mTOR-Related Brain Dysfunctions in Neuropsychiatric Disorders
Abstract
:1. Introduction
2. A Short Overview on Autophagy Impairment in Neurodegenerative Disorders
3. Beyond Classic Neurodegeneration
4. Bridging Neurodegeneration and Psychiatric Disorders: The Paradigm of Methamphetamine-Addicted Brains
5. Cytoskeletal Abnormalities and Neuronal Inclusions in Schizophrenia
6. mTOR Modulation of Dopamine Transmission in Methamphetamine and Schizophrenia
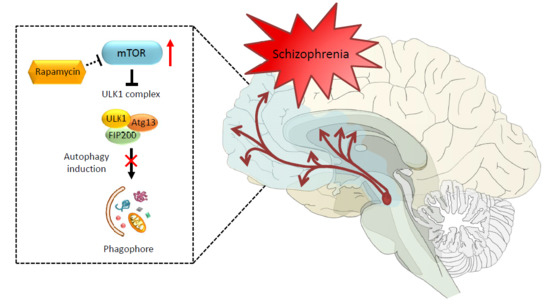
7. A Step Forward about a Role of Autophagy in the Pathophysiology of Schizophrenia
8. Conclusions and Future Perspectives
Funding
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| ADNP | activity-dependent neuroprotective protein |
| Akt | protein kinase B |
| ALS | Amyotrophic lateral sclerosis |
| Aβ | amyloid-β |
| BA 22 | Broadman area 22 |
| BA41 | Broadman area 41 |
| CDK5 | cyclin-dependent kinase 5 |
| CMA | chaperone-mediated autophagy |
| CNS | central nervous system |
| CRMP2 | collapsin response mediator protein 2 |
| D1R | dopamine receptor type 1 |
| DA | dopamine/dopaminergic |
| DAT | dopamine transporter |
| DISC1 | disrupted in schizophrenia 1 |
| DLB | Dementia with Lewy bodies |
| DNR | di-nucleotide repeat |
| DPYSL2 | dihydropyrimidinase-like 2 |
| ErbB4 | avian erythroblastosis oncogene B4-like protein |
| FRKBP12 | FK506-binding protein 12 |
| FTD | Frontotemporal dementia |
| FTLD | Frontotemporal lobar degeneration |
| GFP | green fluorescent protein |
| GPCRs | G-protein coupled receptors |
| HD | Huntington’s disease |
| MAP | microtubule-associated protein |
| MAPT | microtubule-associated protein tau |
| METH | methamphetamine |
| mTOR | mammalian Target Of Rapamycin |
| mTORC1 | mammalian Target Of Rapamycin complex 1 |
| mTORC2 | mammalian Target Of Rapamycin complex 2 |
| NDDs | neurodegenerative disorders |
| NFT | neurofibrillary tangles |
| NRG1 | neuregulin-1 |
| PD | Parkinson’s disease |
| PI3k | phosphatidylinositol-3-Kinase |
| PIP2 | phosphatidylinositol-4,5-phosphate |
| PIP3 | phosphatidylinositol-3,4,5-phosphate |
| PNS | prenatal stress |
| ROS | reactive oxygen species |
| RTKs | receptor tyrosine kinase receptors |
| SNARE | Soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptor |
| SNCA | alpha synuclein gene |
| SNpc | substantia nigra pars compacta |
| SNPs | single-nucleotide polymorphisms |
| SOD1 | superoxide dismutase 1 |
| TDP-43 | TAR DNA-binding protein 43 |
| TEM | transmission electron microscopy |
| TORC | target of rapamycin complex |
| TSC | tuberous sclerosis complex |
| TSC1 | hamartin |
| TSC2 | tuberin |
| VMAT-2 | vesicular monoamine transporter type 2 |
References
- Vezina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Calne, R.Y.; Collier, D.S.; Lim, S.; Pollard, S.G.; Samaan, A.; White, D.J.; Thiru, S. Rapamycin for immunosuppression in organ allografting. Lancet 1989, 2, 227. [Google Scholar] [CrossRef]
- Starzl, T.E.; Schreiber, S.L.; Albers, M.W.; Porter, K.A.; Foglieni, C.S.; Francavilla, A. Hepatotrophic properties in dogs of human FKBP, the binding protein for FK506 and rapamycin. Transplantation 1991, 52, 751–753. [Google Scholar] [PubMed]
- Watson, C.J.; Friend, P.J.; Jamieson, N.V.; Frick, T.W.; Alexander, G.; Gimson, A.E.; Calne, R. Sirolimus: A potent new immunosuppressant for liver transplantation. Transplantation 1999, 67, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Sehgal, S.N. Sirolimus: Its discovery, biological properties, and mechanism of action. Transplant. Proc. 2003, 35, 7S–14S. [Google Scholar] [CrossRef]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Stallock, J.P.; Ng, J.C.; Reinhard, C.; Neufeld, T.P. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000, 14, 2712–2724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garber, K. Rapamycin’s resurrection: A new way to target the cancer cell cycle. J. Natl. Cancer Inst. 2001, 93, 1517–1519. [Google Scholar] [CrossRef] [PubMed]
- Hay, N.; Sonenberg, N. Upstream and downstream of mTOR. Genes Dev. 2004, 18, 1926–1945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fingar, D.C.; Blenis, J. Target of rapamycin (TOR): An integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 2004, 23, 3151–3171. [Google Scholar] [CrossRef] [PubMed]
- Soulard, A.; Cohen, A.; Hall, M.N. TOR signaling in invertebrates. Curr. Opin. Cell Biol. 2009, 21, 825–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz, L.A. Origin and diversification of eukaryotes. Annu. Rev. Microbiol. 2012, 66, 411–427. [Google Scholar] [CrossRef] [PubMed]
- Koltin, Y.; Faucette, L.; Bergsma, D.J.; Levy, M.A.; Cafferkey, R.; Koser, P.L.; Johnson, R.K.; Livi, G.P. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell. Biol. 1991, 11, 1718–1723. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M.C.; Heitman, J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J. Biol. Chem. 1995, 270, 27531–27537. [Google Scholar] [CrossRef] [PubMed]
- Guertin, D.A.; Sabatini, D.M. An expanding role for mTOR in cancer. Trends Mol. Med. 2005, 11, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Sabatini, D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005, 17, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. MTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Guertin, D.A.; Stevens, D.M.; Thoreen, C.C.; Burds, A.A.; Kalaany, N.Y.; Moffat, J.; Brown, M.; Fitzgerald, K.J.; Sabatini, D.M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell 2006, 11, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, T.R.; Laplante, M.; Thoreen, C.C. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 2009, 137, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Kim, D.H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Frias, M.A.; Thoreen, C.C.; Jaffe, J.D.; Schroder, W.; Sculley, T.; Carr, S.A.; Sabatini, D.M. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 2006, 16, 1865–1870. [Google Scholar] [CrossRef] [PubMed]
- Pearce, L.R.; Huang, X.; Boudeau, J.; Pawłowski, R.; Wullschleger, S.; Deak, M.; Ibrahim, A.F.M.; Gourlay, R.; Magnuson, M.A.; Alessi, D.R. Identification of protor as a novel Rictor-binding component ofmTOR complex-2. Biochem. J. 2007, 405, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Thoreen, C.C.; Peterson, T.R.; Lindquist, R.A.; Kang, S.A.; Spooner, E.; Carr, S.A.; Sabatini, D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 2007, 25, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Harris, T.E.; Roth, R.A.; Lawrence, J.C., Jr. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 2007, 282, 20036–20044. [Google Scholar] [CrossRef] [PubMed]
- Kaizuka, T.; Hara, T.; Oshiro, N.; Kikkawa, U.; Yonezawa, K.; Takehana, K.; Iemura, S.; Natsume, T.; Mizushima, N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J. Biol. Chem. 2010, 285, 20109–20116. [Google Scholar] [CrossRef] [PubMed]
- Zoncu, R.; Efeyan, A.; Sabatini, D.M. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 2011, 12, 21–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haissaguerre, M.; Saucisse, N.; Cota, D. Influence of mTOR in energy and metabolic homeostasis. Mol. Cell. Endocrinol. 2014, 397, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.M.; Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009, 10, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Meijer, A.J.; Codogno, P. Signalling and autophagy regulation in health, aging and disease. Mol. Asp. Med. 2006, 27, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Ahlberg, J.; Marzella, L.; Glaumann, H. Uptake and degradation of proteins by isolated rat liver lysosomes. Suggestion of a microautophagic pathway of proteolysis. Lab. Investig. 1982, 47, 523–532. [Google Scholar] [PubMed]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Dice, J.F. Chaperone-mediated autophagy. Autophagy 2007, 3, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Takamura, A.; Kishi, C.; Iemura, S.; Natsume, T.; Guan, J.L.; Mizushima, N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008, 181, 497–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganley, I.G.; Lam du, H.; Wang, J.; Ding, X.; Chen, S.; Jiang, X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009, 284, 12297–12305. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Jun, C.B.; Ro, S.H.; Kim, Y.M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.H. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef] [PubMed]
- Bernard, A.; Klionsky, D.J. Defining the membrane precursor supporting the nucleation of the phagophore. Autophagy 2014, 10, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Burré, J.; Vivona, S.; Diao, J.; Sharma, M.; Brunger, A.T.; Südhof, T.C. Properties of native brain α-synuclein. Nature 2013, 498, E4–E6; discussion E6–E7. [Google Scholar] [CrossRef] [PubMed]
- Nixon, R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013, 19, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Mariño, G.; López-Otín, C. Autophagy: Molecular mechanisms, physiological functions and relevance in human pathology. Cell. Mol. Life Sci. 2004, 61, 1439–1454. [Google Scholar] [CrossRef] [PubMed]
- Fornai, F.; Longone, P.; Ferrucci, M.; Lenzi, P.; Isidoro, C.; Ruggieri, S.; Paparelli, A. Autophagy and amyotrophic lateral sclerosis: The multiple roles of lithium. Autophagy 2008, 4, 527–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornai, F.; Longone, P.; Cafaro, L.; Kastsiuchenka, O.; Ferrucci, M.; Manca, M.L.; Lazzeri, G.; Spalloni, A.; Bellio, N.; Lenzi, P.; et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA 2008, 105, 2052–2057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carloni, S.; Buonocore, G.; Balduini, W. Protective role of autophagy in neonatal hypoxia-ischemia induced brain injury. Neurobiol. Dis. 2008, 32, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, D.; Cloughesy, T.F.; Mischel, P.S. mTOR signaling in glioblastoma: Lessons learned from bench to bedside. Neuro Oncol. 2010, 12, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.W.; Weiss, W.A. Inhibition of PI3K-Akt-mTOR signaling in glioblastoma by mTORC1/2 inhibitors. Methods Mol. Biol. 2012, 821, 349–359. [Google Scholar] [PubMed]
- Arcella, A.; Biagioni, F.; Antonietta Oliva, M.; Bucci, D.; Frati, A.; Esposito, V.; Cantore, G.; Giangaspero, F.; Fornai, F. Rapamycin inhibits the growth of glioblastoma. Brain Res. 2013, 1495, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Heras-Sandoval, D.; Pérez-Rojas, J.M.; Hernández-Damián, J.; Pedraza-Chaverri, J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell. Signal. 2014, 26, 2694–2701. [Google Scholar] [CrossRef] [PubMed]
- Giorgi, F.S.; Biagioni, F.; Lenzi, P.; Frati, A.; Fornai, F. The role of autophagy in epileptogenesis and in epilepsy-induced neuronal alterations. J. Neural Transm. 2015, 122, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Tramutola, A.; Triplett, J.C.; Di Domenico, F.; Niedowicz, D.M.; Murphy, M.P.; Coccia, R.; Perluigi, M.; Butterfield, D.A. Alteration of mTOR signaling occurs early in the progression of Alzheimer disease (AD): Analysis of brain from subjects with pre-clinical AD, amnestic mild cognitive impairment and late-stage AD. J. Neurochem. 2015, 133, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, M.; Biagioni, F.; Lenzi, P.; Gambardella, S.; Ferese, R.; Calierno, M.T.; Falleni, A.; Grimaldi, A.; Frati, A.; Esposito, V.; et al. Rapamycin promotes differentiation increasing βIII-tubulin, NeuN, and NeuroD while suppressing nestin expression in glioblastoma cells. Oncotarget 2017, 8, 29574–29599. [Google Scholar] [CrossRef] [PubMed]
- Ryskalin, L.; Lazzeri, G.; Flaibani, M.; Biagioni, F.; Gambardella, S.; Frati, A.; Fornai, F. mTOR-Dependent Cell Proliferation in the Brain. Biomed. Res. Int. 2017, 2017, 7082696. [Google Scholar] [CrossRef] [PubMed]
- Ryskalin, L.; Limanaqi, F.; Biagioni, F.; Frati, A.; Esposito, V.; Calierno, M.T.; Lenzi, P.; Fornai, F. The emerging role of m-TOR up-regulation in brain Astrocytoma. Histol. Histopathol. 2017, 32, 413–431. [Google Scholar] [PubMed]
- Williams, R.S.; Cheng, L.; Mudge, A.W.; Harwood, A.J. A common mechanism of action for three mood-stabilizing drugs. Nature 2002, 417, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Floto, R.A.; Berger, Z.; Imarisio, S.; Cordenier, A.; Pasco, M.; Cook, L.J.; Rubinsztein, D.C. Lithium induces autophagy by inhibiting inositol monophosphatase. J. Cell Biol. 2005, 170, 1101–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Yu, J.; Pan, H.; Hu, P.; Hao, Y.; Cai, W.; Zhu, H.; Yu, A.D.; Xie, X.; Ma, D.; et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl. Acad. Sci. USA 2007, 104, 19023–19028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kara, N.Z.; Toker, L.; Agam, G.; Anderson, G.W.; Belmaker, R.H.; Einat, H. Trehalose induced antidepressant-like effects and autophagy enhancement in mice. Psychopharmacology 2013, 229, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y.; Lee, K.S.; Choi, Y.K.; Lim, H.J.; Lee, H.G.; Lim, Y.; Lee, Y.H. The antipsychotic agent chlorpromazine induces autophagic cell death by inhibiting the Akt/mTOR pathway in human U-87MG glioma cells. Carcinogenesis 2013, 34, 2080–2089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vucicevic, L.; Misirkic-Marjanovic, M.; Paunovic, V.; Kravic-Stevovic, T.; Martinovic, T.; Ciric, D.; Maric, N.; Petricevic, S.; Harhaji-Trajkovic, L.; Bumbasirevic, V.; et al. Autophagy inhibition uncovers the neurotoxic action of the antipsychotic drug olanzapine. Autophagy 2014, 10, 2362–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; McGreal, S.; Zhao, J.; Huang, R.; Zhou, Y.; Zhong, H.; Xia, M.; Ding, W.X. A cell-based quantitative high-throughput image screening identified novel autophagy modulators. Pharmacol. Res. 2016, 110, 35–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gould, T.D.; O’Donnell, K.C.; Dow, E.R.; Du, J.; Chen, G.; Manji, H.K. Involvement of AMPA receptors in the antidepressant-like effects of lithium in the mouse tail suspension test and forced swim test. Neuropharmacology 2008, 54, 577–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Einat, H.; Yuan, P.; Szabo, S.T.; Dogra, S.; Manji, H.K. Protein kinase C inhibition by tamoxifen antagonizes manic-like behavior in rats: Implications for the development of novel therapeutics for bipolar disorder. Neuropsychobiology 2007, 55, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Zarate, C.A., Jr.; Singh, J.B.; Carlson, P.J.; Quiroz, J.; Jolkovsky, L.; Luckenbaugh, D.A.; Manji, H.K. Efficacy of a protein kinase C inhibitor (tamoxifen) in the treatment of acute mania: A pilot study. Bipolar Disord. 2007, 9, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Anglade, P.; Vyas, S.; Javoy-Agid, F.; Herrero, M.T.; Michel, P.P.; Marquez, J.; Mouatt-Prigent, A.; Ruberg, M.; Hirsch, E.C.; Agid, Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol. Histopathol. 1997, 12, 25–31. [Google Scholar] [PubMed]
- Nixon, R.A.; Wegiel, J.; Kumar, A.; Yu, W.H.; Peterhoff, C.; Cataldo, A.; Cuervo, A.M. Extensive involvement of autophagy in Alzheimer disease: An immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 2005, 64, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Rudnicki, D.D.; Pletnikova, O.; Vonsattel, J.P.; Ross, C.A.; Margolis, R.L. A comparison of huntington disease and huntington disease-like 2 neuropathology. J. Neuropathol. Exp. Neurol. 2008, 67, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Madeo, F.; Eisenberg, T.; Kroemer, G. Autophagy for the avoidance of neurodegeneration. Genes Dev. 2009, 23, 2253–2259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasquali, L.; Longone, P.; Isidoro, C.; Ruggieri, S.; Paparelli, A.; Fornai, F. Autophagy, lithium, and amyotrophic lateral sclerosis. Muscle Nerve 2009, 40, 173–194. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, L.; Ruggieri, S.; Murri, L.; Paparelli, A.; Fornai, F. Does autophagy worsen or improve the survival of dopaminergic neurons? Parkinsonism Relat. Disord. 2009, 15, S24–S27. [Google Scholar] [CrossRef]
- Ferrucci, M.; Fulceri, F.; Toti, L.; Soldani, P.; Siciliano, G.; Paparelli, A.; Fornai, F. Protein clearing pathways in ALS. Arch. Ital. Biol. 2011, 149, 121–149. [Google Scholar] [PubMed]
- Sasaki, S. Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2011, 70, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Natale, G.; Lenzi, P.; Lazzeri, G.; Falleni, A.; Biagioni, F.; Ryskalin, L.; Fornai, F. Compartment-dependent mitochondrial alterations in experimental ALS, the effects of mitophagy and mitochondriogenesis. Front. Cell. Neurosci. 2015, 9, 434. [Google Scholar] [CrossRef] [PubMed]
- Spillantini, M.G.; Schmidt, M.L.; Lee, V.M.; Trojanowski, J.Q.; Jakes, R.; Goedert, M. α-synuclein in Lewy bodies. Nature 1997, 388, 839–840. [Google Scholar] [CrossRef] [PubMed]
- Shults, C.W. Lewy bodies. Proc. Natl. Acad. Sci. USA 2006, 103, 1661–1668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, R.F.; Wade-Martins, R.; Alegre-Abarrategui, J. Direct visualization of alpha-synuclein oligomers reveals previously undetected pathology in Parkinson’s disease brain. Brain 2015, 138, 1642–1657. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Hattori, N. Dopaminergic Neuron-Specific Autophagy-Deficient Mice. Methods Mol. Biol. 2018. [Google Scholar] [CrossRef]
- Hyman, B.T.; Phelps, C.H.; Beach, T.G.; Bigio, E.H.; Cairns, N.J.; Carrillo, M.C.; Dickson, D.W.; Duyckaerts, C.; Frosch, M.P.; Masliah, E.; et al. National institute on aging-Alzheimer’s association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Grossman, M.; Wood, E.M.; Moore, P.; Neumann, M.; Kwong, L.; Forman, M.S.; Clark, C.M.; McCluskey, L.F.; Miller, B.L.; Lee, V.M.; et al. TDP-43 pathologic lesions and clinical phenotype in frontotemporal lobar degeneration with ubiquitin-positive inclusions. Arch. Neurol. 2007, 64, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar, B.; Vacher, C.; Berger, Z.; Davies, J.E.; Luo, S.; Oroz, L.G.; Scaravilli, F.; Easton, D.F.; Duden, R.; O’Kane, C.J.; et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 2004, 36, 585–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, Z.; Ravikumar, B.; Menzies, F.M.; Oroz, L.G.; Underwood, B.R.; Pangalos, M.N.; Schmitt, I.; Wullner, U.; Evert, B.O.; O’Kane, C.J.; et al. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006, 15, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Spencer, B.; Potkar, R.; Trejo, M.; Rockenstein, E.; Patrick, C.; Gindi, R.; Adame, A.; Wyss-Coray, T.; Masliah, E. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson’s and Lewy body diseases. J. Neurosci. 2009, 29, 13578–13588. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Chang, J.C.; Fan, E.Y.; Flajolet, M.; Greengard, P. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer’s APP-CTF for terminal degradation via autophagy. Proc. Natl. Acad. Sci. USA 2013, 110, 17071–17076. [Google Scholar] [CrossRef] [PubMed]
- Jo, C.; Gundemir, S.; Pritchard, S.; Jin, Y.N.; Rahman, I.; Johnson, G.V. Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat. Commun. 2014, 5, 3496. [Google Scholar] [CrossRef] [PubMed]
- Higashi, S.; Iseki, E.; Yamamoto, R.; Minegishi, M.; Hino, H.; Fujisawa, K.; Togo, T.; Katsuse, O.; Uchikado, H.; Furukawa, Y.; et al. Concurrence of TDP-43, tau and alpha-synuclein pathology in brains of Alzheimer’s disease and dementia with Lewy bodies. Brain Res. 2007, 1184, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Colom-Cadena, M.; Gelpi, E.; Charif, S.; Belbin, O.; Blesa, R.; Martí, M.J.; Clarimón, J.; Lleó, A. Confluence of α-synuclein, tau, and β-amyloid pathologies in dementia with Lewy bodies. J. Neuropathol. Exp. Neurol. 2013, 72, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Charles, V.; Mezey, E.; Reddy, P.H.; Dehejia, A.; Young, T.A.; Polymeropoulos, M.H.; Brownstein, M.J.; Tagle, D.A. α-synuclein immunoreactivity of huntingtin polyglutamine aggregates in striatum and cortex of Huntington’s disease patients and transgenic mouse models. Neurosci. Lett. 2000, 289, 29–32. [Google Scholar] [CrossRef]
- Takei, Y.; Oguchi, K.; Koshihara, H.; Hineno, A.; Nakamura, A.; Ohara, S. α-Synuclein coaggregation in familial amyotrophic lateral sclerosis with SOD1 gene mutation. Hum. Pathol. 2013, 44, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Gong, C.X. From chronic cerebral hypoperfusion to Alzheimer-like brain pathology and neurodegeneration. Cell. Mol. Neurobiol. 2015, 35, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Spilman, P.; Podlutskaya, N.; Hart, M.J.; Debnath, J.; Gorostiza, O.; Bredesen, D.; Richardson, A.; Strong, R.; Galvan, V. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS ONE 2010, 5, e9979. [Google Scholar] [CrossRef] [PubMed]
- Bové, J.; Martínez-Vicente, M.; Vila, M. Fighting neurodegeneration with rapamycin: Mechanistic insights. Nat. Rev. Neurosci. 2011, 12, 437–452. [Google Scholar] [CrossRef] [PubMed]
- Roscic, A.; Baldo, B.; Crochemore, C.; Marcellin, D.; Paganetti, P. Induction of autophagy with catalytic mTOR inhibitors reduces huntingtin aggregates in a neuronal cell model. J. Neurochem. 2011, 119, 398–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryor, W.M.; Biagioli, M.; Shahani, N.; Swarnkar, S.; Huang, W.C.; Page, D.T.; MacDonald, M.E.; Subramaniam, S. Huntingtin promotes mTORC1 signaling in the pathogenesis of Huntington’s disease. Sci. Signal. 2014, 7, ra103. [Google Scholar] [CrossRef] [PubMed]
- An, W.L.; Cowburn, R.F.; Li, L.; Braak, H.; Alafuzoff, I.; Iqbal, K.; Iqbal, I.G.; Winblad, B.; Pei, J.J. Up-regulation of phosphorylated/activated p70 S6 kinase and its relationship to neurofibrillary pathology in Alzheimer’s disease. Am. J. Pathol. 2003, 163, 591–607. [Google Scholar] [CrossRef]
- Li, X.; Alafuzoff, I.; Soininen, H.; Winblad, B.; Pei, J.J. Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer’s disease brain. FEBS J. 2005, 272, 4211–4220. [Google Scholar] [CrossRef] [PubMed]
- Caccamo, A.; Maldonado, M.A.; Majumder, S.; Medina, D.X.; Holbein, W.; Magri, A.; Oddo, S. Naturally secreted amyloid-β increases mammalian target of rapamycin (mTOR) activity via a PRAS40-mediated mechanism. J. Biol. Chem. 2011, 286, 8924–8932. [Google Scholar] [CrossRef] [PubMed]
- Ozcelik, S.; Fraser, G.; Castets, P.; Schaeffer, V.; Skachokova, Z.; Breu, K.; Clavaguera, F.; Sinnreich, M.; Kappos, L.; Goedert, M.; et al. Rapamycin attenuates the progression of tau pathology in P301S tau transgenic mice. PLoS ONE 2013, 8, e62459. [Google Scholar] [CrossRef] [PubMed]
- Barmada, S.J.; Serio, A.; Arjun, A.; Bilican, B.; Daub, A.; Ando, D.M.; Tsvetkov, A.; Pleiss, M.; Li, X.; Peisach, D.; et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat. Chem. Biol. 2014, 10, 677–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, T.; Yu, J.T.; Zhu, X.C.; Tan, M.S.; Wang, H.F.; Cao, L.; Zhang, Q.Q.; Shi, J.Q.; Gao, L.; Qin, H.; et al. Temsirolimus promotes autophagic clearance of amyloid-β and provides protective effects in cellular and animal models of Alzheimer’s disease. Pharmacol. Res. 2014, 81, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Frederick, C.; Ando, K.; Leroy, K.; Heraud, C.; Suain, V.; Buee, L.; Brion, J.P. Rapamycin ester analog CCI-779/Temsirolimus alleviates tau pathology and improves motor deficit in mutant tau transgenic mice. J. Alzheimers Dis. 2015, 44, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Lang, U.E.; Heger, J.; Willbring, M.; Domula, M.; Matschke, K.; Tugtekin, S.M. Immunosuppression using the mammalian target of rapamycin (mTOR) inhibitor everolimus: Pilot study shows significant cognitive and affective improvement. Transplant. Proc. 2009, 41, 4285–4288. [Google Scholar] [CrossRef] [PubMed]
- Ehninger, D.; Silva, A.J. Rapamycin for treating Tuberous sclerosis and Autism spectrum disorders. Trends Mol. Med. 2011, 17, 78–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, S.K.; Lee, J.H.; Yang, J.E.; Lim, C.S.; Lee, J.A.; Lee, Y.S.; Lee, K.; Kaang, B.K. Everolimus improves neuropsychiatric symptoms in a patient with tuberous sclerosis carrying a novel TSC2 mutation. Mol. Brain 2016, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Kilincaslan, A.; Kok, B.E.; Tekturk, P.; Yalcinkaya, C.; Ozkara, C.; Yapici, Z. Beneficial Effects of Everolimus on Autism and Attention-Deficit/Hyperactivity Disorder Symptoms in a Group of Patients with Tuberous Sclerosis Complex. J. Child Adolesc. Psychopharmacol. 2017, 27, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Cleary, C.; Linde, J.A.; Hiscock, K.M.; Hadas, I.; Belmaker, R.H.; Agam, G.; Flaisher-Grinberg, S.; Einat, H. Antidepressive-like effects of rapamycin in animal models: Implications for mTOR inhibition as a new target for treatment of affective disorders. Brain Res. Bull. 2008, 76, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Bou Khalil, R. Is there any place for macrolides in mood disorders? Med. Hypotheses 2012, 78, 86–87. [Google Scholar] [CrossRef] [PubMed]
- Ehninger, D.; Han, S.; Shilyansky, C.; Zhou, Y.; Li, W.; Kwiatkowski, D.J.; Ramesh, V.; Silva, A.J. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat. Med. 2008, 14, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Kasai, S.; Kobayashi, T.; Takamatsu, Y.; Hino, O.; Ikeda, K.; Mizuguchi, M. Rapamycin reverses impaired social interaction in mouse models of tuberous sclerosis complex. Nat. Commun. 2012, 3, 1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, G.; Gudsnuk, K.; Kuo, S.H.; Cotrina, M.L.; Rosoklija, G.; Sosunov, A.; Sonders, M.S.; Kanter, E.; Castagna, C.; Yamamoto, A.; et al. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron 2014, 83, 1131–1143. [Google Scholar] [CrossRef] [PubMed]
- Kara, N.Z.; Flaisher-Grinberg, S.; Anderson, G.W.; Agam, G.; Einat, H. Mood-stabilizing effects of rapamycin and its analog temsirolimus: Relevance to autophagy. Behav. Pharmacol. 2018, 29, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, K.R.; Healy, D.G.; Schapira, A.H.; National Institute for Clinical Excellence. Non-motor symptoms of Parkinson’s disease: Diagnosis and management. Lancet Neurol. 2006, 5, 235–245. [Google Scholar] [CrossRef]
- Velakoulis, D.; Walterfang, M.; Mocellin, R.; Pantelis, C.; McLean, C. Frontotemporal dementia presenting as schizophrenia-like psychosis in young people: Clinicopathological series and review of cases. Br. J. Psychiatry 2009, 194, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Lyketsos, C.G.; Carrillo, M.C.; Ryan, J.M.; Khachaturian, A.S.; Trzepacz, P.; Amatniek, J.; Cedarbaum, J.; Brashear, R.; Miller, D.S. Neuropsychiatric symptoms in Alzheimer’s disease. Alzheimers Dement. 2011, 7, 532–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornai, F.; Frati, A.; Gesi, M.; Fulceri, F.; Paparelli, S.; Falleni, A.; Ruggieri, S. Neurobiology and neuroanatomy of psychiatric symptoms in parkinsonism. Arch. Ital. Biol. 2013, 151, 179–191. [Google Scholar] [PubMed]
- Aarsland, D.; Kramberger, M.G. Neuropsychiatric Symptoms in Parkinson’s Disease. J. Parkinsons Dis. 2015, 5, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, R.; Kawakami, I.; Onaya, M.; Higashi, S.; Arai, N.; Akiyama, H.; Hasegawa, M.; Arai, T. Frontotemporal dementia with trans-activation response DNA-binding protein 43 presenting with catatonic syndrome. Neuropathology 2018, 38, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Gururajan, A.; van den Buuse, M. Is the mTOR-signalling cascade disrupted in Schizophrenia? J. Neurochem. 2014, 129, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Maas, D.A.; Vallès, A.; Martens, G.J.M. Oxidative stress, prefrontal cortex hypomyelination and cognitive symptoms in schizophrenia. Transl. Psychiatry 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Khan, B.K.; Woolley, J.D.; Chao, S.; See, T.; Karydas, A.M.; Miller, B.L.; Rankin, K.P. Schizophrenia or neurodegenerative disease prodrome? Outcome of a first psychotic episode in a 35-year-old woman. Psychosomatics 2012, 53, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.; Ishikawa, T.; Michiue, T.; Li, D.R.; Zhao, D.; Oritani, S.; Zhu, B.L.; Maeda, H. Ubiquitin-immunoreactive structures in the midbrain of methamphetamine abusers. Leg. Med. 2005, 3, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Fornai, F.; Lenzi, P.; Gesi, M.; Soldani, P.; Ferrucci, M.; Lazzeri, G.; Capobianco, L.; Battaglia, G.; De Blasi, A.; Nicoletti, F.; et al. Methamphetamine produces neuronal inclusions in the nigrostriatal system and in PC12 cells. J. Neurochem. 2004, 88, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Castino, R.; Lazzeri, G.; Lenzi, P.; Bellio, N.; Follo, C.; Ferrucci, M.; Fornai, F.; Isisoro, C. Suppression of autophagy precipitates neuronal cell death following low doses of methamphetamine. J. Neurochem. 2008, 106, 1426–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yui, K.; Ikemoto, S.; Goto, K.; Nishijima, K.; Yoshino, T.; Ishiguro, T. Spontaneous recurrence of methamphetamine-induced paranoid-hallucinatory states in female subjects: Susceptibility to psychotic states and implications for relapse of schizophrenia. Pharmacopsychiatry 2002, 35, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Fasihpour, B.; Molavi, S.; Shariat, S.V. Clinical features of inpatients with methamphetamine induced psychosis. J. Ment. Health 2013, 22, 341–349. [Google Scholar] [CrossRef] [PubMed]
- McKetin, R. Methamphetamine psychosis: Insights from the past. Addiction 2018, 113, 1522–1527. [Google Scholar] [CrossRef] [PubMed]
- Limanaqi, F.; Gambardella, S.; Biagioni, F.; Busceti, C.; Fornai, F. Epigenetic effects induced by methamphetamine and methamphetamine-dependent oxidative stress. Oxid. Med. Cell. Longev. 2018, 2018, 4982453. [Google Scholar] [CrossRef]
- Abi-Dargham, A.; Moore, H. Prefrontal DA transmission at D1 receptors and the pathology of schizophrenia. Neuroscientist 2003, 9, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Perreault, M.L.; Hasbi, A.; Alijaniaram, M.; Fan, T.; Varghese, G.; Fletcher, P.J.; Seeman, P.; O’Dowd, B.F.; George, S.R. The dopamine D1-D2 receptor heteromer localizes in dynorphin/enkephalin neurons: Increased high affinity state following amphetamine and in schizophrenia. J. Biol. Chem. 2010, 285, 36625–36634. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Ji, X.; Liu, J.; Li, Z.; Zhang, X. Dopamine Receptor Subtypes Differentially Regulate Autophagy. Int. J. Mol. Sci. 2018, 19, 1540. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, L.; Lazzeri, G.; Isidoro, C.; Ruggieri, S.; Paparelli, A.; Fornai, F. Role of autophagy during methamphetamine neurotoxicity. Ann. N. Y. Acad. Sci. 2008, 1139, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Chandramani-Shivalingappa, P.; Jin, H.; Ghosh, A.; Anantharam, V.; Ali, S.; Kanthasamy, A.G.; Kanthasamy, A. Methamphetamine-induced neurotoxicity linked to ubiquitin-proteasome system dysfunction and autophagy-related changes that can be modulated by protein kinase C delta in dopaminergic neuronal cells. Neuroscience 2012, 210, 308–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aki, T.; Funakoshi, T.; Unuma, K.; Uemura, K. Impairment of autophagy: From hereditary disorder to drug intoxication. Toxicology 2013, 311, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.H.; Wu, W.R.; Lee, L.M.; Huang, P.R.; Chen, J.C. mTOR signaling in the nucleus accumbens mediates behavioral sensitization to methamphetamine. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 86, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Meredith, C.W.; Jaffe, C.; Ang-Lee, K.; Saxon, A.J. Implications of chronic methamphetamine use: A literature review. Harv. Rev. Psychiatry 2005, 13, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Homer, B.D.; Solomon, T.M.; Moeller, R.W.; Mascia, A.; DeRaleau, L.; Halkitis, P.N. Methamphetamine abuse and impairment of social functioning: A review of the underlying neurophysiological causes and behavioral implications. Psychol. Bull. 2008, 134, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.D.; Fowler, J.S.; Wang, G.J.; Shumay, E.; Telang, F.; Thanos, P.K.; Alexoff, D. Distribution and Pharmacokinetics of Methamphetamine in the Human Body: Clinical Implications. PLoS ONE 2010, 5, e15269. [Google Scholar] [CrossRef] [PubMed]
- Laruelle, M.; Abi-Dargham, A.; Gil, R.; Kegeles, L.; Innis, R. Increased dopamine transmission in schizophrenia: Relationship to illness phases. Biol. Psychiatry 1999, 46, 56–72. [Google Scholar] [CrossRef]
- Weidenauer, A.; Bauer, M.; Sauerzopf, U.; Bartova, L.; Praschak-Rieder, N.; Sitte, H.H.; Kasper, S.; Willeit, M. Making Sense of: Sensitization in Schizophrenia. Int. J. Neuropsychopharmacol. 2016, 20, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, B. Neurochemistry of schizophrenia: The contribution of neuroimaging postmortem pathology and neurochemistry in schizophrenia. Curr. Top. Med. Chem. 2012, 12, 2375–2392. [Google Scholar] [CrossRef] [PubMed]
- Howes, O.D.; Kambeitz, J.; Kim, E.; Stahl, D.; Slifstein, M.; Abi-Dargham, A.; Kapur, S. The nature of dopamine dysfunction in schizophrenia and what this means for treatment. Arch. Gen. Psychiatry 2012, 69, 776–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abekawa, T.; Ohmori, T.; Koyama, T. Effects of repeated administration of a high dose of methamphetamine on dopamine and glutamate release in rat striatum and nucleus accumbens. Brain Res. 1994, 643, 276–281. [Google Scholar] [CrossRef]
- Stephans, S.E.; Yamamoto, B.Y. Effect of repeated methamphetamine administrations on dopamine and glutamate efflux in rat prefrontal cortex. Brain Res. 1995, 700, 99–106. [Google Scholar] [CrossRef]
- Nishijima, K.; Kashiwa, A.; Hashimoto, A.; Iwama, H.; Umino, A.; Nishikawa, T. Differential effects of phencyclidine and methamphetamine on dopamine metabolism in rat frontal cortex and striatum as revealed by in vivo dialysis. Synapse 1996, 22, 304–312. [Google Scholar] [CrossRef]
- Larsen, K.E.; Fon, E.A.; Hastings, T.G.; Edwards, R.H.; Sulzer, D. Methamphetamine-induced degeneration of dopaminergic neurons involves autophagy and upregulation of dopamine synthesis. J. Neurosci. 2002, 22, 8951–8960. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Sumiyoshi, T.; Itoh, H.; Kurachi, M. Inhibition of dopamine synthesis with alpha-methyl-p-tyrosine abolishes the enhancement of methamphetamine-induced extracellular dopamine levels in the amygdala of rats with excitotoxic lesions of the entorhinal cortex. Neurosci. Lett. 2004, 356, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Purves-Tyson, T.D.; Owens, S.J.; Rothmond, D.A.; Halliday, G.M.; Double, K.L.; Stevens, J.; McCrossin, T.; Shannon Weickert, C. Putative presynaptic dopamine dysregulation in schizophrenia is supported by molecular evidence from post-mortem human midbrain. Transl. Psychiatry 2017, 7, e1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Volkow, N.D.; Chang, L.; Wang, G.J.; Fowler, J.S.; Leonido-Yee, M.; Franceschi, D.; Sedler, M.J.; Gatley, S.J.; Hitzemann, R.; Ding, Y.S.; et al. Association of dopamine transporter reduction with psychomotor impairment in methamphetamine abusers. Am. J. Psychiatry 2001, 158, 377–382. [Google Scholar] [CrossRef] [PubMed]
- German, C.L.; Hanson, G.R.; Fleckenstein, A.E. Amphetamine and methamphetamine reduce striatal dopamine transporter function without concurrent dopamine transporter relocalization. J. Neurochem. 2012, 123, 288–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, M.; Li, W.; Huang, S.; Song, J.; Kim, J.Y.; Tian, X.; Kang, E.; Sano, Y.; Liu, C.; Balaji, J.; et al. mTOR Inhibition ameliorates cognitive and affective deficits caused by Disc1 knockdown in adult-born dentate granule neurons. Neuron 2013, 77, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, H.; Elgersma, Y.; Bahn, S. A brain proteomic investigation of rapamycin effects in the Tsc1+/− mouse model. Mol. Autism 2017, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Beaulieu, J.M.; Sotnikova, T.D.; Yao, W.D.; Kockeritz, L.; Woodgett, J.R.; Gainetdinov, R.R.; Caron, M.G. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase 3 signaling cascade. Proc. Natl. Acad. Sci. USA 2004, 101, 5099–5104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, D.; Torres, C.A.; Setlik, W.; Cebrián, C.; Mosharov, E.V.; Tang, G.; Cheng, H.C.; Kholodilov, N.; Yarygina, O.; Burke, R.E.; et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron 2012, 74, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Hogan, K.A.; Staal, R.G.; Sonsalla, P.K. Analysis of VMAT2 binding after methamphetamine or MPTP treatment: Disparity between homogenates and vesicle preparations. J. Neurochem. 2000, 74, 2217–2220. [Google Scholar] [CrossRef] [PubMed]
- Guillot, T.S.; Shepherd, K.R.; Richardson, J.R.; Wang, M.Z.; Li, Y.; Emson, P.C.; Miller, G.W. Reduced vesicular storage of dopamine exacerbates methamphetamine-induced neurodegeneration and astrogliosis. J. Neurochem. 2008, 106, 2205–2217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cubells, J.F.; Rayport, S.; Rajendran, G.; Sulzer, D. Methamphetamine neurotoxicity involves vacuolation of endocytic organelles and dopamine-dependent intracellular oxidative stress. J. Neurosci. 1994, 14, 2260–2271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornai, F.; Chen, K.; Giorgi, F.S.; Gesi, M.; Alessandri, M.G.; Shih, J.C. Striatal dopamine metabolism in monoamine oxidase B-deficient mice: A brain dialysis study. J. Neurochem. 1999, 73, 2434–2440. [Google Scholar] [CrossRef] [PubMed]
- Gesi, M.; Santinami, A.; Ruffoli, R.; Conti, G.; Fornai, F. Novel aspects of dopamine oxidative metabolism (confounding outcomes take place of certainties). Pharmacol. Toxicol. 2001, 89, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Lazzeri, G.; Lenzi, P.; Busceti, C.L.; Ferrucci, M.; Falleni, A.; Bruno, V.; Paparelli, A.; Fornai, F. Mechanisms involved in the formation of dopamine-induced intracellular bodies within striatal neurons. J. Neurochem. 2007, 101, 1414–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fornai, F.; Lenzi, P.; Frenzilli, G.; Gesi, M.; Ferrucci, M.; Lazzeri, G.; Biagioni, F.; Nigro, M.; Falleni, A.; Giusiani, M.; et al. DNA damage and ubiquitinated neuronal inclusions in the substantia nigra and striatum of mice following MDMA (ecstasy). Psychopharmacology 2004, 173, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Butler, B.; Gamble-George, J.; Prins, P.; North, A.; Clarke, J.T.; Khoshbouei, H. Chronic Methamphetamine Increases Alpha-Synuclein Protein Levels in the Striatum and Hippocampus but not in the Cortex of Juvenile Mice. J. Addict. Prev. 2014, 2, 6. [Google Scholar] [PubMed]
- Ferrucci, M.; Ryskalin, L.; Biagioni, F.; Gambardella, S.; Busceti, C.L.; Falleni, A.; Lazzeri, G.; Fornai, F. Methamphetamine increases Prion Protein and induces dopamine-dependent expression of protease resistant PrPsc. Arch. Ital. Biol. 2017, 155, 81–97. [Google Scholar] [PubMed]
- Kobayashi, H.; Ide, S.; Hasegawa, J.; Ujike, H.; Sekine, Y.; Ozaki, N.; Inada, T.; Harano, M.; Komiyama, T.; Yamada, M.; et al. Study of association between α-synuclein gene polymorphism and methamphetamine psychosis/dependence. Ann. N. Y. Acad. Sci. 2004, 1025, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Nilsson, C.; Kachergus, J.; Munz, M.; Larsson, E.M.; Schüle, B.; Langston, J.W.; Middleton, F.A.; Ross, O.A.; Hulihan, M.; et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology 2007, 68, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Kolomeets, N.S.; Uranova, N.A. Synaptic contacts in schizophrenia: Study with immunocytochemical identification of dopaminergic neurons. Zhurnal Nevrol. Psikhiatrii Imeni S. S. Korsakova 1997, 97, 39–43. [Google Scholar] [CrossRef]
- Uranova, N.A.; Levité, O.I. Ultrastructure of the substantia nigra in schizophrenia. Zhurnal Nevrol. Psikhiatrii Imeni S. S. Korsakova 1987, 87, 1017–1024. [Google Scholar]
- Arnold, S.E.; Lee, V.M.; Gur, R.E.; Trojanowski, J.Q. Abnormal expression of two microtubule-associated proteins (MAP2 and MAP5) in specific subfields of the hippocampal formation in schizophrenia. Proc. Natl. Acad. Sci. USA 1991, 88, 10850–10854. [Google Scholar] [CrossRef] [PubMed]
- Cotter, D.; Kerwin, R.; Doshi, B.; Martin, C.S.; Everall, I.P. Alterations in hippocampal non-phosphorylated MAP2 protein expression in schizophrenia. Brain Res. 1997, 765, 238–246. [Google Scholar] [CrossRef]
- Rosoklija, G.; Keilp, J.G.; Toomayan, G.; Mancevski, B.; Haroutunian, V.; Liu, D.; Malespina, D.; Hays, A.P.; Sadiq, S.; Latov, N.; et al. Altered subicular MAP2 immunoreactivity in schizophrenia. Prilozi 2005, 26, 13–34. [Google Scholar] [PubMed]
- Somenarain, L.; Jones, L.B. A comparative study of MAP2 immunostaining in areas 9 and 17 in schizophrenia and Huntington chorea. J. Psychiatr. Res. 2010, 44, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Shelton, M.A.; Newman, J.T.; Gu, H.; Sampson, A.R.; Fish, K.N.; MacDonald, M.L.; Moyer, C.E.; DiBitetto, J.V.; Dorph-Petersen, K.A.; Penzes, P.; et al. Loss of Microtubule-Associated Protein 2 Immunoreactivity Linked to Dendritic Spine Loss in Schizophrenia. Biol. Psychiatry 2015, 78, 374–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soustek, Z. Ultrastructure of cortical synapses in the brain of schizophrenics. Zentralbl. Allg. Pathol. 1989, 135, 25–32. [Google Scholar] [PubMed]
- Prohovnik, I.; Dwork, A.J.; Kaufman, M.A.; Willson, N. Alzheimer-type neuropathology in elderly schizophrenia patients. Schizophr. Bull. 1993, 19, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, H.M.; Constantinidis, J.; Wegiel, J.; Bobinski, M.; Tarnawski, M. Neurofibrillary pathology in brains of elderly schizophrenics treated with neuroleptics. Alzheimer Dis. Assoc. Disord. 1994, 8, 211–227. [Google Scholar] [CrossRef] [PubMed]
- Arnold, S.E.; Trojanowski, J.Q.; Gur, R.E.; Blackwell, P.; Han, L.Y.; Choi, C. Absence of neurodegeneration and neural injury in the cerebral cortex in a sample of elderly patients with schizophrenia. Arch. Gen. Psychiatry 1998, 55, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Purohit, D.P.; Perl, D.P.; Haroutunian, V.; Powchik, P.; Davidson, M.; Davis, K.L. Alzheimer disease and related neurodegenerative diseases in elderly patients with schizophrenia: A postmortem neuropathologic study of 100 cases. Arch. Gen. Psychiatry 1998, 55, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Falke, E.; Han, L.Y.; Arnold, S.E. Absence of neurodegeneration in the thalamus and caudate of elderly patients with schizophrenia. Psychiatry Res. 2000, 93, 103–110. [Google Scholar] [CrossRef]
- Bozikas, V.P.; Kövari, E.; Bouras, C.; Karavatos, A. Neurofibrillary tangles in elderly patients with late onset schizophrenia. Neurosci. Lett. 2002, 324, 109–112. [Google Scholar] [CrossRef]
- Momeni, P.; Wickremaratchi, M.M.; Bell, J.; Arnold, R.; Beer, R.; Hardy, J.; Revesz, T.; Neal, J.W.; Morris, H.R. Familial early onset frontotemporal dementia caused by a novel S356T MAPT mutation, initially diagnosed as schizophrenia. Clin. Neurol. Neurosurg. 2010, 112, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Uranova, N.A.; Vikhreva, O.V.; Rachmanova, V.I.; Orlovskaya, D.D. Ultrastructural alterations of myelinated fibers and oligodendrocytes in the prefrontal cortex in schizophrenia: A postmortem morphometric study. Schizophr. Res. Treat. 2011, 2011, 325789. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.K.; Roche, J.K.; Sinha, V.; Roberts, R.C. Substantia nigra ultrastructural pathology in schizophrenia. Schizophr. Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Leliveld, S.R.; Bader, V.; Hendriks, P.; Prikulis, I.; Sajnani, G.; Requena, J.R.; Korth, C. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J. Neurosci. 2008, 28, 3839–3845. [Google Scholar] [CrossRef] [PubMed]
- Ottis, P.; Bader, V.; Trossbach, S.V.; Kretzschmar, H.; Michel, M.; Leliveld, S.R.; Korth, C. Convergence of two independent mental disease genes on the protein level: Recruitment of dysbindin to cell-invasive disrupted-in-schizophrenia 1 aggresomes. Biol. Psychiatry 2011, 70, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Atkin, T.; Kittler, J. DISC1 and the aggresome: A disruption to cellular function? Autophagy 2012, 8, 851–852. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Sun, Y.; Ye, H.; Zhu, L.; Liu, J.; Wu, X.; Wang, L.; He, T.; Shen, Y.; Wu, J.Y.; et al. Increased dysbindin-1B isoform expression in schizophrenia and its propensity in aggresome formation. Cell Discov. 2015, 1, 15032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camargo, L.M.; Collura, V.; Rain, J.C.; Mizuguchi, K.; Hermjakob, H.; Kerrien, S.; Bonnert, T.P.; Whiting, P.J.; Brandon, N.J. Disrupted in Schizophrenia 1 Interactome: Evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol. Psychiatry 2007, 12, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Martins-de-Souza, D.; Cassoli, J.S.; Nascimento, J.M.; Hensley, K.; Guest, P.C.; Pinzon-Velasco, A.M.; Turck, C.W. The protein interactome of collapsin response mediator protein-2 (CRMP2/DPYSL2) reveals novel partner proteins in brain tissue. Proteom. Clin. Appl. 2015, 9, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Edgar, P.F.; Douglas, J.E.; Cooper, G.J.; Dean, B.; Kydd, R.; Faull, R.L. Comparative proteome analysis of the hippocampus implicates chromosome 6q in schizophrenia. Mol. Psychiatry 2000, 5, 85–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston-Wilson, N.L.; Sims, C.D.; Hofmann, J.P.; Anderson, L.; Shore, A.D.; Torrey, E.F.; Yolken, R.H. Disease-specific alterations in frontal cortex brain proteins in schizophrenia, bipolar disorder, and major depressive disorder. The Stanley Neuropathology Consortium. Mol. Psychiatry 2000, 5, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Beasley, C.L.; Pennington, K.; Behan, A.; Wait, R.; Dunn, M.J.; Cotter, D. Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: Evidence for disease-associated changes. Proteomics 2006, 6, 3414–3425. [Google Scholar] [CrossRef] [PubMed]
- Martins-de-Souza, D.; Gattaz, W.F.; Schmitt, A.; Maccarrone, G.; Hunyadi-Gulyás, E.; Eberlin, M.N.; Souza, G.H.; Marangoni, S.; Novello, J.C.; Turck, C.W.; et al. Proteomic analysis of dorsolateral prefrontal cortex indicates the involvement of cytoskeleton, oligodendrocyte, energy metabolism and new potential markers in schizophrenia. J. Psychiatr. Res. 2009, 43, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, V.; Verstreken, P. Autophagy in the presynaptic compartment in health and disease. J. Cell Biol. 2017, 216, 1895–1906. [Google Scholar] [CrossRef] [PubMed]
- Merenlender-Wagner, A.; Shemer, Z.; Touloumi, O.; Lagoudaki, R.; Giladi, E.; Andrieux, A.; Grigoriadis, N.C.; Gozes, I. New horizons in schizophrenia treatment: Autophagy protection is coupled with behavioral improvements in a mouse model of schizophrenia. Autophagy 2014, 10, 2324–2332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merenlender-Wagner, A.; Malishkevich, A.; Shemer, Z.; Udawela, M.; Gibbons, A.; Scarr, E.; Dean, B.; Levine, J.; Agam, G.; Gozes, I. Autophagy has a key role in the pathophysiology of schizophrenia. Mol. Psychiatry 2015, 20, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Morfini, G.A.; Burns, M.; Binder, L.I.; Kanaan, N.M.; LaPointe, N.; Bosco, D.A.; Brown, R.H., Jr.; Brown, H.; Tiwari, A.; Hayward, L.; et al. Axonal transport defects in neurodegenerative diseases. J. Neurosci. 2009, 29, 12776–12786. [Google Scholar] [CrossRef] [PubMed]
- Daoust, A.; Bohic, S.; Saoudi, Y.; Debacker, C.; Gory-Fauré, S.; Andrieux, A.; Barbier, E.L.; Deloulme, J.C. Neuronal transport defects of the MAP6 KO mouse—A model of schizophrenia- and alleviation by Epothilone D treatment, as observed using MEMRI. Neuroimage 2014, 96, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, F.; Riva, M.A.; Molteni, R. Synaptic alterations associated with depression and schizophrenia: Potential as a therapeutic target. Expert Opin. Ther. Targets 2016, 20, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Bridi, J.C.; Hirth, F. Mechanisms of α-Synuclein Induced Synaptopathy in Parkinson’s Disease. Front. Neurosci. 2018, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- St Clair, D.; Blackwood, D.; Muir, W.; Carothers, A.; Walker, M.; Spowart, G.; Gosden, C.; Evans, H.J. Association within a family of a balanced autosomal translocation with major mental illness. Lancet 1990, 336, 13–16. [Google Scholar] [CrossRef]
- Schurov, I.L.; Handford, E.J.; Brandon, N.J.; Whiting, B.J. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol. Psychiatry 2004, 9, 1100–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandon, N.J.; Sawa, A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat. Rev. Neurosci. 2011, 12, 707–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hikida, T.; Gamo, N.J.; Sawa, A. DISC1 as a therapeutic target for mental illnesses. Expert Opin. Ther. Targets 2012, 16, 1151–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipska, B.K.; Mitkus, S.N.; Mathew, S.V.; Fatula, R.; Hyde, T.M.; Weinberger, D.R.; Kleinman, J.E. Functional genomics in postmortem human brain: Abnormalities in a DISC1 molecular pathway in schizophrenia. Dialogues Clin. Neurosci. 2006, 8, 353–357. [Google Scholar] [PubMed]
- Dahoun, T.; Trossbach, S.V.; Brandon, N.J.; Korth, C.; Howes, O.D. The impact of Disrupted-in-Schizophrenia 1 (DISC1) on the dopaminergic system: A systematic review. Transl. Psychiatry 2017, 7, e1015. [Google Scholar] [CrossRef] [PubMed]
- Nakai, T.; Nagai, T.; Wang, R.; Yamada, S.; Kuroda, K.; Kaibuchi, K.; Yamada, K. Alterations of GABAergic and dopaminergic systems in mutant mice with disruption of exons 2 and 3 of the Disc1 gene. Neurochem. Int. 2014, 74, 74–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pogorelov, V.M.; Nomura, J.; Kim, J.; Kannan, G.; Ayhan, Y.; Yang, C.; Taniguchi, Y.; Abazyan, B.; Valentine, H.; Krasnova, I.N.; et al. Mutant DISC1 affects methamphetamine-induced sensitization and conditioned place preference: A comorbidity model. Neuropharmacology 2012, 62, 1242–1251. [Google Scholar] [CrossRef] [PubMed]
- Niwa, M.; Lee, R.S.; Tanaka, T.; Okada, K.; Kano, S.; Sawa, A. A critical period of vulnerability to adolescent stress: Epigenetic mediators in mesocortical dopaminergic neurons. Hum. Mol. Genet. 2016, 25, 1370–1381. [Google Scholar] [CrossRef] [PubMed]
- Owen, M.J.; Sawa, A.; Mortensen, P.B. Schizophrenia. Lancet 2016, 388, 86–97. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Duan, X.; Liu, C.Y.; Jang, M.H.; Guo, J.U.; Pow-anpongkul, N.; Kang, E.; Song, H.; Ming, G.L. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 2009, 63, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, D.H.; Fordyce, A.; Walker, M.T.; St Clair, D.M.; Porteous, D.J.; Muir, W.J. Schizophrenia and affective disorders—Cosegregation with a translocation at chromosome 1q42 that directly disrupts brain-expressed genes: Clinical and P300 findings in a family. Am. J. Hum. Genet. 2001, 69, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Clapcote, S.J.; Lipina, T.V.; Millar, J.K.; Mackie, S.; Christie, S.; Ogawa, F.; Lerch, J.P.; Trimble, K.; Uchiyama, M.; Sakuraba, Y.; et al. Behavioral phenotypes of Disc1 missense mutations in mice. Neuron 2007, 54, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Chubb, J.E.; Bradshaw, N.J.; Soares, D.C.; Porteous, D.J.; Millar, J.K. The DISC locus in psychiatric illness. Mol. Psychiatry 2008, 13, 36–64. [Google Scholar] [CrossRef] [PubMed]
- Gangarossa, G.; Ceolin, L.; Paucard, A.; Lerner-Natoli, M.; Perroy, J.; Fagni, L.; Valjent, E. Repeated stimulation of dopamine D1-like receptor and hyperactivation of mTOR signaling lead to generalized seizures, altered dentate gyrus plasticity, and memory deficits. Hippocampus 2014, 24, 1466–1481. [Google Scholar] [CrossRef] [PubMed]
- Siuta, M.A.; Robertson, S.D.; Kocalis, H.; Saunders, C.; Gresch, P.J.; Khatri, V.; Shiota, C.; Kennedy, J.P.; Lindsley, C.W.; Daws, L.C.; et al. Dysregulation of the norepinephrine transporter sustains cortical hypodopaminergia and schizophrenia-like behaviors in neuronal rictor null mice. PLoS Biol. 2010, 8, e1000393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emamian, E.S.; Hall, D.; Birnbaum, M.J.; Karayiorgou, M.; Gogos, J.A. Convergent evidence for impaired AKT1-GSK3β signaling in schizophrenia. Nat. Genet. 2004, 36, 131–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, M.; Iwata, N.; Suzuki, T.; Kitajima, T.; Yamanouchi, Y.; Kinoshita, Y.; Inada, T.; Ozaki, N. Association of AKT1 with schizophrenia confirmed in a Japanese population. Biol. Psychiatry 2004, 56, 698–700. [Google Scholar] [CrossRef] [PubMed]
- Schwab, S.G.; Hoefgen, B.; Hanses, C.; Hassenbach, M.B.; Albus, M.; Lerer, B.; Trixler, M.; Maier, W.; Wildenauer, D.B. Further evidence for association of variants in the AKT1 gene with schizophrenia in a sample of European sib-pair families. Biol. Psychiatry 2005, 58, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Bajestan, S.N.; Sabouri, A.H.; Nakamura, M.; Takashima, H.; Keikhaee, M.R.; Behdani, F.; Fayyazi, M.R.; Sargolzaee, M.R.; Bajestan, M.N.; Sabouri, Z.; et al. Association of AKT1 haplotype with the risk of schizophrenia in Iranian population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006, 141, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.Q.; Xing, Q.H.; Zheng, Y.L.; Li, S.; Gao, J.J.; He, G.; Guo, T.W.; Feng, G.Y.; Xu, F.; He, L. Association of AKT1 gene polymorphisms with risk of schizophrenia and with response to antipsychotics in the Chinese population. J. Clin. Psychiatry 2007, 68, 1358–1367. [Google Scholar] [CrossRef] [PubMed]
- Thiselton, D.L.; Vladimirov, V.I.; Kuo, P.H.; McClay, J.; Wormley, B.; Fanous, A.; O’Neill, F.A.; Walsh, D.; Van den Oord, E.J.; Kendler, K.S.; et al. AKT1 is associated with schizophrenia across multiple symptom dimensions in the Irish study of high density schizophrenia families. Biol. Psychiatry 2008, 63, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Karege, F.; Méary, A.; Perroud, N.; Jamain, S.; Leboyer, M.; Ballmann, E.; Fernandez, R.; Malafosse, A.; Schürhoff, F. Genetic overlap between schizophrenia and bipolar disorder: A study with AKT1 gene variants and clinical phenotypes. Schizophr. Res. 2012, 135, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Nohesara, S.; Ghadirivasfi, M.; Barati, M.; Ghasemzadeh, M.R.; Narimani, S.; Mousavi-Behbahani, Z.; Joghataei, M.; Soleimani, M.; Taban, M.; Mehrabi, S.; et al. Methamphetamine-induced psychosis is associated with DNA hypomethylation and increased expression of AKT1 and key dopaminergic genes. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2016, 171, 1180–1189. [Google Scholar] [CrossRef] [PubMed]
- Petryshen, T.L.; Middleton, F.A.; Kirby, A.; Aldinger, K.A.; Purcell, S.; Tahl, A.R.; Morley, C.P.; McGann, L.; Gentile, K.L.; Rockwell, G.N.; et al. Support for involvement of neuregulin 1 in schizophrenia pathophysiology. Mol. Psychiatry 2005, 10, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Law, A.J.; Kleinman, J.E.; Weinberger, D.R.; Weickert, C.S. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum. Mol. Genet. 2007, 16, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Hahn, C.G.; Wang, H.Y.; Cho, D.S.; Talbot, K.; Gur, R.E.; Berrettini, W.H.; Bakshi, K.; Kamins, J.; Borgmann-Winter, K.E.; Siegel, S.J.; et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat. Med. 2006, 12, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Xiong, W.C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. Nat. Rev. 2008, 9, 437–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seshadri, S.; Kamiya, A.; Yokota, Y.; Prikulis, I.; Kano, S.; Hayashi-Takagi, A.; Stanco, A.; Eom, T.Y.; Rao, S.; Ishizuka, K.; et al. Disrupted-in-Schizophrenia-1 expression is regulated by beta-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Proc. Natl. Acad. Sci. USA 2010, 107, 5622–5627. [Google Scholar] [CrossRef] [PubMed]
- Skirzewski, M.; Karavanova, I.; Shamir, A.; Erben, L.; Garcia-Olivares, J.; Shin, J.H.; Vullhorst, D.; Alvarez, V.A.; Amara, S.G.; Buonanno, A. ErbB4 Signaling in Dopaminergic Axonal Projections Increases Extracellular Dopamine Levels and Regulates Spatial/Working Memory Behaviors. Mol. Psychiatry 2017. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Murtie, J.C.; El-Khodor, B.F.; Edgar, N.; Sardi, S.P.; Hooks, B.M.; Benoit-Marand, M.; Chen, C.; Moore, H.; O’Donnell, P.; et al. Loss of erbB signaling in oligodendrocytes alters myelin and dopaminergic function, a potential mechanism for neuropsychiatric disorders. Proc. Natl. Acad. Sci. USA 2007, 104, 8131–8136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Tang, R.; Xiao, Z.; Shi, Y.; Feng, G.; Gu, N.; Shi, J.; Xing, Y.; Yan, L.; Sang, H.; et al. An investigation of the dihydropyrimidinase-like 2 (DPYSL2) gene in schizophrenia: Genetic association study and expression analysis. Int. J. Neuropsychopharmacol. 2006, 9, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Nakata, K.; Ujike, H.; Sakai, A.; Takaki, M.; Imamura, T.; Tanaka, Y.; Kuroda, S. The human dihydropyrimidinase-related protein 2 gene on chromosome 8p21 is associated with paranoid-type schizophrenia. Biol. Psychiatry 2003, 53, 571–576. [Google Scholar] [CrossRef]
- Fallin, M.D.; Lasseter, V.K.; Liu, Y.; Avramopoulos, D.; McGrath, J.; Wolyniec, P.S.; Nestadt, G.; Liang, K.Y.; Chen, P.L.; Valle, D.; et al. Linkage and association on 8p21.2-p21.1 in schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011, 156, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Pham, X.; Zhang, L.; Chen, P.L.; Burzynski, G.; McGaughey, D.M.; He, S.; McGrath, J.A.; Wolyniec, P.; Fallin, M.D.; et al. Functional variants in DPYSL2 sequence increase risk of schizophrenia and suggest a link to mTOR signaling. G3 2014, 5, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Chong, S.A.; Keefe, R. Psychosocial factors in the neurobiology of schizophrenia: A selective review. Ann. Acad. Med. Singap. 2009, 38, 402–406. [Google Scholar] [PubMed]
- Lee, H.; Joo, J.; Nah, S.S.; Kim, J.W.; Kim, H.K.; Kwon, J.T.; Lee, H.Y.; Kim, Y.O.; Kim, H.J. Changes in Dpysl2 expression are associated with prenatally stressed rat offspring and susceptibility to schizophrenia in humans. Int. J. Mol. Med. 2015, 35, 1574–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobeissy, F.H.; Warren, M.W.; Ottens, A.K.; Sadasivan, S.; Zhang, Z.; Gold, M.S.; Wang, K.K. Psychoproteomic analysis of rat cortex following acute methamphetamine exposure. J. Proteome Res. 2008, 7, 1971–1983. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Yamashita, N.; Kimura, A.; Kimura, Y.; Hirano, H.; Makihara, H.; Kawamoto, Y.; Jitsuki-Takahashi, A.; Yonezaki, K.; Takase, K.; et al. Comprehensive behavioral study and proteomic analyses of CRMP2-deficient mice. Genes Cells 2016, 21, 1059–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, X.; Sasamoto, K.; Nagai, J.; Yamazaki, Y.; Saito, K.; Goshima, Y.; Inoue, T.; Ohshima, T. Phosphorylation of CRMP2 by Cdk5 Regulates Dendritic Spine Development of Cortical Neuron in the Mouse Hippocampus. Neural Plast. 2016, 2016, 6790743. [Google Scholar] [CrossRef] [PubMed]
- Benavides, D.R.; Bibb, J.A. Role of Cdk5 in drug abuse and plasticity. Ann. N. Y. Acad. Sci. 2004, 1025, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Mlewski, E.C.; Arias, C.; Paglini, G. Association between the expression of amphetamine-induced behavioral sensitization and Cdk5/p35 activity in dorsal striatum. Behav. Neurosci. 2016, 130, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Lebel, M.; Patenaude, C.; Allyson, J.; Massicotte, G.; Cyr, M. Dopamine D1 receptor activation induces tau phosphorylation via Cdk5 and GSK3 signaling pathways. Neuropharmacology 2009, 57, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Cantrup, R.; Sathanantham, K.; Rushlow, W.J.; Rajakumar, N. Chronic hyperdopaminergic activity of schizophrenia is associated with increased ΔFosB levels and cdk-5 signaling in the nucleus accumbens. Neuroscience 2012, 222, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Ferreras, S.; Fernández, G.; Danelon, V.; Pisano, M.V.; Masseroni, L.; Chapleau, C.A.; Krapacher, F.A.; Mlewski, E.C.; Mascó, D.H.; Arias, C.; et al. Cdk5 Is Essential for Amphetamine to Increase Dendritic Spine Density in Hippocampal Pyramidal Neurons. Front. Cell. Neurosci. 2017, 11, 372. [Google Scholar] [CrossRef] [PubMed]
- Engmann, O.; Hortobágyi, T.; Pidsley, R.; Troakes, C.; Bernstein, H.G.; Kreutz, M.R.; Mill, J.; Nikolic, M.; Giese, K.P. Schizophrenia is associated with dysregulation of a Cdk5 activator that regulates synaptic protein expression and cognition. Brain 2011, 134, 2408–2421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Eissa, M.M.; Fiorentino, A.; Sharp, S.I.; O’Brien, N.L.; Wolfe, K.; Giaroli, G.; Curtis, D.; Bass, N.J.; McQuillin, A. Exome sequence analysis and follow up genotyping implicates rare ULK1 variants to be involved in susceptibility to schizophrenia. Ann. Hum. Genet. 2018, 82, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Horesh, Y.; Katsel, P.; Haroutunian, V.; Domany, E. Gene expression signature is shared by patients with Alzheimer’s disease and schizophrenia at the superior temporal gyrus. Eur. J. Neurol. 2011, 18, 410–424. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, P.W.; Wright, I.C.; Bullmore, E.T.; Brammer, M.; Howard, R.J.; Williams, S.C.; Shapleske, J.; Rossell, S.; David, A.S.; McGuire, P.K.; et al. Auditory hallucinations and the temporal cortical response to speech in schizophrenia: A functional magnetic resonance imaging study. Am. J. Psychiatry 1997, 154, 1676–1682. [Google Scholar] [CrossRef] [PubMed]
- Allen, P.; Larøi, F.; McGuire, P.K.; Aleman, A. The hallucinating brain: A review of structural and functional neuroimaging studies of hallucinations. Neurosci. Biobehav. Rev. 2008, 32, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.R.; Huxley-Jones, J.; Maycox, P.R.; Lennon, M.; Thornber, A.; Kelly, F.; Bates, S.; Taylor, A.; Reid, J.; Jones, N.; et al. Transcription and pathway analysis of the superior temporal cortex and anterior prefrontal cortex in schizophrenia. J. Neurosci. Res. 2011, 89, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Vinayagam, A.; Stelzl, U.; Foulle, R.; Plassmann, S.; Zenkner, M.; Timm, J.; Assmus, H.E.; Andrade-Navarro, M.A.; Wanker, E.E. A directed protein interaction network for investigating intracellular signal transduction. Sci. Signal. 2011, 4, rs8. [Google Scholar] [CrossRef] [PubMed]
- Dresner, E.; Agam, G.; Gozes, I. Activity-dependent neuroprotective protein (ADNP) expression level is correlated with the expression of the sister protein ADNP2: Deregulation in schizophrenia. Eur. Neuropsychopharmacol. 2011, 21, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Javitt, D.C.; Buchanan, R.W.; Keefe, R.S.; Kern, R.; McMahon, R.P.; Green, M.F.; Lieberman, J.; Goff, D.C.; Csernansky, J.G.; McEvoy, J.P.; et al. Effect of the neuroprotective peptide davunetide (AL-108) on cognition and functional capacity in schizophrenia. Schizophr. Res. 2012, 136, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Krishna, G.; Imarisio, S.; Saili, S.; O’Kane, C.J.; Rubinsztein, D.C. A rational mechanism for combination treatment of Huntington’s disease using lithium and rapamycin. Hum. Mol. Genet. 2008, 17, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.C.; Lin, C.C.; Chen, T.T.; Chen, J.Y.; Tsai, H.J.; Wang, C.Y.; Chen, S.Y. Clozapine induces autophagic cell death in non-small cell lung cancer cells. Cell. Physiol. Biochem. 2015, 35, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Sade, Y.; Toker, L.; Kara, N.Z.; Einat, H.; Rapoport, S.; Moechars, D.; Berry, G.T.; Bersudsky, Y.; Agam, G. IP3 accumulation and/or inositol depletion: Two downstream lithium’s effects that may mediate its behavioral and cellular changes. Transl. Psychiatry 2016, 6, e968. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Park, S.; Yu, H.S.; Ko, K.H.; Park, H.G.; Kim, Y.S. The antipsychotic agent clozapine induces autophagy via the AMPK-ULK1-Beclin1 signaling pathway in the rat frontal cortex. Prog. Neuropsychopharmacol. Biol. Psychiatry 2018, 81, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.D.; Jeong, E.I.; Nah, J.; Yoo, S.M.; Lee, W.J.; Kim, Y.; Moon, S.; Hong, S.H.; Jung, Y.K. Pimozide reduces toxic forms of tau in TauC3 mice via 5′ adenosine monophosphate-activated protein kinase-mediated autophagy. J. Neurochem. 2017, 142, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Park, S.J.; Kim, E.S.; Jo, Y.K.; Hong, J.; Cho, D.H. Sertindole, a potent antagonist at dopamine D₂ receptors, induces autophagy by increasing reactive oxygen species in SH-SY5Y neuroblastoma cells. Biol. Pharm. Bull. 2012, 35, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Charvin, D.; Roze, E.; Perrin, V.; Deyts, C.; Betuing, S.; Pagès, C.; Régulier, E.; Luthi-Carter, R.; Brouillet, E.; Déglon, N.; et al. Haloperidol protects striatal neurons from dysfunction induced by mutated huntingtin in vivo. Neurobiol. Dis. 2008, 29, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Höllerhage, M.; Goebel, J.N.; de Andrade, A.; Hildebrandt, T.; Dolga, A.; Culmsee, C.; Oertel, W.H.; Hengerer, B.; Höglinger, G.U. Trifluoperazine rescues human dopaminergic cells from wild-type α-synuclein-induced toxicity. Neurobiol. Aging 2014, 35, 1700–1711. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Jeong, H.J.; Liu, Q.F.; Oh, S.T.; Koo, B.S.; Kim, Y.; Chung, I.W.; Kim, Y.S.; Jeon, S. Clozapine Improves Memory Impairment and Reduces Aβ Level in the Tg-APPswe/PS1dE9 Mouse Model of Alzheimer’s Disease. Mol. Neurobiol. 2017, 54, 450–460. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Chung, S.; An, H.; Kim, J.; Seo, J.; Kim, D.H.; Yoon, S.Y. Haloperidol and clozapine block formation of autophagolysosomes in rat primary neurons. Neuroscience 2012, 209, 64–73. [Google Scholar] [CrossRef] [PubMed]



| Protein | Schizophrenia | Methamphetamine |
|---|---|---|
| DISC1 | [153,212,213,214,215] | [208,209] |
| Akt | [218,219,220,221,222,223,224] | [225] |
| NRG1/ErbB4 | [228,230,232] | [231] |
| CRMP2 | [236,238] | [239,240] |
| CDK5/p35 | [247] | [130,242,243] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryskalin, L.; Limanaqi, F.; Frati, A.; Busceti, C.L.; Fornai, F. mTOR-Related Brain Dysfunctions in Neuropsychiatric Disorders. Int. J. Mol. Sci. 2018, 19, 2226. https://doi.org/10.3390/ijms19082226
Ryskalin L, Limanaqi F, Frati A, Busceti CL, Fornai F. mTOR-Related Brain Dysfunctions in Neuropsychiatric Disorders. International Journal of Molecular Sciences. 2018; 19(8):2226. https://doi.org/10.3390/ijms19082226
Chicago/Turabian StyleRyskalin, Larisa, Fiona Limanaqi, Alessandro Frati, Carla L. Busceti, and Francesco Fornai. 2018. "mTOR-Related Brain Dysfunctions in Neuropsychiatric Disorders" International Journal of Molecular Sciences 19, no. 8: 2226. https://doi.org/10.3390/ijms19082226