Genetic and Physio-Biochemical Characterization of a Novel Premature Senescence Leaf Mutant in Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Results
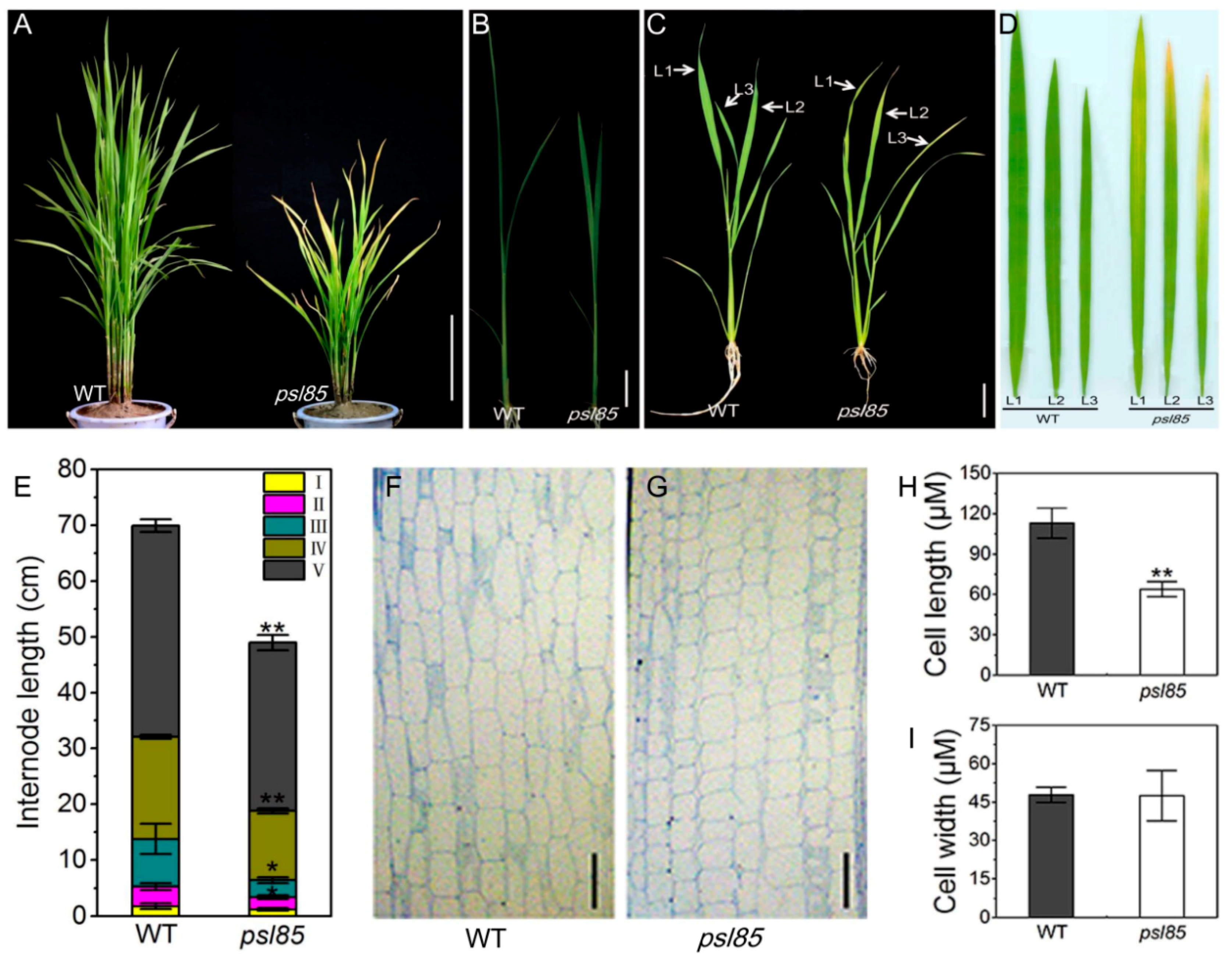
2.1. Phenotypic Performance of psl85
2.2. Alterations of Chlorophyll Contents, Chloroplast Ultrastructure, and Photosynthetic Parameters
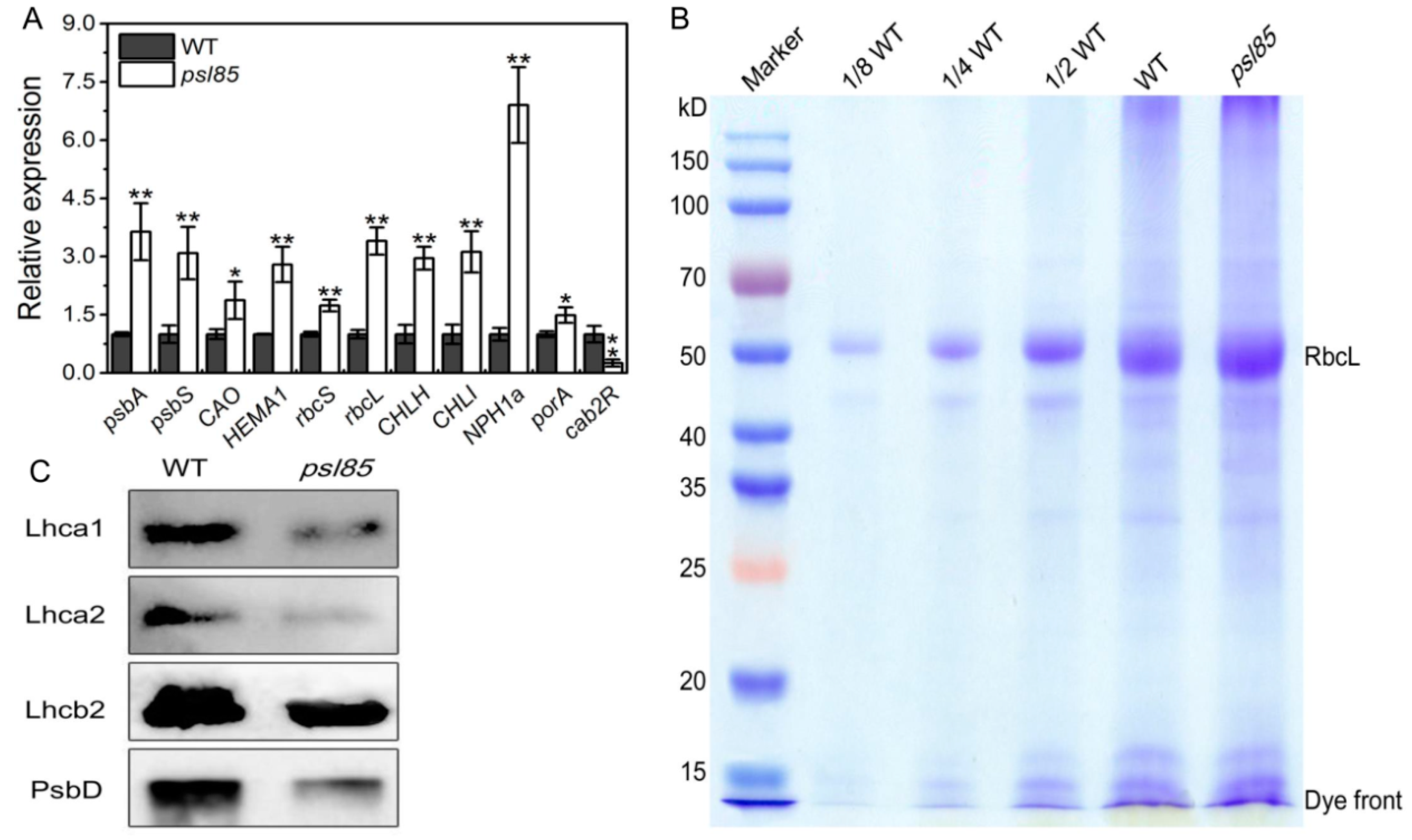
2.3. Impaired Photosynthesis System in psl85 Mutant
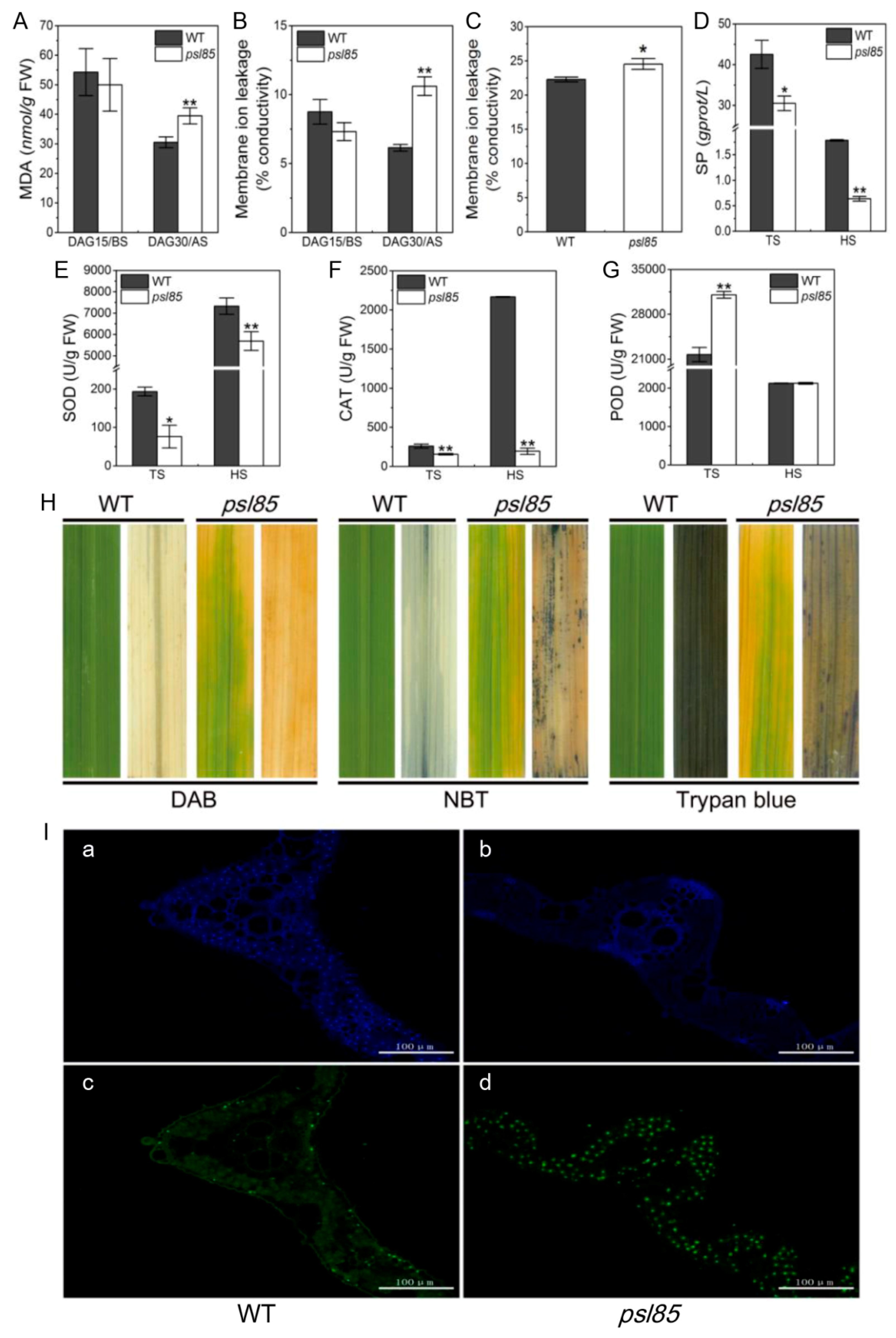
2.4. Alterations of Physio-Biochemical Indicators Related to Senescence at Different Growth Stages
2.5. Elevated Expression of Chlorophyll Metabolism and Senescence-Associated Genes by qRT-PCR
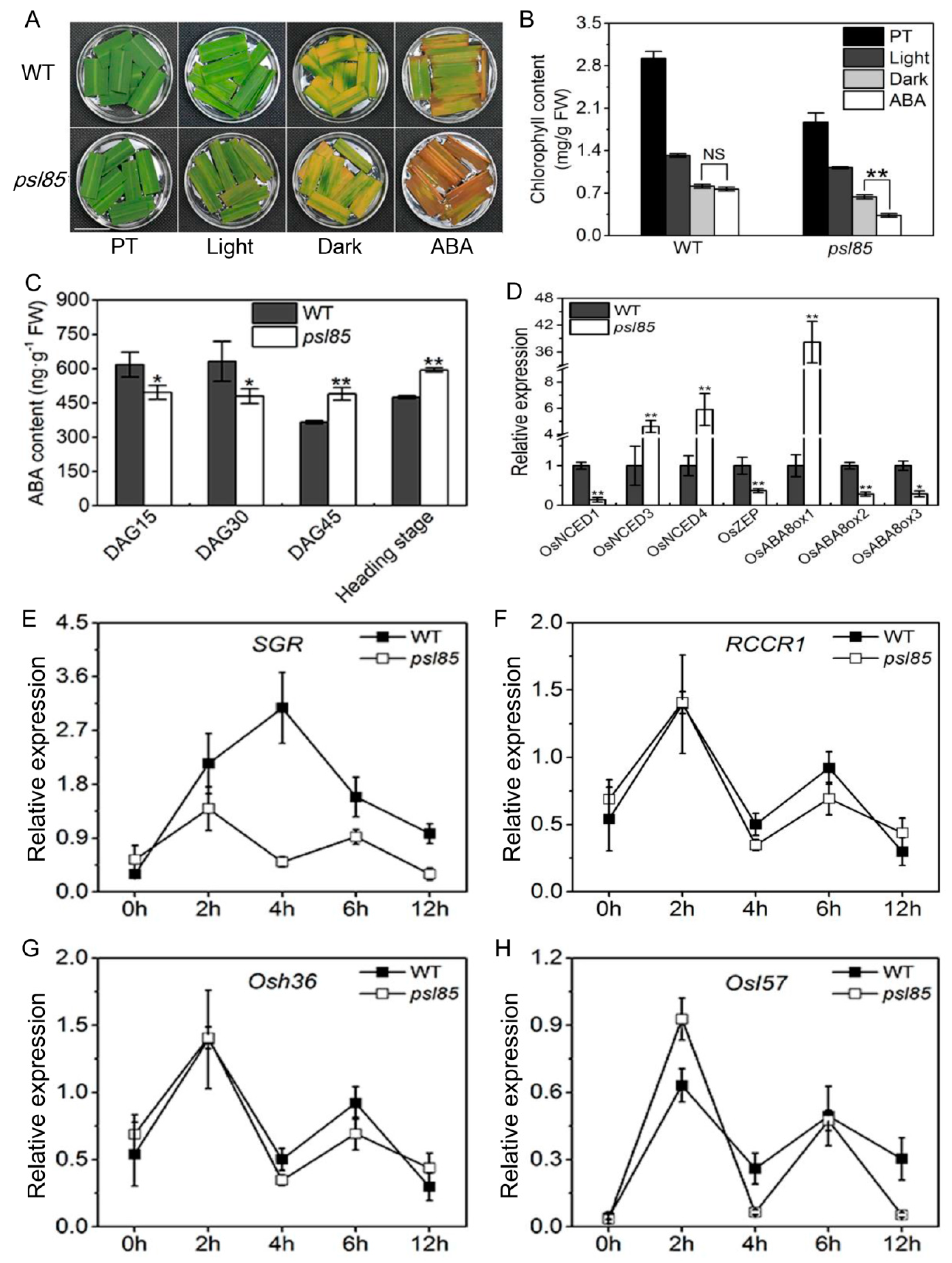
2.6. Induction of Synchronous Senescence by Darkness and ABA
2.7. Genetic Analysis and Mapping of PSL85
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Histochemical Analysis
4.3. Chlorophyll Content and Membrane Ion Leakage Determination
4.4. Senescence-Related and Photosynthesis Parameter Measurement
4.5. TUNEL Assay
4.6. Transmission Electron Microscopy and Paraffin Section
4.7. Darkness- and ABA-Induced Leaf Senescence
4.8. ABA Level Analysis
4.9. Gene Expression Analysis
4.10. SDS-PAGE and Immunoblot Analysis
4.11. Genetic Analysis and Gene Mapping
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Quirino, B.F.; Noh, Y.S.; Himelblau, E.; Amasino, R.M. Molecular aspects of leaf senescence. Trends Plant Sci. 2000, 5, 278–282. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Nam, H.G.; Lin, J.F.; Wu, S.H.; Swidzinski, J.; Ishizaki, K.; et al. Comparative transcriptome analysis reveals significant differences in gene expression and signaling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Chung, K.M.; Park, J.H.; Oh, S.A.; Ahn, T.; Hong, S.H.; Jang, S.K.; Nam, H.G. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 2001, 13, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Ryu, H.; Hong, S.H.; Woo, H.R.; Lim, P.O.; Lee, I.C.; Sheen, J.; Nam, H.G.; Hwang, I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2006, 103, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.D.; Seo, P.J.; Yoon, H.K.; Park, C.M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, L.; Zhang, Z.; Wu, J.L. Identification and comparative analysis of premature senescence leaf mutants in rice (Oryza sativa L.). Int. J. Mol. Sci. 2018, 19, 140. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Masclaux-Daubresse, C.; Lim, P.O. Plant senescence: How plants know when and how to die. J. Exp. Bot. 2018, 69, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, H.; Tremmel, I.; Haase, W.; Kubitscheck, U. Supramolecular photosystem II organization in grana thylakoid membranes: Evidence for a structured arrangement. Biochemistry 2004, 43, 9204–9213. [Google Scholar] [CrossRef] [PubMed]
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hörtensteiner, S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell. 2009, 21, 767–785. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Sakuraba, Y.; Lee, T.; Kim, K.W.; An, G.; Lee, H.Y.; Paek, N.C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015, 57, 562–576. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Han, S.H.; Yang, H.J.; Piao, W.; Paek, N.C. Mutation of Rice Early Flowering3.1 (OsELF3.1) delays leaf senescence in rice. Plant Mol. Biol. 2016, 92, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.; Bagchi, S.; Raychaudhuri, P. Damaged DNA binding protein 2 in reactive oxygen species (ROS) regulation and premature senescence. Int. J. Mol. Sci. 2012, 13, 11012–11026. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ye, C.; Zhao, Y.; Cheng, X.; Wang, Y.; Jiang, Y.Q.; Yang, B. An oilseed rape WRKY-type transcription factor regulates ROS accumulation and leaf senescence in Nicotiana benthamiana and Arabidopsis through modulating transcription of RbohD and RbohF. Planta 2018, 247, 1323–1338. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.N.; Shi, Y.F.; Zhang, X.B.; Song, L.X.; Feng, B.H.; Wang, H.M.; Xu, X.; Li, X.H.; Guo, D.; Wu, J.L. Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J. Integr. Plant Biol. 2016, 58, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.B.; Feng, B.H.; Wang, H.M.; Xu, X.; Shi, Y.F.; He, Y.; Chen, Z.; Sathe, A.P.; Shi, L.; Wu, J.L. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J. Integr. Plant Biol. 2018, 60, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Ren, D.; Hu, S.; Li, G.; Dong, G.; Jiang, L.; Hu, X.; Ye, W.; Cui, Y.; Zhu, L.; et al. Down-regulation of a nicotinate phosphoribosyltransferase gene, OsNaPRT1, leads to withered leaf tips. Plant Physiol. 2016, 171, 1085–1098. [Google Scholar] [CrossRef] [PubMed]
- Buchanan-Wollaston, V.; Earl, S.; Harrison, E.; Mathas, E.; Navabpour, S.; Page, T.; Pink, D. The molecular analysis of leaf senescence–a genomics approach. Plant Biotechnol. J. 2003, 1, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 27, 10013–10018. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Gao, J.; Zhu, X.; Song, Y.; Li, Z.; Ren, G.; Zhou, X.; Kuai, B. ABF2, ABF3, and ABF4 promote ABA mediated chlorophyll degradation and leaf senescence by transcriptional activation of chlorophyll catabolic genes and senescence-associated genes in Arabidopsis. Mol. Plant 2016, 9, 1272–1285. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 17, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef] [PubMed]
- Jiao, B.B.; Wang, J.J.; Zhu, X.D.; Zeng, L.J.; Li, Q.; He, Z.H. A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol. Plant 2012, 5, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Piao, W.; Kim, E.Y.; Han, S.H.; Sakuraba, Y.; Paek, N.C. Rice phytochrome B (OsPhyB) negatively regulates dark- and starvation-induced leaf senescence. Plants 2015, 4, 644–663. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.C.; Nam, H.G. Leaf senescence in plants: From model plants to crops, still so many unknowns. J. Integr. Plant Biol. 2012, 54, 514–515. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhang, L.; Li, R.; Wang, X.; Liu, S.; Liu, X.; Jing, Y.; Xiao, J. SKL1 is essential for chloroplast development in Arabidopsis. Front. Plant Sci. 2018, 9, 179. [Google Scholar] [CrossRef] [PubMed]
- Schlimme, M.; Blaschke, L.; Lagrimini, L.M.; Polle, A. Growth performance and lignification in tobacco with suppressed apoplastic anionic peroxidase activity under ambient and elevated CO2 concentrations. Int. J. Plant Sci. 2002, 163, 749–754. [Google Scholar] [CrossRef]
- Breusegem, F.V.; Dat, J.F. Reactive oxygen species in plant cell death. Plant Physiol. 2006, 141, 384–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef] [PubMed]
- Mittova, V.; Guy, M.; Tal, M.; Volokita, M. Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt tolerant tomato species Lycopersicon pennellii. J. Exp. Bot. 2004, 55, 1105–1113. [Google Scholar] [CrossRef] [PubMed]
- Affenzeller, M.J.; Darehshouri, A.; Andosch, A.; Lütz, C.; Lütz-Meindl, U. Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J. Exp. Bot. 2009, 60, 939–954. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.; Yang, Y.; Ren, D.; Huang, L.; Dai, L.; Wang, Y.; Chen, L.; Tu, Z.; Gao, Y.; Li, X.; et al. A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol. 2017, 174, 1151–1166. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y. Varietal difference in the correlation between leaf nitrogen content and photosynthesis in rice (Oryza sativa L.) plants is related to specific leaf weight. J. Integr. Agric. 2016, 15, 2002–2011. [Google Scholar] [CrossRef]
- Ma, X.; Sun, X.; Li, C.; Huan, R.; Sun, C.; Wang, Y.; Xiao, F.; Wang, Q.; Chen, P.; Ma, F.; et al. Map-based cloning and characterization of the novel yellow-green leaf gene ys83 in rice (Oryza sativa). Plant Physiol. Biochem. 2017, 111, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Qin, X.; Zeevaart, J.A. Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 2003, 131, 1591–1601. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D.; Sakata, K.; Mizutani, M. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004, 134, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.C.; Qin, P.; Liu, Z.; Wang, G.; Chen, W.; Tong, J.; Xiao, L.; Tu, B.; Sun, Y.; Yan, W.; et al. Characterization and fine-mapping of a novel premature leaf senescence mutant yellow leaf and dwarf 1 in rice. Plant Physiol. Biochem. 2017, 111, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ruan, B.; Wang, Z.; Ren, D.; Zhang, Y.; Leng, Y.; Zeng, D.; Hu, J.; Zhang, G.; Zhu, L.; et al. Fine mapping of a novel defective glume 1 (dg1) mutant, which affects vegetative and spikelet development in rice. Front. Plant Sci. 2017, 8, 486. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Li, M.; Liang, N.; Yan, H.; Wei, Y.; Xu, X.; Liu, J.; Xu, Z.; Chen, F.; Wu, G. Molecular cloning and function analysis of the stay green gene in rice. Plant J. 2007, 52, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Li, M.; Chen, Y.; Wu, P.; Wu, G.; Jiang, H. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J. Plant Physiol. 2011, 168, 1952–1959. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.H.; Wang, C.H.; Huang, L.T.; Chen, S.C. Leaf senescence in rice plants: cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 2001, 52, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yan, Y.; Li, Y.; Chu, X.; Wu, C.; Guo, X. GhWRKY40, a multiple stress-responsive cotton WRKY Gene, plays an important role in the wounding response and enhances susceptibility to Ralstonia solanacearum infection in transgenic Nicotiana benthamiana. PLoS ONE 2014, 9, e93577. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Chen, J.; Zeng, L.; Goh, M.; Leung, H.; Khush, G.S.; Wang, G.L. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 2000, 13, 869–876. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Shi, Y.F.; Zhang, X.B.; Wang, H.M.; Xu, X.; Wu, J.L. Identification of a gravitropism-deficient mutant in rice. Rice Sci. 2017, 24, 109–118. [Google Scholar] [CrossRef]


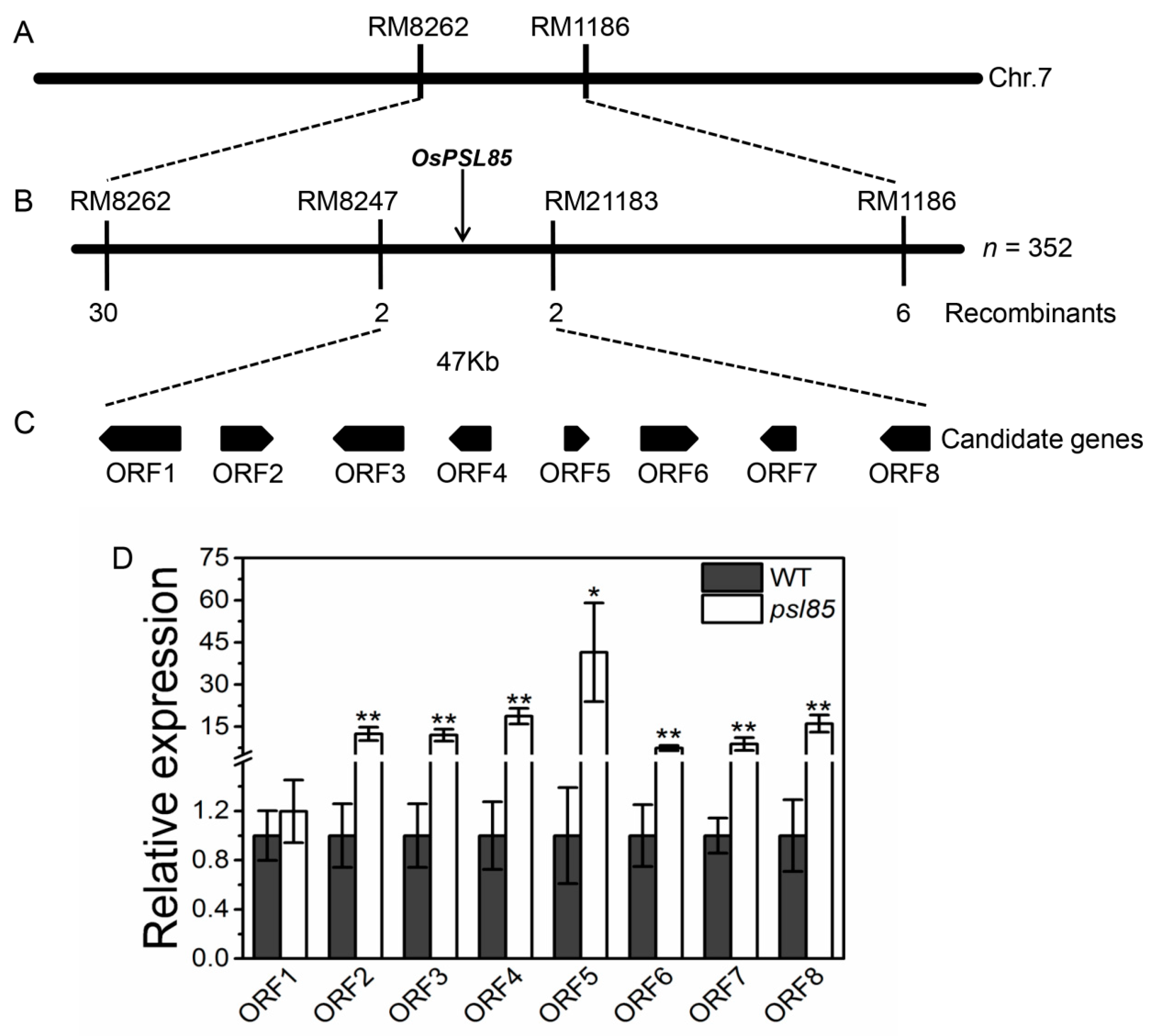
| ORF | Gene ID | Annotation |
|---|---|---|
| ORF1 | LOC_Os07g10610 | Expressed protein |
| ORF2 | LOC_Os07g10620 | Expressed protein |
| ORF3 | LOC_Os07g10630 | Expressed protein |
| ORF4 | LOC_Os07g10640 | Hypothetical protein |
| ORF5 | LOC_Os07g10650 | Hypothetical protein |
| ORF6 | LOC_Os07g10660 | Ribosomal protein, putative, expressed |
| ORF7 | LOC_Os07g10670 | Expressed protein |
| ORF8 | LOC_Os07g10680 | Polygalacturonase, putative, expressed |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Zhang, Z.; Li, L.; Tang, S.; Wu, J.-L. Genetic and Physio-Biochemical Characterization of a Novel Premature Senescence Leaf Mutant in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2018, 19, 2339. https://doi.org/10.3390/ijms19082339
He Y, Zhang Z, Li L, Tang S, Wu J-L. Genetic and Physio-Biochemical Characterization of a Novel Premature Senescence Leaf Mutant in Rice (Oryza sativa L.). International Journal of Molecular Sciences. 2018; 19(8):2339. https://doi.org/10.3390/ijms19082339
Chicago/Turabian StyleHe, Yan, Zhihong Zhang, Liangjian Li, Shaoqing Tang, and Jian-Li Wu. 2018. "Genetic and Physio-Biochemical Characterization of a Novel Premature Senescence Leaf Mutant in Rice (Oryza sativa L.)" International Journal of Molecular Sciences 19, no. 8: 2339. https://doi.org/10.3390/ijms19082339





