Identification of an Essential Cytoplasmic Region of Interleukin-7 Receptor α Subunit in B-Cell Development
Abstract
:1. Introduction
2. Results
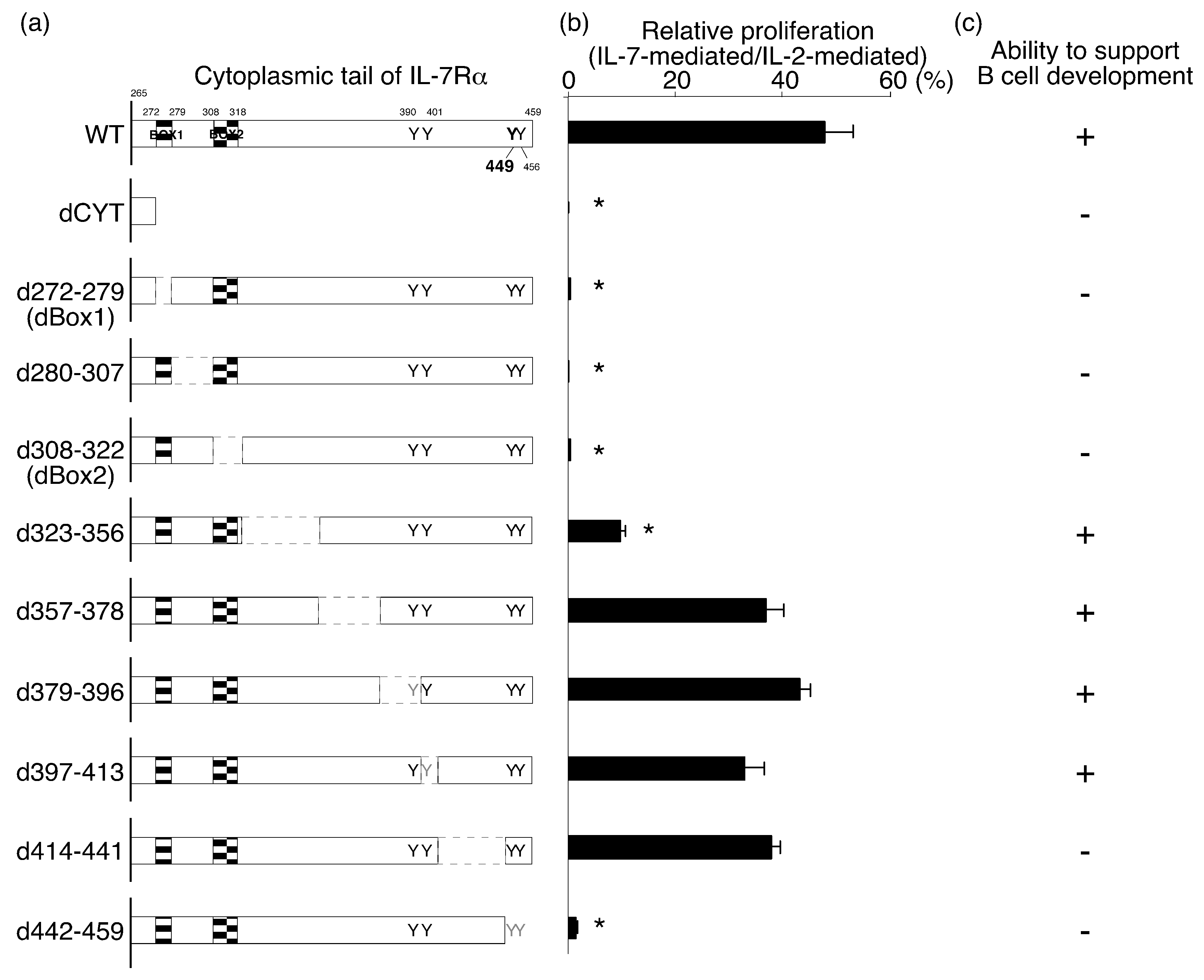
2.1. Identification of the IL-7Rα Cytoplasmic Regions that Are Necessary for IL-7-Mediated Cell Proliferation
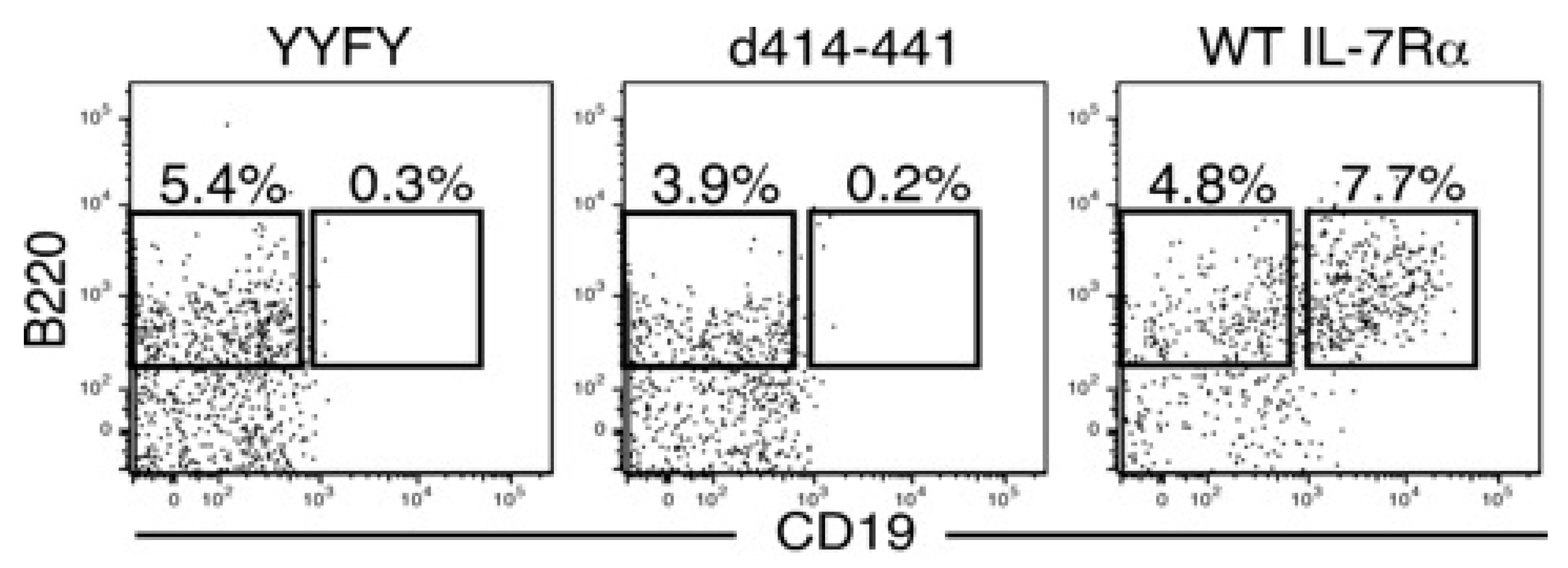
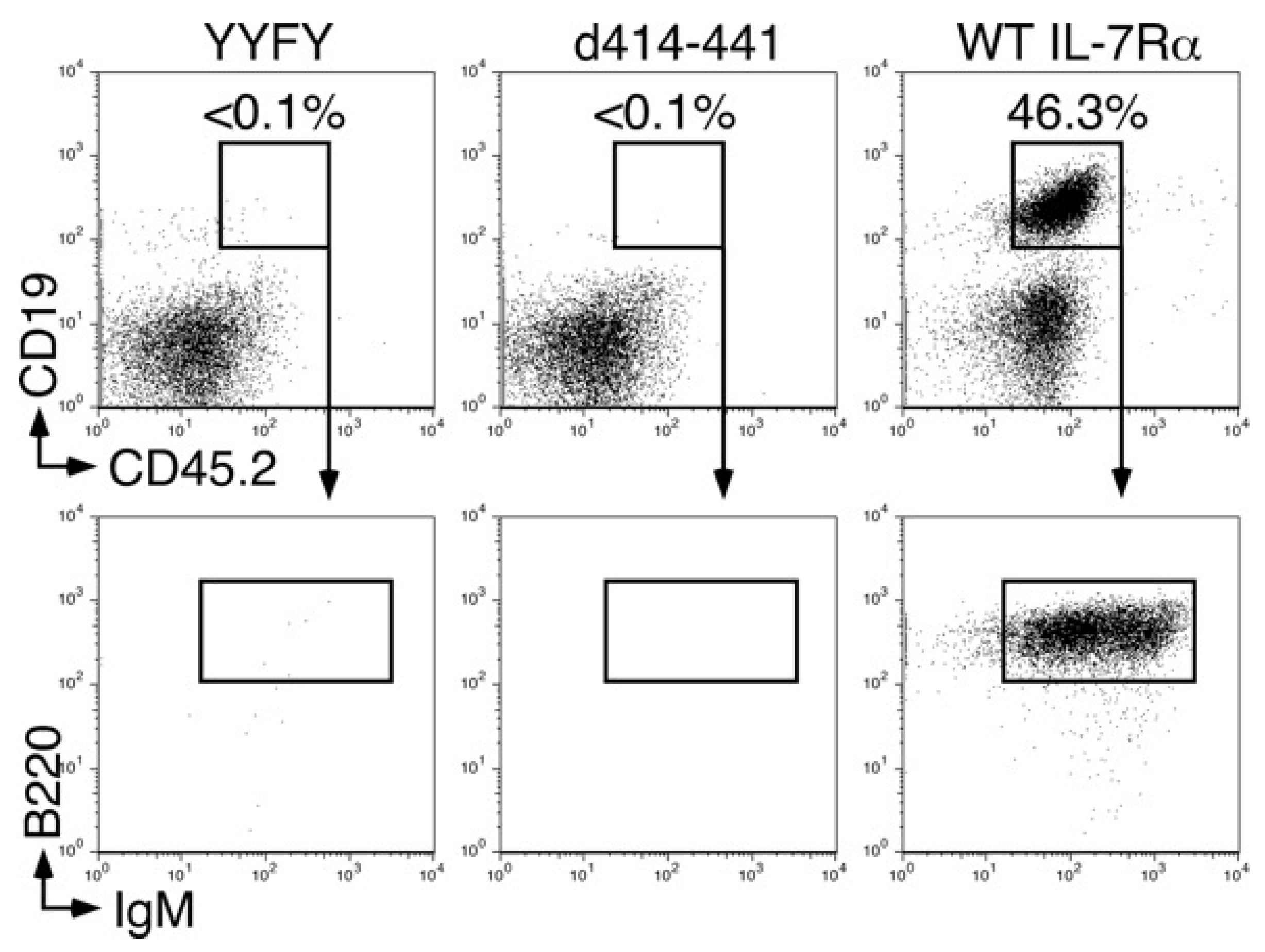
2.2. The Amino Acids Region from Positions 414 to 441 of the IL-7Rα Subunit Was Necessary for B-Cell Development
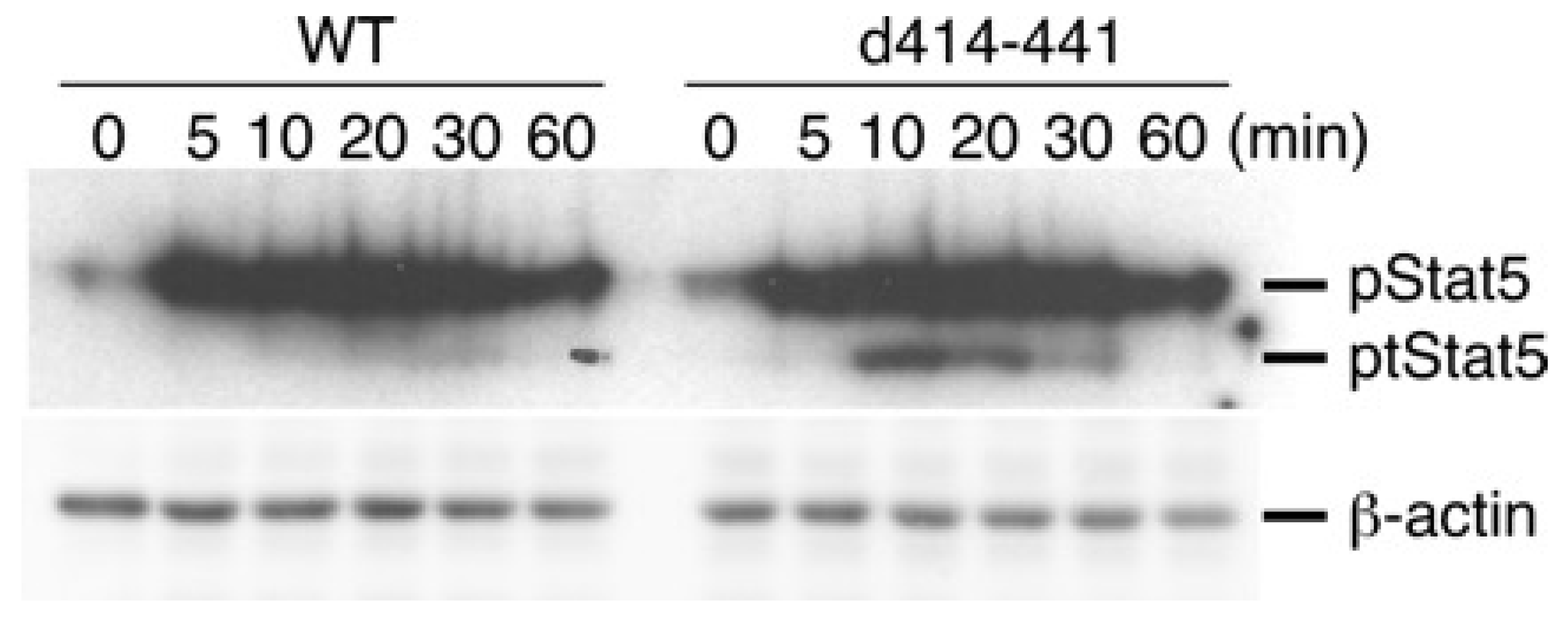
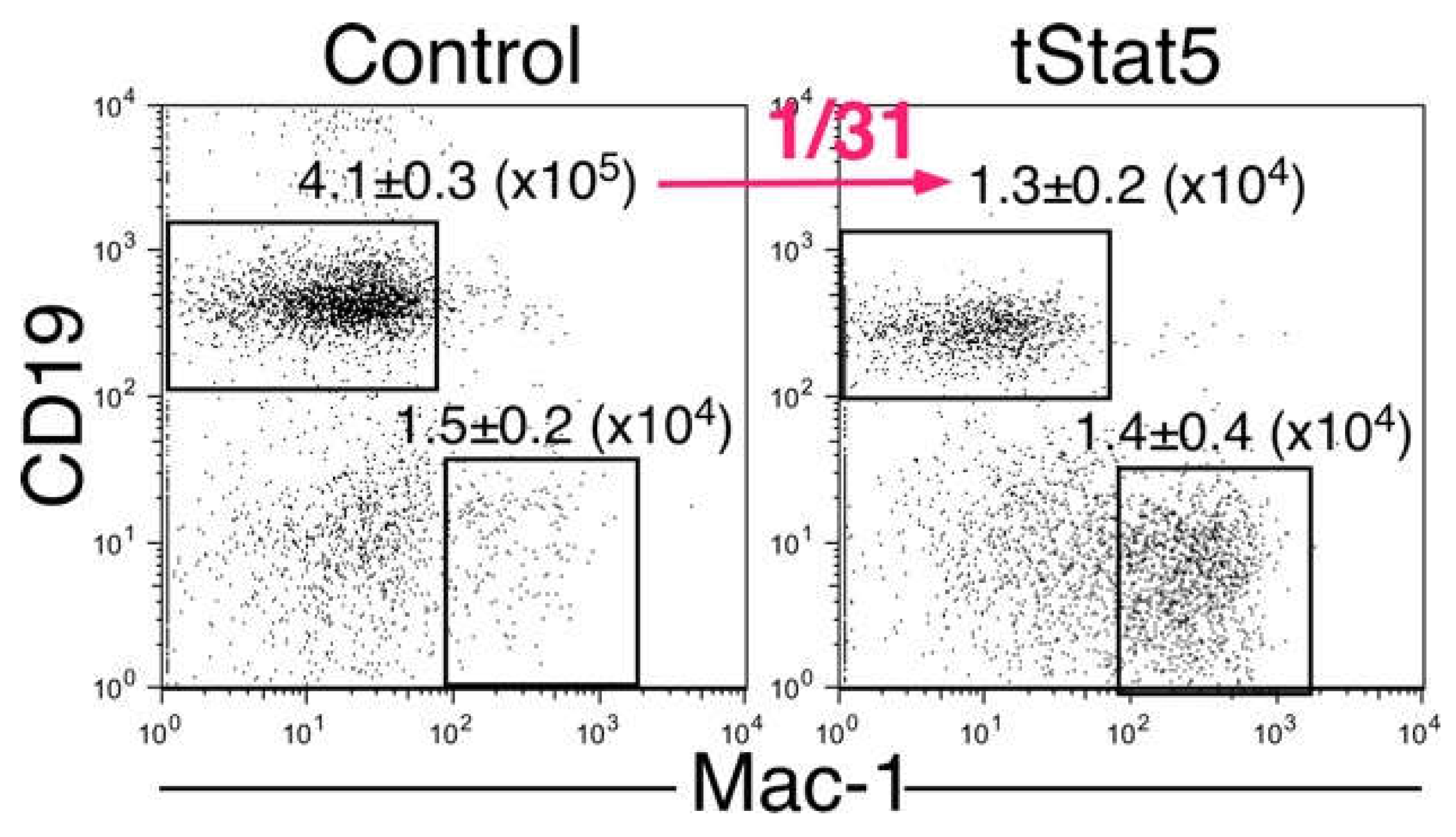
2.3. The Truncated Form of STAT5 (tSTAT5) Was Upregulated in CTLL-2 Cells with the d414–441 Mutant Compared with that in WT Cells after IL-7 Stimulation
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Construction of Mutant IL-7Rα Subunits and Retrovirus Production
4.3. Establishment of CTLL-2 Transfectants
4.4. Proliferation Assays
4.5. Cell Sorting and FACS Analysis
4.6. Immunoblotting
4.7. In Vitro and In Vivo Differentiation Assays
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CBP | CREB-binding protein |
| CD | Cluster of differentiation |
| EBF | Early B cell factor |
| ETP | Early T cell progenitor |
| γc | Common γ |
| HPC | Hematopoietic progenitor cell |
| Lck | Lymphocyte-specific protein tyrosine kinase |
| IL | Interleukin |
| JAK | Janus kinase |
| KLS | c-Kit+ Lineage− Sca-1+ |
| RAG | Recombination activating gene |
| Src | Rat sarcoma |
| SCF | Stem cell factor |
| STAT | Signal transducer and activator of transcription |
References
- Hofmeister, R.; Khaled, A.R.; Benbernou, N.; Rajnavolgyi, E.; Muegge, K.; Durum, S.K. Interleukin-7: Physiological roles and mechanisms of action. Cytokine Growth Factor Rev. 1999, 10, 41–60. [Google Scholar] [CrossRef]
- Ceredig, R.; Rolink, A.G. The key role of IL-7 in lymphopoiesis. Semin. Immunol. 2012, 24, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Sugamura, K.; Asao, H.; Kondo, M.; Tanaka, N.; Ishii, N.; Ohbo, K.; Nakamura, M.; Takeshita, T. The interleukin-2 receptor gamma chain: Its role in the multiple cytokine receptor complexes and T cell development in XSCID. Annu. Rev. Immunol. 1996, 14, 179–205. [Google Scholar] [CrossRef] [PubMed]
- Leonard, W.J. The molecular basis of X-linked severe combined immunodeficiency: Defective cytokine receptor signaling. Annu. Rev. Med. 1996, 47, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Rochman, Y.; Spolski, R.; Leonard, W.J. New insights into the regulation of T cells by γc family cytokines. Nat. Rev. Immunol. 2009, 9, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Corfe, S.A.; Paige, C.J. The many roles of IL-7 in B cell development; mediator of survival, proliferation and differentiation. Semin. Immunol. 2012, 24, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Puel, A.; Ziegler, S.F.; Buckley, R.H.; Leonard, W.J. Defective IL7R expression in T−B+NK+ severe combined immunodeficiency. Nat. Genet. 1998, 20, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Peschon, J.J.; Morrissey, P.J.; Grabstein, K.H.; Ramsdell, F.J.; Maraskovsky, E.; Gliniak, B.C.; Park, L.S.; Ziegler, S.F.; Williams, D.E.; Ware, C.B.; et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 1994, 180, 1955–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Freeden-Jeffry, U.; Vieira, P.; Lucian, L.A.; McNeil, T.; Burdach, S.E.; Murray, R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 1995, 181, 1519–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kittipatarin, C.; Khaled, A.R. Interlinking interleukin-7. Cytokine 2007, 39, 75–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, W.J. Role of Jak kinases and STATs in cytokine signal transduction. Int. J. Hematol. 2001, 73, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Benbernou, N.; Chertov, O.; Khaled, A.R.; Wooters, J.; Durum, S.K. IL-7 induces tyrosine phosphorylation of clathrin heavy chain. Cell Signal 2004, 16, 281–286. [Google Scholar] [CrossRef]
- Lin, J.X.; Migone, T.S.; Tsang, M.; Friedmann, M.; Weatherbee, J.A.; Zhou, L.; Yamauchi, A.; Bloom, E.T.; Mietz, J.; John, S.; et al. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity 1995, 2, 331–339. [Google Scholar] [CrossRef]
- Kikuchi, K.; Lai, A.Y.; Hsu, C.L.; Kondo, M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J. Exp. Med. 2005, 201, 1197–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanner, J.W.; Chen, W.; Young, R.L.; Longmore, G.D.; Shaw, A.S. The conserved box 1 motif of cytokine receptors is required for association with JAK kinases. J. Biol. Chem. 1995, 270, 6523–6530. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Li, W.Q.; Hofmeister, R.R.; Young, H.A.; Hodge, D.R.; Keller, J.R.; Khaled, A.R.; Durum, S.K. Distinct regions of the interleukin-7 receptor regulate different Bcl2 family members. Mol. Cell Biol. 2004, 24, 6501–6513. [Google Scholar] [CrossRef] [PubMed]
- Page, T.H.; Lali, F.V.; Foxwell, B.M. Interleukin-7 activates p56lck and p59fyn, two tyrosine kinases associated with the p90 interleukin-7 receptor in primary human T cells. Eur. J. Immunol. 1995, 25, 2956–2960. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Narazaki, M.; Hibi, M.; Yawata, H.; Yasukawa, K.; Hamaguchi, M.; Taga, T.; Kishimoto, T. Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc. Natl. Acad. Sci. USA 1991, 88, 11349–11353. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.H.; Berry, J.A.; Russell, S.M.; Leonard, W.J. Delineation of the regions of interleukin-2 (IL-2) receptor β chain important for association of Jak1 and Jak3. Jak1-independent functional recruitment of Jak3 to Il-2Rβ. J. Biol. Chem. 1998, 273, 10719–10725. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.J.; Mahajan, V.S.; Trajman, L.C.; Irvine, D.J.; Lauffenburger, D.A.; Chen, J. Interleukin-7 receptor signaling network: An integrated systems perspective. Cell Mol. Immunol. 2008, 5, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, T.; Kasai, H.; Kondo, M. Acetylation Modulates IL-2 Receptor Signaling in T. Cells. J. Immunol. 2016, 197, 4334–4343. [Google Scholar] [CrossRef] [PubMed]
- Leonard, W.J.; O’Shea, J.J. Jaks and STATs: Biological implications. Annu. Rev. Immunol. 1998, 16, 293–322. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Gao, J.S.; Guan, Y.J.; McLane, K.E.; Yuan, Z.L.; Ramratnam, B.; Chin, Y.E. Acetylation-dependent signal transduction for type I interferon receptor. Cell 2007, 131, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Hirano, T. Molecular basis of the cell specificity of cytokine action. Biochim. Biophys. Acta 2002, 1592, 281–296. [Google Scholar] [CrossRef]
- Minami, Y.; Taniguchi, T. IL-2 signaling: Recruitment and activation of multiple protein tyrosine kinases by the components of the IL-2 receptor. Curr. Opin. Cell Biol. 1995, 7, 156–162. [Google Scholar] [CrossRef]
- Okuda, K.; Foster, R.; Griffin, J.D. Signaling domains of the βc chain of the GM-CSF/IL-3/IL-5 receptor. Ann. N. Y. Acad. Sci. 1999, 872, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Grosschedl, R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 1995, 376, 263–267. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, K.; Kasai, H.; Watanabe, A.; Lai, A.Y.; Kondo, M. IL-7 specifies B cell fate at the common lymphoid progenitor to pre-proB transition stage by maintaining early B cell factor expression. J. Immunol. 2008, 181, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, E.V. Transcriptional control of early T and B cell developmental choices. Annu. Rev. Immunol. 2014, 32, 283–321. [Google Scholar] [CrossRef] [PubMed]
- Kondo, M.; Scherer, D.C.; Miyamoto, T.; King, A.G.; Akashi, K.; Sugamura, K.; Weissman, I.L. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature 2000, 407, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.L.; King-Fleischman, A.G.; Lai, A.Y.; Matsumoto, Y.; Weissman, I.L.; Kondo, M. Antagonistic effect of CCAAT enhancer-binding protein-α and Pax5 in myeloid or lymphoid lineage choice in common lymphoid progenitors. Proc. Natl. Acad. Sci. USA 2006, 103, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.L.; Kikuchi, K.; Kondo, M. Activation of mitogen-activated protein kinase kinase (MEK)/extracellular signal regulated kinase (ERK) signaling pathway is involved in myeloid lineage commitment. Blood 2007, 110, 1420–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adolfsson, J.; Mansson, R.; Buza-Vidas, N.; Hultquist, A.; Liuba, K.; Jensen, C.T.; Bryder, D.; Yang, L.; Borge, O.J.; Thoren, L.A.; et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 2005, 121, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.L.; Weissman, I.L. Flk-2 is a marker in hematopoietic stem cell differentiation: A simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA 2001, 98, 14541–14546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
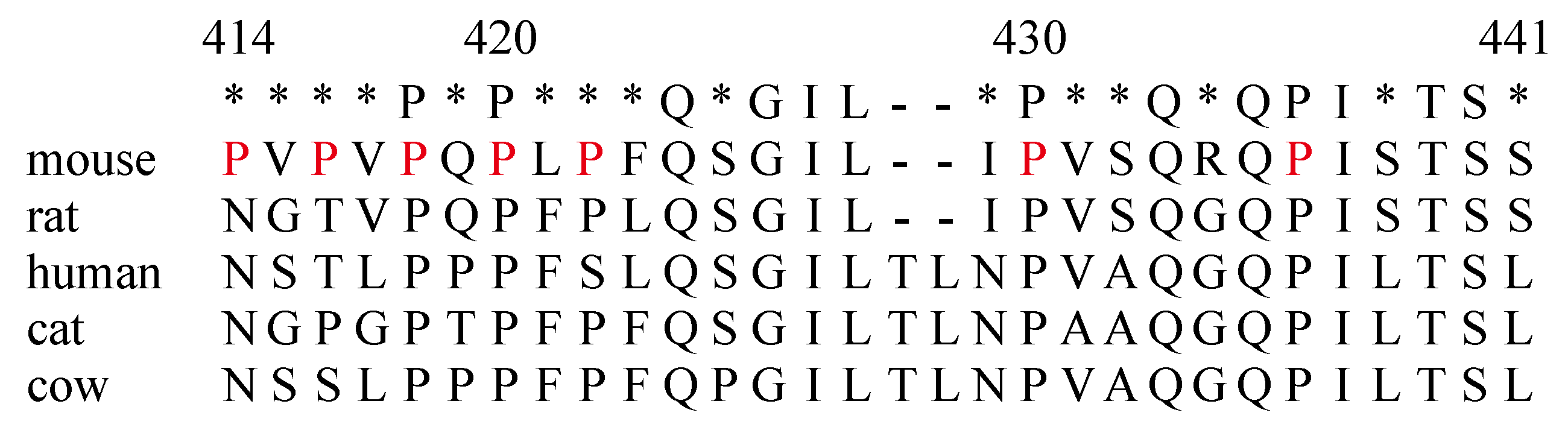
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasai, H.; Kuwabara, T.; Matsui, Y.; Nakajima, K.; Kondo, M. Identification of an Essential Cytoplasmic Region of Interleukin-7 Receptor α Subunit in B-Cell Development. Int. J. Mol. Sci. 2018, 19, 2522. https://doi.org/10.3390/ijms19092522
Kasai H, Kuwabara T, Matsui Y, Nakajima K, Kondo M. Identification of an Essential Cytoplasmic Region of Interleukin-7 Receptor α Subunit in B-Cell Development. International Journal of Molecular Sciences. 2018; 19(9):2522. https://doi.org/10.3390/ijms19092522
Chicago/Turabian StyleKasai, Hirotake, Taku Kuwabara, Yukihide Matsui, Koichi Nakajima, and Motonari Kondo. 2018. "Identification of an Essential Cytoplasmic Region of Interleukin-7 Receptor α Subunit in B-Cell Development" International Journal of Molecular Sciences 19, no. 9: 2522. https://doi.org/10.3390/ijms19092522




