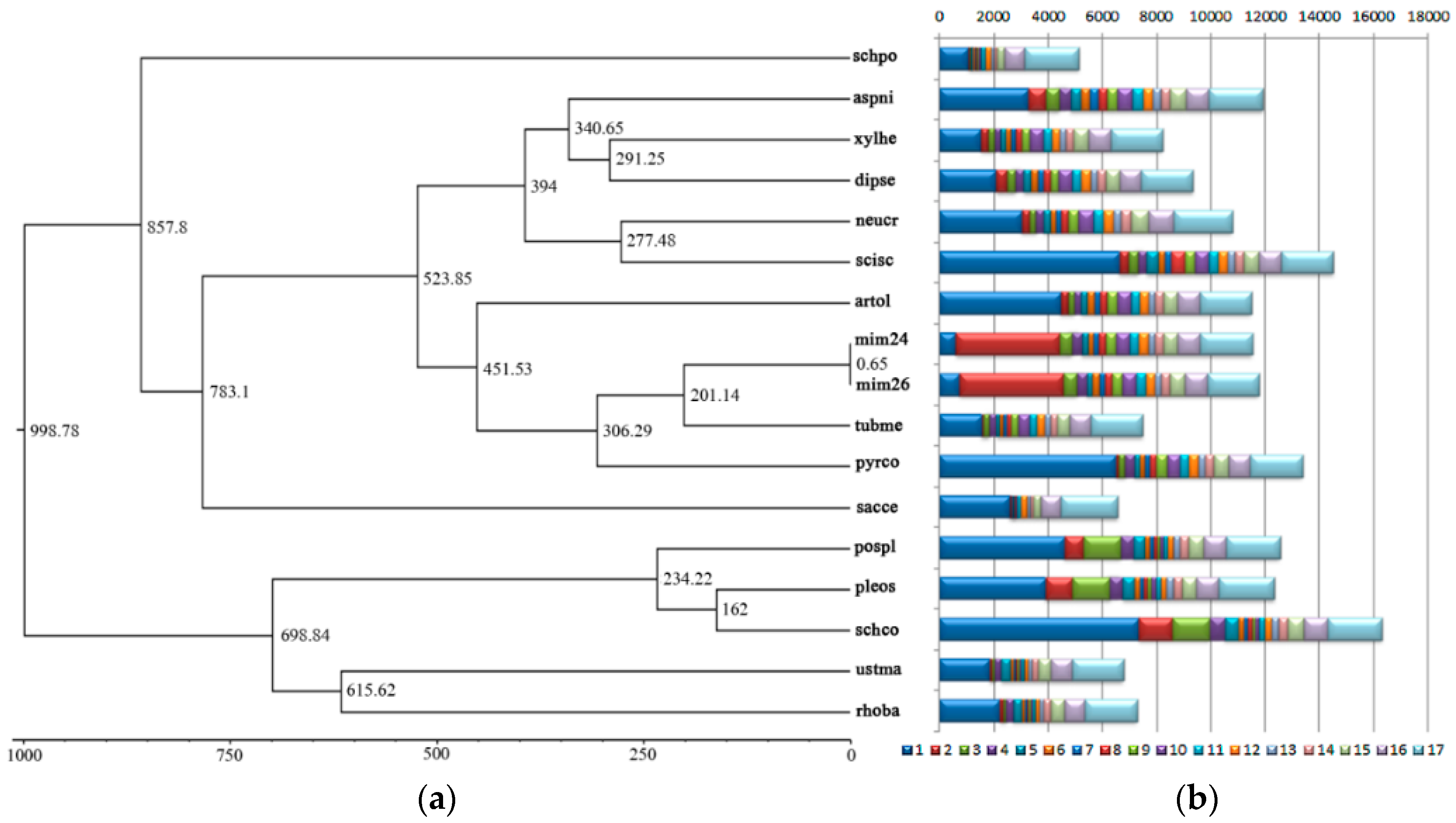
2.6. Phylogenetic Tree Construction and Evolutionary Divergence Time Analysis
3. Discussion
3.1. De Novo Genomic Sequencing Assembly and Annotation
3.2. Mating-Type Locus of the Two Opposite-Polarity Monospore Strains of M. Importuna
3.3. C.omparative Genome Analysis
3.4. The Two Monospore-Specific Genes Reveal the Possible Heterothallic Characteristics of M. Importuna
4. Materials and Methods
4.1. Strain Selection and Material Preparation
4.2. Genome and Transcriptome Sequencing
4.3. Genome Assembly and Annotation
4.4. Phylogenetic Tree Construction and Evolutionary Divergence Time Analysis
4.5. Comparative Analysis of the Two Mating-Type Monospore Genomes
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Traeger, S.; Altegoer, F.; Freitag, M.; Gabaldon, T.; Kempken, F.; Kumar, A.; Marcet-Houben, M.; Pöggeler, S.; Stajich, J.E.; Nowrousian, M. The Genome and Development-Dependent Transcriptomes of Pyronema confluens: A Window into Fungal Evolution. PLOS Genet. 2013, 9, e1003820. [Google Scholar] [CrossRef] [PubMed]
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M.; Cannon, P.F.; Eriksson, O.E.; Huhndorf, S.; James, T.; Kirk, P.M.; Lücking, R.; et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007, 111, 509–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Ya, Z.; Peixin, H. Morel Biology and Cultivation; Jilin Science and Technology Press: Jilin, China, 2017. [Google Scholar]
- He, P.X.; Wang, K.; Cai, Y.L.; Liu, W. Live cell confocal laser imaging studies on the nuclear behavior during meiosis and ascosporogenesis in Morchella importuna under artificial cultivation. Micron 2017, 101, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Ower, R. Notes on the Development of the Morel Ascocarp: Morchella esculenta. Mycologia 1982, 74, 142–144. [Google Scholar] [CrossRef]
- Pilz, D.; McLain, R.; Alexander, S.; Villarreal-Ruiz, L.; Berch, S.; Wurtz, T.L.; Smith, J.E. Ecology and Management of Morels Harvested from the Forests of Western North America; DIANE Publishing: Collingdale, PA, USA, 2008. [Google Scholar]
- Masaphy, S. Biotechnology of morel mushrooms: Successful fruiting body formation and development in a soilless system. Biotechnol. Lett. 2010, 32, 1523–1527. [Google Scholar] [CrossRef] [PubMed]
- Ower, R.D.; Mills, G.L.; Malachowski, J.A. Cultivation of Morchella. U.S. Patent 4,866,878, 19 September 1989. [Google Scholar]
- Ower, R.D.; Mills, G.L.; Malachowski, J.A. Cultivation of Morchella. U.S. Patent 4,757,640, 19 July 1988. [Google Scholar]
- Ower, R.D.; Mills, G.L.; Malachowski, J.A. Cultivation of Morchella. U.S. Patent 4,594,809, 17 June 1986. [Google Scholar]
- Masaphy, S. External ultrastructure of fruit body initiation in Morchella. Mycol. Res. 2005, 109, 508–512. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Wang, K.; Cai, Y.; Hu, X.; Zheng, Y.; Zhang, J.; Liu, W. Involvement of autophagy and apoptosis and lipid accumulation in sclerotial morphogenesis of Morchella importuna. Micron 2018, 109, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Yingli, C.; Ya, Z. The resolution of key technologies for rapid development of Morchella cultivation in China. Edible Med. Mushrooms 2018, 26, 142–147. [Google Scholar]
- Volk, T.J.; Leonard, T.J. Cytology of the life-cycle of Morchella. Mycol. Res. 1990, 94, 399–406. [Google Scholar] [CrossRef]
- Buscot, F.; Roux, J. Association between living roots and ascocarps of Morchella rotunda. Trans. Br. Mycol. Soc. 1987, 89, 249–252. [Google Scholar] [CrossRef]
- Solak, M.H.; Ersel, F.Y.; Isiloglu, M. Five new records of Morchella species for Turkey. Mikol. Fitopatol. 2004, 38, 60–66. [Google Scholar]
- Bunyard, B.A.; Nicholson, M.S.; Royse, D.J. A Systematic Assessment of Morchella Using RFLP Analysis of the 28S ribosomal RNA gene. Mycologia 1994, 86, 762–772. [Google Scholar] [CrossRef]
- Winder, R.S. Cultural studies of Morchella elata. Mycol. Res. 2006, 110, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Amir, R.; Levanon, D.; Hadar, Y.; Chet, I. Morphology and physiology of Morchella esculenta during sclerotial formation. Mycol. Res. 1993, 97, 683–689. [Google Scholar] [CrossRef]
- Amir, R.; Steudle, E.; Levanon, D.; Hadar, Y.; Chet, I. Turgor Changes in Morchella esculenta during Translocation and Sclerotial Formation. Exp. Mycol. 1995, 19, 129–136. [Google Scholar] [CrossRef]
- Amir, R.; Levanon, D.; Hadar, Y.; Chet, I. Formation of sclerotia by Morchella esculenta: Relationship between media composition and turgor potential in the mycelium. Mycol. Res. 1992, 96, 943–948. [Google Scholar] [CrossRef]
- Turkoglu, A.; Kivrak, I.; Mercan, N.; Duru, M.E.; Gezer, K.; Turkoglu, H. Antioxidant and antimicrobial activities of Morchella conica Pers. Afr. J. Biotechnol. 2006, 5, 1146–1150. [Google Scholar]
- Rotzoll, N.; Dunkel, A.; Hofmann, T. Quantitative studies, taste reconstitution, and omission experiments on the key taste compounds in Morel mushrooms (Morchella deliciosa fr.). J. Agric. Food Chem. 2006, 54, 2705–2711. [Google Scholar] [CrossRef] [PubMed]
- Elmastas, M.; Turkekul, I.; Ozturk, L.; Gulcin, I.; Isildak, O.; Aboul-Enein, H.Y. Antioxidant activity of two wild edible mushrooms (Morchella vulgaris and Morchella esculanta) from North Turkey. Comb. Chem. High Throughput Screen. 2006, 9, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Dalgleish, H.J.; Jacobson, K.M. A first assessment of genetic variation among Morchella esculenta (Morel) populations. J. Hered. 2005, 96, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Du, X.H.; Zhao, Q.; Xia, E.H.; Gao, L.Z.; Richard, F.; Yang, Z.L. Mixed-reproductive strategies, competitive mating-type distribution and life cycle of fourteen black morel species. Sci. Rep. 2017, 7, 1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, H.M.; Chen, L.J.; Chen, W.M.; Zhao, Q.; Zhang, X.L.; Su, K.M.; Zhao, Y.C. Characterization of mating-type idiomorphs suggests that Morchella importuna, Mel-20 and = M.sextelata are heterothallic. Mycol. Prog. 2017, 16, 743–752. [Google Scholar] [CrossRef]
- Loizides, M.; Alvarado, P.; Clowez, P.; Moreau, P.A.; de la Osa, L.R.; Palazon, A. Morchella tridentina, M. rufobrunnea, and M. kakiicolor: A study of three poorly known Mediterranean morels, with nomenclatural updates in section Distantes. Mycol. Prog. 2015, 14, 13. [Google Scholar]
- Taskin, H.; Buyukalaca, S.; Hansen, K.; O’Donnell, K. Multilocus phylogenetic analysis of true morels (Morchella) reveals high levels of endemics in Turkey relative to other regions of Europe. Mycologia 2012, 104, 446–461. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.; Dewsbury, D.R.; ODonnell, K.; Carter, M.C.; Rehner, S.A.; Moore, J.D.; Moncalvo, J.M.; Canfield, S.A.; Stephenson, S.L.; Methven, A.S.; et al. Taxonomic revision of true morels (Morchella) in Canada and the United States. Mycologia 2012, 104, 1159–1177. [Google Scholar] [CrossRef] [PubMed]
- Du, X.H.; Zhao, Q.; O’Donnell, K.; Rooney, A.P.; Yang, Z.L. Multigene molecular phylogenetics reveals true morels (Morchella) are especially species-rich in China. Fungal Genet. Biol. 2012, 49, 455–469. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Rooney, A.P.; Mills, G.L.; Kuo, M.; Weber, N.S.; Rehner, S.A. Phylogeny and historical biogeography of true morels (Morchella) reveals an early Cretaceous origin and high continental endemism and provincialism in the Holarctic. Fungal Genet. Biol. 2011, 48, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Taskin, H.; Buyukalaca, S.; Dogan, H.H.; Rehner, S.A.; O’Donnell, K. A multigene molecular phylogenetic assessment of true morels (Morchella) in Turkey. Fungal Genet. Biol. 2010, 47, 672–682. [Google Scholar] [CrossRef] [PubMed]
- He, P.X.; Cai, Y.L.; Liu, S.M.; Han, L.; Huang, L.N.; Liu, W. Morphological and ultrastructural examination of senescence in Morchella Elata. Micron 2015, 78, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Kohler, A.; Murat, C.; Balestrini, R.; Coutinho, P.M.; Jaillon, O.; Montanini, B.; Morin, E.; Noel, B.; Percudani, R.; et al. Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 2010, 464, 1033. [Google Scholar] [CrossRef] [PubMed]
- Derbyshire, M.; Denton-Giles, M.; Hegedus, D.; Seifbarghy, S.; Rollins, J.; van Kan, J.; Seidl, M.F.; Faino, L.; Mbengue, M.; Navaud, O.; et al. The Complete Genome Sequence of the Phytopathogenic Fungus Sclerotinia sclerotiorum Reveals Insights into the Genome Architecture of Broad Host Range Pathogens. Genome Boil. Evol. 2017, 9, 593–618. [Google Scholar] [CrossRef] [PubMed]
- Trappe, M.J.; Trappe, J.M.; Bonito, G.M. Kalapuya brunnea gen. & sp. nov. and its relationship to the other sequestrate genera in Morchellaceae. Mycologia 2010, 102, 1058–1065. [Google Scholar] [PubMed]
- Hoff, K.J.; Lange, S.; Lomsadze, A.; Borodovsky, M.; Stanke, M. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 2016, 32, 767–769. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.-I.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157. [Google Scholar] [CrossRef] [PubMed]
- Nowrousian, M.; Stajich, J.E.; Chu, M.; Engh, I.; Espagne, E.; Halliday, K.; Kamerewerd, J.; Kempken, F.; Knab, B.; Kuo, H.-C.; et al. De novo Assembly of a 40 Mb Eukaryotic Genome from Short Sequence Reads: Sordaria macrospora, a Model Organism for Fungal Morphogenesis. PLOS Genet. 2010, 6, e1000891. [Google Scholar] [CrossRef] [PubMed]
- Chibucos, M.C.; Crabtree, J.; Nagaraj, S.; Chaturvedi, S.; Chaturvedi, V. Draft Genome Sequences of Human Pathogenic Fungus Geomyces pannorum Sensu Lato and Bat White Nose Syndrome Pathogen Geomyces(Pseudogymnoascus) destructans. Genome Announc. 2013, 1, e01045-13. [Google Scholar] [CrossRef] [PubMed]
- Shelest, E. Transcription factors in fungi. FEMS Microbiol. Lett. 2008, 286, 145–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daboussi, M.-J.; Capy, P. Transposable Elements in Filamentous Fungi. Annu. Rev. Microbiol. 2003, 57, 275–299. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Waalwijk, C.; de Wit, P.J.G.M.; Tang, D.; van der Lee, T. RNA-Seq analysis reveals new gene models and alternative splicing in the fungal pathogen Fusarium Graminearum. BMC Genome 2013, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 2.3.1–2.3.22. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol. Boil. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farman, M.L. Telomeres in the rice blast fungus Magnaporthe oryzae: The world of the end as we know it. FEMS Microbiol. Lett. 2007, 273, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Bakkeren, G.; Kämper, J.; Schirawski, J. Sex in smut fungi: Structure, function and evolution of mating-type complexes. Fungal Genet. Biol. 2008, 45, S15–S21. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Kubatova, A.; Novakova, A.; Minnis, A.M.; Kolarik, M.; Lindner, D.L. Molecular Characterization of a Heterothallic Mating System in Pseudogymnoascus destructans, the Fungus Causing White-Nose Syndrome of Bats. G3 Genes Genomes Genet. 2014, g3-114. [Google Scholar] [CrossRef] [PubMed]
- Gazis, R.; Kuo, A.; Riley, R.; LaButti, K.; Lipzen, A.; Lin, J.; Amirebrahimi, M.; Hesse, C.N.; Spatafora, J.W.; Henrissat, B.; et al. The genome of Xylona heveae provides a window into fungal endophytism. Fungal Biol. 2016, 120, 26–42. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.F.; Stielow, J.B.; de Vries, M.; Weiss, V.A.; Vicente, V.A.; de Hoog, S. Draft Genome Sequence of the Ant-Associated Fungus Phialophora attae (CBS 131958). Genome Announc. 2015, 3, e01099-15. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.H.; Moore, G.G.; Horn, B.W.; Carbone, I. Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasit. Fungal Genet. Biol. 2008, 45, 1292–1299. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Xiao, G.; Zheng, P.; Cen, K.; Zhan, S.; Wang, C. Divergent and Convergent Evolution of Fungal Pathogenicity. Genome Boil. Evol. 2016, 8, 1374–1387. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, L.; Ji, X.; Feng, Y.; Li, X.; Zou, C.; Xu, J.; Ren, Y.; Mi, Q.; Wu, J.; et al. Genomic and Proteomic Analyses of the Fungus Arthrobotrys oligospora Provide Insights into Nematode-Trap Formation. PLoS Pathog. 2011, 7, e1002179. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Limpens, E.; Zhang, Z.; Ivanov, S.; Saunders, D.G.O.; Mu, D.; Pang, E.; Cao, H.; Cha, H.; Lin, T.; et al. Single Nucleus Genome Sequencing Reveals High Similarity among Nuclei of an Endomycorrhizal Fungus. PLoS Genet. 2014, 10, e1004078. [Google Scholar] [CrossRef] [PubMed]
- Gehrmann, T.; Pelkmans, J.F.; Ohm, R.A.; Vos, A.M.; Sonnenberg, A.S.M.; Baars, J.J.P.; Wösten, H.A.B.; Reinders, M.J.T.; Abeel, T. Nucleus-specific expression in the multinuclear mushroom-forming fungus Agaricus bisporus reveals different nuclear regulatory programs. Proc. Natl. Acad. Sci. USA 2018, 115, 4429–4434. [Google Scholar] [CrossRef] [PubMed]
- Belfiori, B.; Riccioni, C.; Paolocci, F.; Rubini, A. Characterization of the reproductive mode and life cycle of the whitish truffle T. borchii. Mycorrhiza 2016, 26, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.; MacCallum, I.; Kleber, M.; Shlyakhter, I.A.; Belmonte, M.K.; Lander, E.S.; Nusbaum, C.; Jaffe, D.B. ALLPATHS: De novo assembly of whole-genome shotgun microreads. Genome Res. 2008, 18, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Hill, C.M.; Wu, S.; Ruan, J.; Ma, Z. DBG2OLC: Efficient Assembly of Large Genomes Using Long Erroneous Reads of the Third Generation Sequencing Technologies. Sci. Rep. 2016, 6, 31900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, M.; Baldwin-Brown, J.G.; Long, A.D.; Emerson, J.J. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 2016, 44, e147. [Google Scholar] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhu, H.; Ruan, J.; Qian, W.; Fang, X.; Shi, Z.; Li, Y.; Li, S.; Shan, G.; Kristiansen, K.; et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2009, 20, 265–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, K.-K.; LaButti, K.; Khalak, A.; Tse, D. FinisherSC: A repeat-aware tool for upgrading de novo assembly using long reads. Bioinformatics 2015, 31, 3207–3209. [Google Scholar] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Nansheng, C. Using RepeatMasker to Identify Repetitive Elements in Genomic Sequences. Curr. Protoc. Bioinform. 2004, 5, 4.10.1–4.10.14. [Google Scholar]
- RepeatMasker Open-3.0. Available online: http://www.repeatmasker.org (accessed on 25 August 2018).
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Boil. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [PubMed]
- Sanderson, M.J. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 2003, 19, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.W.; Berbee, M.L. Dating divergences in the Fungal Tree of Life: Review and new analyses. Mycologia 2006, 98, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Douzery, E.J.P.; Snell, E.A.; Bapteste, E.; Delsuc, F.; Philippe, H. The timing of eukaryotic evolution: Does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl. Acad. Sci. USA 2004, 101, 15386–15391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Phillippy, A.; Carlton, J.; Salzberg, S.L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002, 30, 2478–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]


| Description | M04M26 | M04M24 |
|---|---|---|
| Genome scaffold number (n) | 106 | 394 |
| Genome contig number (n) | 111 | 763 |
| Longest length (bp) | 3,559,784 | 2,650,670 |
| Shortest length (bp) | 13,107 | 990 |
| Genome scaffold size (bp) | 51,078,152 | 48,978,911 |
| Genome contig size (bp) | 51,061,582 | 48,728,179 |
| Rate of N | 0.000324405 | 0.005119183 |
| Rate of GC | 0.473300338 | 0.472875746 |
| Scaffold N50 (bp) | 978,718 | 653,797 |
| Contig N50 (bp) | 951,626 | 198,406 |
| Scaffold N90 (bp) | 265,726 | 167,990 |
| Contig N90 (bp) | 261,029 | 47,755 |
| Genome completeness (%) | 94.31 | 90.45 |
| Number of sequences ≥ 1 kb (n) | 106 | 392 |
| Number of sequences ≥ 2 kb (n) | 106 | 252 |
| Number of sequences ≥ 3 kb (n) | 106 | 215 |
| Database | M04M24 | Percentage (%) | M04M26 | Percentage (%) |
|---|---|---|---|---|
| Nr | 8357 | 72.55 | 8517 | 72.36 |
| InterPro | 10364 | 89.97 | 10,577 | 89.86 |
| KOG | 5990 | 52.00 | 6000 | 50.98 |
| COG | 5795 | 50.31 | 5816 | 49.41 |
| SwissProt | 6292 | 54.62 | 6300 | 53.53 |
| GO | 6173 | 53.59 | 6219 | 52.84 |
| KAAS | 3459 | 30.03 | 3436 | 29.19 |
| CDD | 7162 | 62.18 | 7154 | 60.78 |
| Total | 10,497 | 91.13 | 10,947 | 93.01 |
| Whole genome | 11,519 | 100.00 | 11,770 | 100.00 |
| M04M24 | ||||||
|---|---|---|---|---|---|---|
| ID | Pop Total | Pop Term | Study Total | Study Term | p | Name |
| GO:0044421 | 6171 | 54 | 104 | 41 | 1.37 × 10−66 | Extracellular region part |
| GO:0043230 | 6171 | 46 | 104 | 36 | 5.28 × 10−54 | Extracellular organelle |
| GO:0005576 | 6171 | 133 | 104 | 41 | 2.48 × 10−44 | Extracellular region |
| GO:0030054 | 6171 | 29 | 104 | 24 | 2.52 × 10−39 | Cell junction |
| GO:0032501 | 6171 | 57 | 104 | 29 | 4.64 × 10−38 | Multicellular organismal process |
| GO:0031982 | 6171 | 130 | 104 | 38 | 2.59 × 10−35 | Vesicle |
| GO:0099080 | 6171 | 54 | 104 | 23 | 1.38 × 10−27 | Supramolecular complex |
| GO:0032502 | 6171 | 151 | 104 | 30 | 5.10 × 10−25 | Developmental process |
| GO:0005886 | 6171 | 135 | 104 | 27 | 3.68 × 10−22 | Plasma membrane |
| GO:1903561 | 6171 | 46 | 104 | 36 | 1.44 × 10−20 | Extracellular vesicle |
| GO:0071944 | 6171 | 211 | 104 | 31 | 1.56 × 10−20 | Cell periphery |
| GO:0022610 | 6171 | 32 | 104 | 15 | 5.82 × 10−19 | Biological adhesion |
| GO:0008219 | 6171 | 32 | 104 | 15 | 6.81 × 10−18 | Cell death |
| GO:0034330 | 6171 | 15 | 104 | 13 | 2.97 × 10−16 | Cell junction organization |
| GO:0009888 | 6171 | 24 | 104 | 22 | 6.76 × 10−16 | Tissue development |
| GO:0009611 | 6171 | 16 | 104 | 14 | 7.52 × 10−16 | Response to wounding |
| GO:0048869 | 6171 | 125 | 104 | 22 | 1.44 × 10−15 | Cellular developmental process |
| GO:0032879 | 6171 | 60 | 104 | 17 | 4.61 × 10−15 | Regulation of localization |
| GO:0040011 | 6171 | 11 | 104 | 9 | 5.78 × 10−15 | Locomotion |
| GO:0048471 | 6171 | 13 | 104 | 10 | 4.70 × 10−14 | Perinuclear region of cytoplasm |
| M04M26 | ||||||
| ID | Pop Total | Pop Term | Study Total | Study Term | p | Name |
| GO:0003676 | 6216 | 981 | 167 | 69 | 2.08 × 10−8 | Nucleic-acid binding |
| GO:1901363 | 6216 | 1829 | 167 | 84 | 5.76 × 10−7 | Heterocyclic-compound binding |
| GO:0097159 | 6216 | 1830 | 167 | 84 | 5.94 × 10−7 | Organic-cyclic-compound binding |
| GO:0015074 | 6216 | 39 | 167 | 7 | 1.26 × 10−4 | DNA integration |
| GO:0005488 | 6216 | 3400 | 167 | 110 | 0.00130 | Binding |
| GO:0006259 | 6216 | 282 | 167 | 11 | 0.00661 | DNA metabolic process |
| GO:0019843 | 6216 | 26 | 167 | 2 | 0.00770 | Ribosomal RNA (rRNA) binding |
| GO:0010257 | 6216 | 1 | 167 | 1 | 0.00913 | NADH dehydrogenase complex assembly |
| GO:0000256 | 6216 | 3 | 167 | 1 | 0.00977 | Allantoin catabolic process |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Chen, L.; Cai, Y.; Zhang, Q.; Bian, Y. Opposite Polarity Monospore Genome De Novo Sequencing and Comparative Analysis Reveal the Possible Heterothallic Life Cycle of Morchella importuna. Int. J. Mol. Sci. 2018, 19, 2525. https://doi.org/10.3390/ijms19092525
Liu W, Chen L, Cai Y, Zhang Q, Bian Y. Opposite Polarity Monospore Genome De Novo Sequencing and Comparative Analysis Reveal the Possible Heterothallic Life Cycle of Morchella importuna. International Journal of Molecular Sciences. 2018; 19(9):2525. https://doi.org/10.3390/ijms19092525
Chicago/Turabian StyleLiu, Wei, LianFu Chen, YingLi Cai, QianQian Zhang, and YinBing Bian. 2018. "Opposite Polarity Monospore Genome De Novo Sequencing and Comparative Analysis Reveal the Possible Heterothallic Life Cycle of Morchella importuna" International Journal of Molecular Sciences 19, no. 9: 2525. https://doi.org/10.3390/ijms19092525







