Nitrogen Source Dependent Changes in Central Sugar Metabolism Maintain Cell Wall Assembly in Mitochondrial Complex I-Defective frostbite1 and Secondarily Affect Programmed Cell Death
Abstract
1. Introduction
2. Results
2.1. Characterization of fro1 Plants Cultured on Different Nitrogen Sources
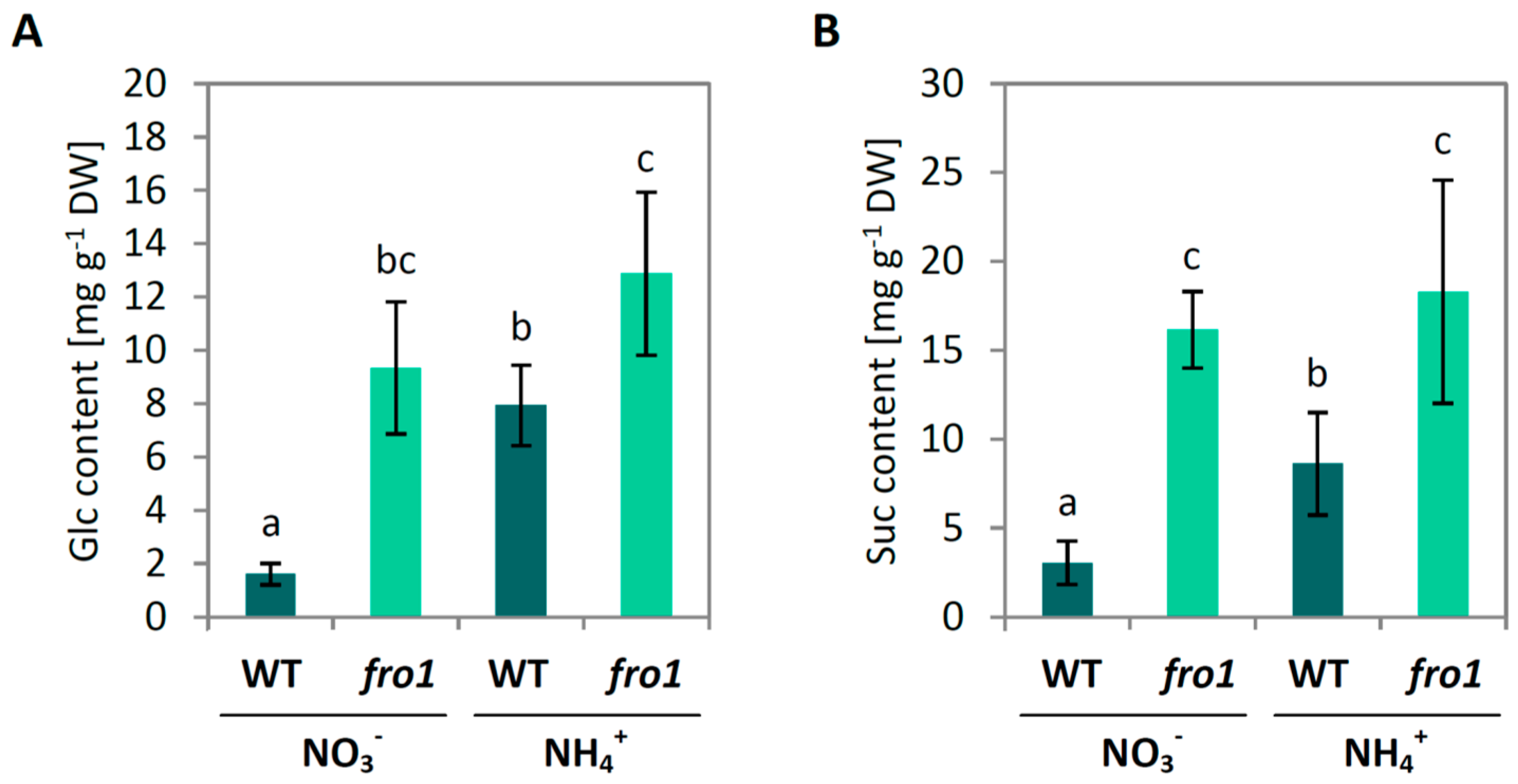
2.2. Sugar Metabolism in fro1
2.3. Analysis of Cell Size and Cell Wall Design in fro1
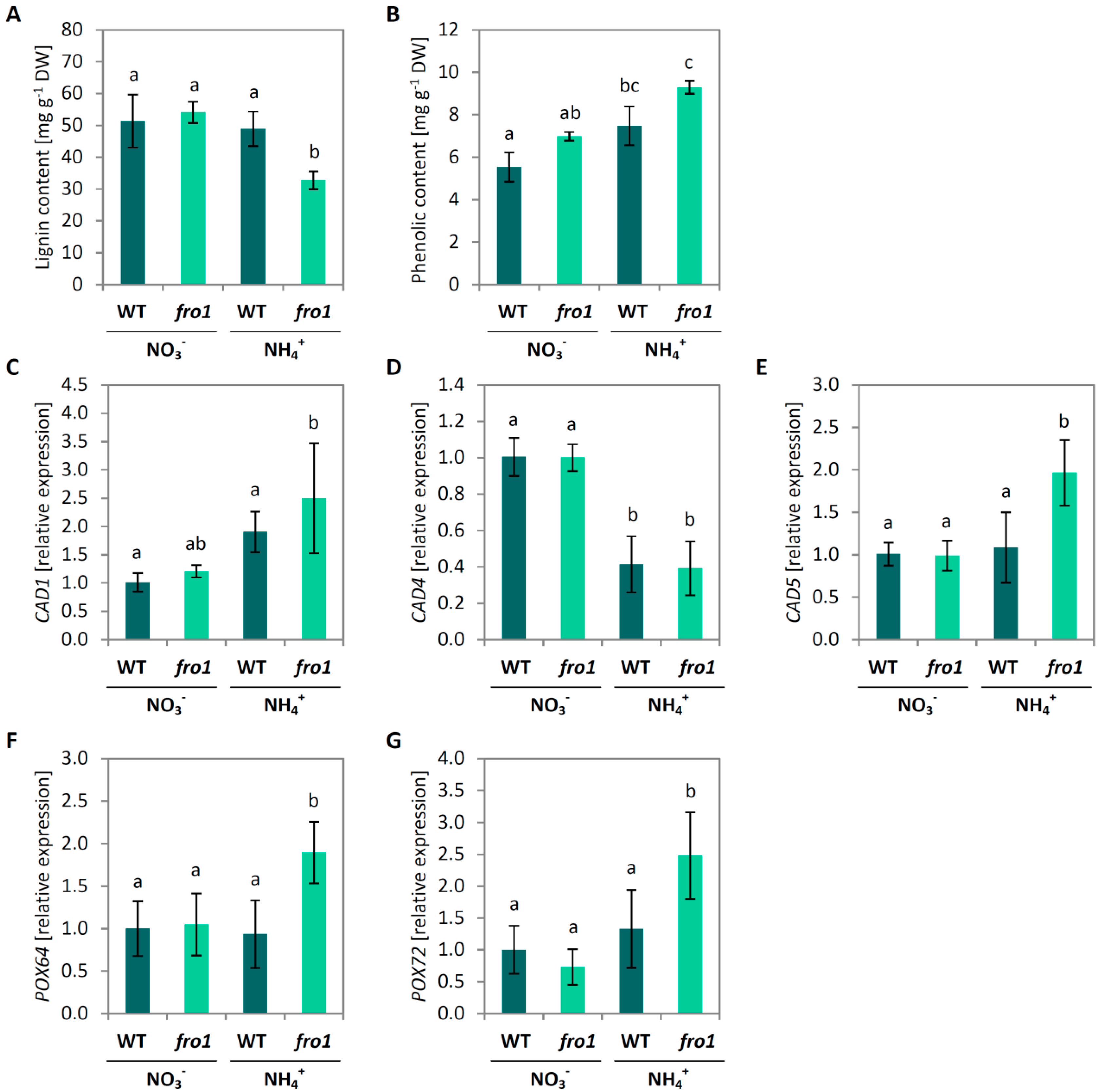
2.4. Cell Wall Composition in fro1
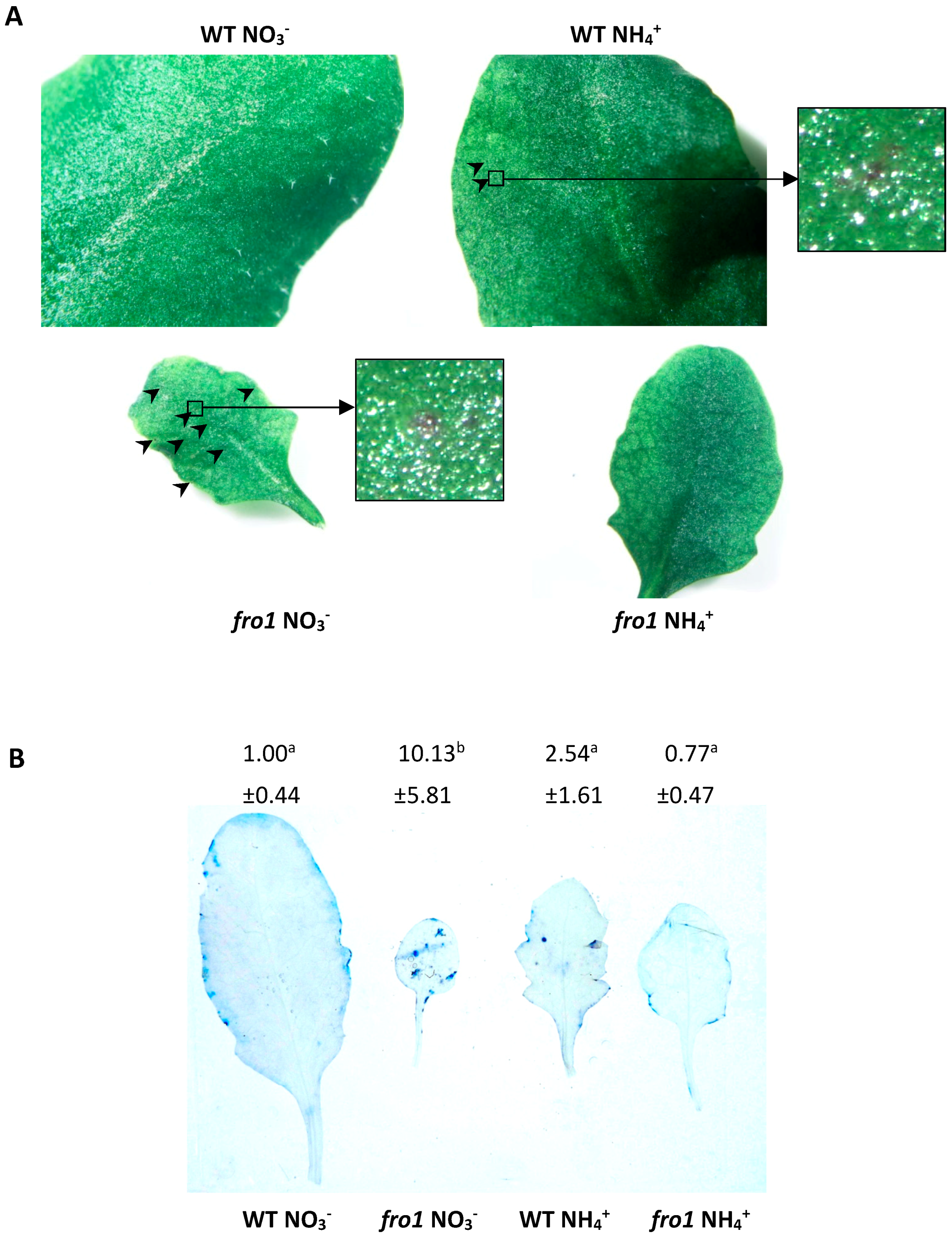
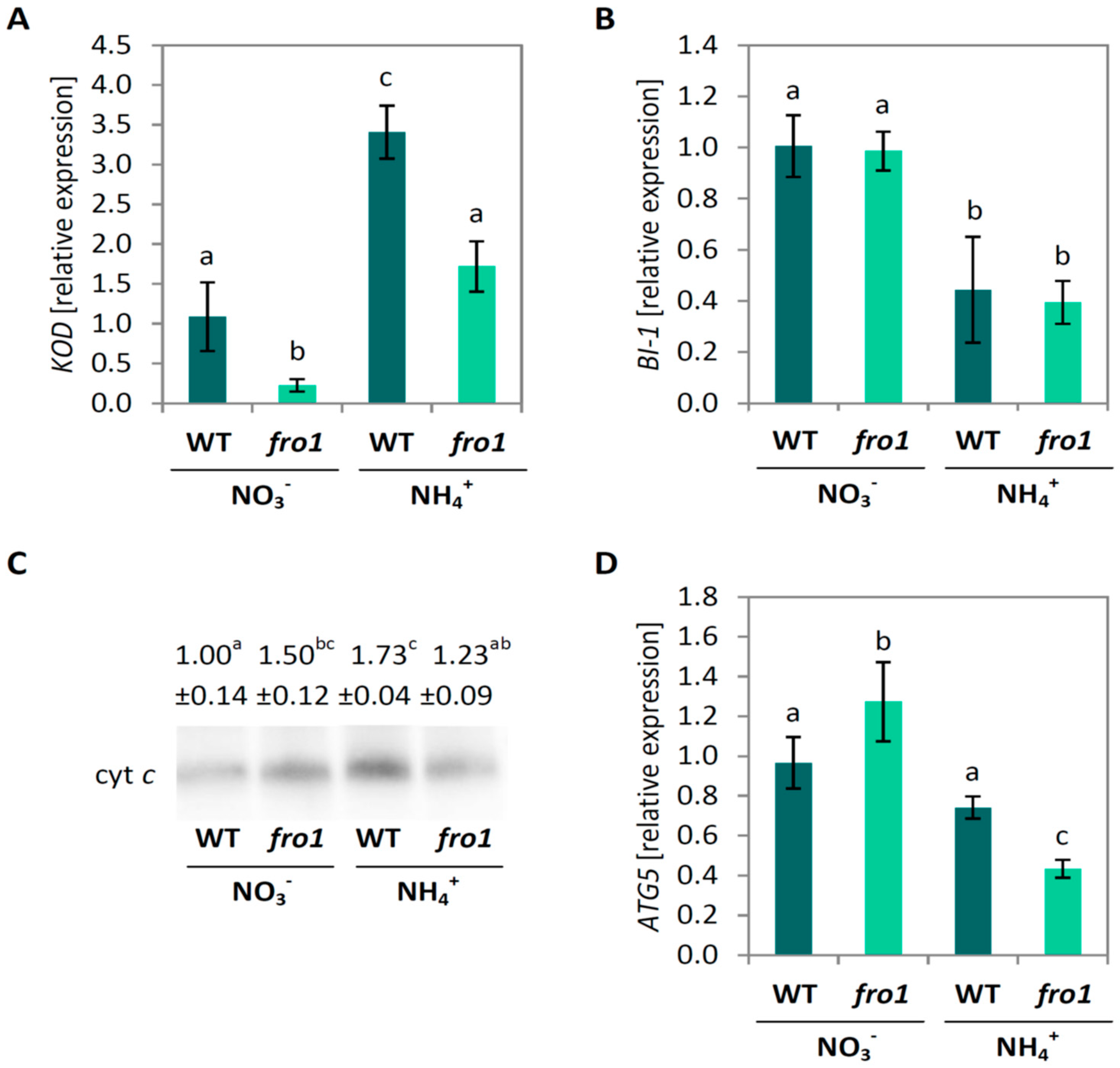
2.5. Analysis of Programmed Cell Death Markers in fro1
2.6. Reactive Oxygen Species Localization in fro1
2.7. Changes in Mitochondria-Related Signaling in fro1
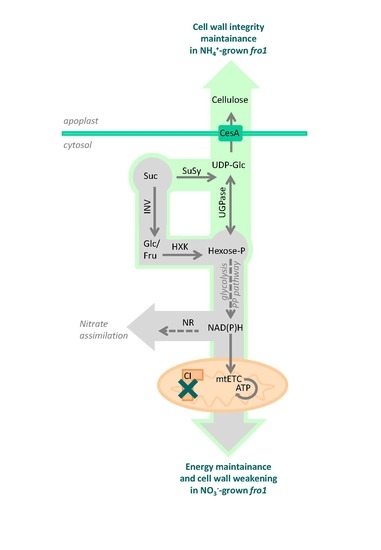
3. Discussion
3.1. Sugar Availability under Ammonium Nutrition May Limit Cell Wall Synthesis in WT but not in fro1 Plants
3.2. Fro1 Does not Show Significant Differences in the Pattern of Sugar Signaling
3.3. Ammonium Nutrition Mitigates PCD Occurrence in fro1 Plants
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Phenotype Analysis
4.3. Lesions Identification
4.4. Cell Wall Preparations, Cellulose, Lignin, and Phenol Content Assay
4.5. Determination of Sugars and Protein Level
4.6. Enzyme Activity Measurement and Protein Level Determination
4.7. Quantitative RT-PCR Analyses
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ATP | adenosine triphosphate |
| cyt c | cytochrome c |
| fro1 | frostbite1 |
| Glc | glucose |
| HXK | hexokinase |
| mtETC | mitochondrial electron transport chain |
| NADH | nicotinamide adenine dinucleotide, reduced |
| NADPH | nicotinamide adenine dinucleotide phosphate, reduced |
| PCD | programmed cell death |
| POX | peroxidase |
| ROS | reactive oxygen species |
| Suc | sucrose |
| TCA | tricarboxylic acid |
| UGPase | UDP-glucose pyrophosphorylase |
| WT | wild-type |
References
- Bloom, A.J. Photorespiration and nitrate assimilation: A major intersection between plant carbon and nitrogen. Photosynth. Res. 2015, 123, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Foyer, C.H. A re-evaluation of the ATP:NADPH budget during C3 photosynthesis: A contribution from nitrate assimilation and its associated respiratory activity? J. Exp. Bot. 1998, 49, 1895–1908. [Google Scholar] [CrossRef]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Fait, A.; Nunes-Nesi, A.; Williams, T.; Fernie, A.R. The mitochondrion: An integration point of cellular metabolism and signalling. CRC Crit. Rev. Plant Sci. 2007, 26, 17–43. [Google Scholar] [CrossRef]
- Gardeström, P.; Igamberdiev, A.U. The origin of cytosolic ATP in photosynthetic cells. Physiol. Plant. 2016, 157, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.; De Clercq, I.; Van Aken, O.; Law, S.R.; Ivanova, A.; Willems, P.; Giraud, E.; Van Breusegem, F.; Whelan, J. Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol. Plant 2014, 7, 1075–1093. [Google Scholar] [CrossRef] [PubMed]
- Crawford, T.; Lehotai, N.; Strand, Å. The role of retrograde signals during plant stress responses. J. Exp. Bot. 2017, 69, 2783–2795. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovska, M.; Vanlerberghe, G.C. Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ. 2013, 36, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Van Aken, O.; Schwarzländer, M.; Belt, K.; Millar, A.H. The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol. 2016, 171, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Schertl, P.; Braun, H.P. Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 2014, 5, 163. [Google Scholar] [CrossRef] [PubMed]
- Meyer, E.H.; Tomaz, T.; Carroll, A.J.; Estavillo, G.; Delannoy, E.; Tanz, S.K.; Small, I.D.; Pogson, B.J.; Millar, A.H. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 2009, 151, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Rasmusson, A.G.; Geisler, D.A.; Møller, I.M. The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 2008, 8, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Klodmann, J.; Braun, H.P. Proteomic approach to characterize mitochondrial complex I from plants. Phytochemistry 2011, 72, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Pla, M.; Mathieu, C.; De Paepe, R.; Chétrit, P.; Vedel, F. Deletion of the last two exons of the mitochondrial nad7 gene results in lack of the NAD7 polypeptide in a Nicotiana sylvestris CMS mutant. Mol. Gen. Genet. 1995, 248, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Gutierres, S.; Sabar, M.; Lelandais, C.; Chetrit, P.; Diolez, P.; Degand, H.; Boutry, M.; Vedel, F.; de Kouchkovsky, Y.; De Paepe, R. Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc. Natl. Acad. Sci. USA 1997, 94, 3436–3441. [Google Scholar] [CrossRef] [PubMed]
- Vidal, G.; Ribas-Carbó, M.; Garmier, M.; Dubertret, G.; Rasmusson, A.G.; Mathieu, C.; Foyer, C.H.; De Paepe, R. Lack of respiratory chain complex I impairs alternative oxidase engagement and modulates redox signaling during elicitor-induced cell death in Tobacco. Plant Cell 2007, 19, 640–655. [Google Scholar] [CrossRef] [PubMed]
- Marienfeld, J.R.; Newton, K.J. The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics 1994, 138, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Yamato, K.T.; Newton, K.J. Heteroplasmy and homoplasmy for maize mitochondrial mutants: A rare homoplasmic nad4 deletion mutant plant. J. Hered. 1999, 90, 369–373. [Google Scholar] [CrossRef]
- Lilly, J.W.; Bartoszewski, G.; Malepszy, S.; Havey, M.J. A major deletion in the cucumber mitochondrial genome sorts with the MSC phenotype. Curr. Genet. 2001, 40, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Juszczuk, I.M.; Flexas, J.; Szal, B.; Dąbrowska, Z.; Ribas-Carbó, M.; Rychter, A.M. Effect of mitochondrial genome rearrangement on respiratory activity, photosynthesis, photorespiration and energy status of MSC16 cucumber (Cucumis sativus) mutant. Physiol. Plant. 2007, 131, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Kühn, K.; Obata, T.; Feher, K.; Bock, R.; Fernie, A.R.; Meyer, E.H. Complete mitochondrial complex I deficiency induces an up-regulation of respiratory fluxes that is abolished by traces of functional complex I. Plant Physiol. 2015, 168, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Fromm, S.; Senkler, J.; Eubel, H.; Peterhänsel, C.; Braun, H.P. Life without complex I: Proteome analyses of an Arabidopsis mutant lacking the mitochondrial NADH dehydrogenase complex. J. Exp. Bot. 2016, 67, 3079–3093. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Qin, G.; Kang, D.; Chen, Z.; Gu, H.; Qu, L.J. A nuclear-encoded mitochondrial gene AtCIB22 is essential for plant development in Arabidopsis. J. Genet. Genom. 2010, 37, 667–683. [Google Scholar] [CrossRef]
- de Longevialle, A.F.; Meyer, E.H.; Andrés, C.; Taylor, N.L.; Lurin, C.; Millar, A.H.; Small, I.D. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 2007, 19, 3256–3265. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, N.; Sakurai, N. A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial NAD4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Bezawork-Geleta, A.; Kolton, M.; Maayan, I.; Belausov, E.; Levy, M.; Mett, A.; Gidoni, D.; Shaya, F.; Ostersetzer-Biran, O. AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 2009, 15, 2299–2311. [Google Scholar] [CrossRef] [PubMed]
- Kühn, K.; Carrie, C.; Giraud, E.; Wang, Y.; Meyer, E.H.; Narsai, R.; des Francs-Small, C.C.; Zhang, B.; Murcha, M.W.; Whelan, J. The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J. 2011, 67, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.W.; Wang, H.J.; Hsieh, M.H.; Hsieh, H.L.; Jauh, G.Y. Arabidopsis mTERF15 is required for mitochondrial nad2 intron 3 splicing and functional complex I activity. PLoS ONE. 2014, 9, e112360. [Google Scholar] [CrossRef] [PubMed]
- Koprivova, A.; des Francs-Small, C.C.; Calder, G.; Mugford, S.T.; Tanz, S.; Lee, B.-R.; Zechmann, B.; Small, I.; Kopriva, S. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J. Biol. Chem. 2010, 285, 32192–32199. [Google Scholar] [CrossRef] [PubMed]
- Brangeon, J.; Sabar, M.; Gutierres, S.; Combettes, B.; Bove, J.; Gendy, C.; Chetrit, P.; des Francs-Small, C.C.; Pla, M.; Vedel, F.; et al. Defective splicing of the first nad4 intron is associated with lack of several complex I subunits in the Nicotiana sylvestris NMS1 nuclear mutant. Plant J. 2000, 21, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Sabar, M.; De Paepe, R.; de Kouchkovsky, Y. Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol. 2000, 124, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Juszczuk, I.M.; Szal, B.; Rychter, A.M. Oxidation–reduction and reactive oxygen species homeostasis in mutant plants with respiratory chain complex I dysfunction. Plant Cell Environ. 2012, 35, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Subrahmanian, N.; Remacle, C.; Hamel, P.P. Plant mitochondrial complex I composition and assembly: A review. Biochim. Biophys. Acta 2016, 1857, 1001–1014. [Google Scholar] [CrossRef] [PubMed]
- Szal, B.; Dąbrowska, Z.; Malmberg, G.; Gardeström, P.; Rychter, A.M. Changes in energy status of leaf cells as the consequence of mitochondrial genome rearrangement. Planta 2008, 227, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Florez-Sarasa, I.; Ostaszewska, M.; Galle, A.; Flexas, J.; Rychter, A.M.; Ribas-Carbó, M. Changes of alternative oxidase activity, capacity and protein content in leaves of Cucumis sativus wild-type and MSC16 mutant grown under different light intensities. Physiol. Plant. 2009, 137, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Dutilleul, C.; Lelarge, C.; Prioul, J.L.; De Paepe, R.; Foyer, C.H.; Noctor, G. Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol. 2005, 139, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Szal, B.; Łukawska, K.; Zdolińska, I.; Rychter, A.M. Chilling stress and mitochondrial genome rearrangement in the MSC16 cucumber mutant affect the alternative oxidase and antioxidant defense system to a similar extent. Physiol. Plant. 2009, 137, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Carrie, C.; Giraud, E.; Elhafez, D.; Narsai, R.; Duncan, O.; Whelan, J.; Murcha, M.W. Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 2012, 24, 2675–2695. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lyu, W.; Berkowitz, O.; Radomiljac, J.D.; Law, S.R.; Murcha, M.W.; Carrie, C.; Teixeira, P.F.; Kmiec, B.; Duncan, O.; et al. Inactivation of mitochondrial complex I induces the expression of a twin cysteine protein that targets and affects cytosolic, chloroplastidic and mitochondrial function. Mol. Plant 2016, 9, 696–710. [Google Scholar] [CrossRef] [PubMed]
- Garmier, M.; Dutilleul, C.; Mathieu, C.; Chétrit, P.; Boccara, M.; De Paepe, R. Changes in antioxidant expression and harpin-induced hypersensitive response in a Nicotiana sylvestris mitochondrial mutant. Plant Physiol. Biochem. 2002, 40, 561–566. [Google Scholar] [CrossRef]
- Dutilleul, C.; Garmier, M.; Noctor, G.; Mathieu, C.; Chétrit, P.; Foyer, C.H.; de Paepe, R. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 2003, 15, 1212–1226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Norberg, F.E.B.; Szilágyi, A.; De Paepe, R.; Åkerlund, H.E.; Rasmusson, A.G. The mitochondrial external NADPH dehydrogenase modulates the leaf NADPH/NADP+ ratio in transgenic Nicotiana sylvestris. Plant Cell Physiol. 2008, 49, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Karpova, O.V.; Kuzmin, E.V.; Elthon, T.E.; Newton, K.J. Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 2002, 14, 3271–3284. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, E.V.; Karpova, O.V.; Elthon, T.E.; Newton, K.J. Mitochondrial respiratory deficiencies signal up-regulation of genes for heat shock proteins. J. Biol. Chem. 2004, 279, 20672–20677. [Google Scholar] [CrossRef] [PubMed]
- Tarasenko, V.I.; Garnik, E.Y.; Konstantinov, Y.M. Characterization of Arabidopsis mutant with inactivated gene coding for Fe-S subunit of mitochondrial respiratory chain complex I. Russ. J. Plant Physiol. 2010, 57, 392–400. [Google Scholar] [CrossRef]
- Lee, B.H.; Lee, H.; Xiong, L.; Zhu, J.K. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 2002, 14, 1235–1251. [Google Scholar] [CrossRef] [PubMed]
- Podgórska, A.; Ostaszewska, M.; Gardeström, P.; Rasmusson, A.G.; Szal, B. In comparison with nitrate nutrition, ammonium nutrition increases growth of the frostbite1 Arabidopsis mutant. Plant Cell Environ. 2015, 38, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Britto, D.T.; Kronzucker, H.J. NH4+ toxicity in higher plants: A critical review. J. Plant Physiol. 2002, 159, 567–584. [Google Scholar] [CrossRef]
- Bittsánszky, A.; Pilinszky, K.; Gyulai, G.; Komives, T. Overcoming ammonium toxicity. Plant Sci. 2015, 231, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Walch-Liu, P.; Neumann, G.; Bangerth, F.; Engels, C. Rapid effects of nitrogen form on leaf morphogenesis in tobacco. J. Exp. Bot. 2000, 51, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; von Wirén, N. Ammonium as a signal for physiological and morphological responses in plants. J. Exp. Bot. 2017, 68, 2581–2592. [Google Scholar] [CrossRef] [PubMed]
- Gerendás, J.; Zhu, Z.; Bendixen, R.; Ratcliffe, R.G.; Sattelmacher, B. Physiological and biochemical processes related to ammonium toxicity in higher plants. J. Plant Nutr. Soil Sci. 1997, 160, 239–251. [Google Scholar] [CrossRef]
- Britto, D.T.; Kronzucker, H.J. Ecological significance and complexity of N-source preference in plants. Ann. Bot. 2013, 112, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Szal, B.; Podgórska, A. The role of mitochondria in leaf nitrogen metabolism. Plant Cell Environ. 2012, 35, 1756–1768. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Schinner, K.; Sattelmacher, B.; Hansen, U.P. Different apparent CO2 compensation points in nitrate-and ammonium-grown Phaseolus vulgaris and the relationship to non-photorespiratory CO2 evolution. Physiol. Plant. 2005, 123, 288–301. [Google Scholar] [CrossRef]
- Escobar, M.A.; Geisler, D.A.; Rasmusson, A.G. Reorganization of the alternative pathways of the Arabidopsis respiratory chain by nitrogen supply: Opposing effects of ammonium and nitrate. Plant J. 2006, 45, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Podgórska, A.; Gieczewska, K.; Łukawska-Kuźma, K.; Rasmusson, A.G.; Gardeström, P.; Szal, B. Long-term ammonium nutrition of Arabidopsis increases the extrachloroplastic NAD(P)H/NAD(P)+ ratio and mitochondrial reactive oxygen species level in leaves but does not impair photosynthetic capacity. Plant Cell Environ. 2013, 36, 2034–2045. [Google Scholar] [CrossRef] [PubMed]
- Podgórska, A.; Burian, M.; Gieczewska, K.; Ostaszewska-Bugajska, M.; Zebrowski, J.; Solecka, D.; Szal, B. Altered cell wall plasticity can restrict plant growth under ammonium nutrition. Front. Plant Sci. 2017, 8, 1344. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.Y.; Wu, H.M. THESEUS 1, FERONIA and relatives: A family of cell wall-sensing receptor kinases? Curr. Opin. Plant Biol. 2011, 14, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Desprez, T.; Juraniec, M.; Crowell, E.F.; Jouy, H.; Pochylova, Z.; Parcy, F.; Hofte, H.; Gonneau, M.; Vernhettes, S. Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 15572–15577. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.H. Functional analysis of a cinnamyl alcohol dehydrogenase involved in lignin biosynthesis in wheat. J. Exp. Bot. 2010, 61, 2735–2744. [Google Scholar] [CrossRef] [PubMed]
- Blanvillain, R.; Young, B.; Cai, Y.; Hecht, V.; Varoquaux, F.; Delorme, V.; Lancelin, J.-M.; Delseny, M.; Gallois, P. The Arabidopsis peptide kiss of death is an inducer of programmed cell death. EMBO J. 2011, 30, 1173–1183. [Google Scholar] [CrossRef] [PubMed]
- Borysiuk, K.; Ostaszewska-Bugajska, M.; Vaultier, M.N.; Hasenfratz-Sauder, M.P.; Szal, B. Enhanced formation of methylglyoxal-derived advanced glycation end products in Arabidopsis under ammonium nutrition. Front. Plant Sci. 2018, 9, 667. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Chung, M.S.; Ju, H.W.; Na, H.S.; Lee, D.J.; Cheong, H.S.; Kim, C.S. Physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 1, a plasma membrane protein involved in oxidative stress. J. Plant Res. 2011, 124, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Lim, J.H.; Ahn, C.S.; Park, K.; Kim, G.T.; Kim, W.T.; Pai, H.S. Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 2006, 18, 2341–2355. [Google Scholar] [CrossRef] [PubMed]
- Jossier, M.; Bouly, J.-P.; Meimoun, P.; Arjmand, A.; Lessard, P.; Hawley, S.; Grahame Hardie, D.; Thomas, M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 2009, 59, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Endler, A.; Persson, S. Cellulose synthases and synthesis in Arabidopsis. Mol. Plant 2011, 4, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Verbančič, J.; Lunn, J.E.; Stitt, M.; Persson, S. Carbon supply and the regulation of cell wall synthesis. Mol. Plant 2018, 11, 75–94. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Paredez, A.; Carroll, A.; Palsdottir, H.; Doblin, M.; Poindexter, P.; Khitrov, N.; Auer, M.; Somerville, C.R. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 15566–15571. [Google Scholar] [CrossRef] [PubMed]
- Kleczkowski, L.A.; Geisler, M.; Ciereszko, I.; Johansson, H. UDP-glucose pyrophosphorylase. An old protein with new tricks. Plant Physiol. 2004, 134, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.G. Cellulose biosynthesis and deposition in higher plants. New Phytol. 2008, 178, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Timmers, J.; Vernhettes, S.; Desprez, T.; Vincken, J.P.; Visser, R.G.; Trindade, L.M. Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. FEBS Lett. 2009, 583, 978–982. [Google Scholar] [CrossRef] [PubMed]
- Marjamaa, K.; Kukkola, E.M.; Fagerstedt, K.V. The role of xylem class III peroxidases in lignification. J. Exp. Bot. 2009, 60, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, N.; Kaneta, T.; Sato, S.; Sato, Y. Analysis of expression profiles of three peroxidase genes associated with lignification in Arabidopsis thaliana. Physiol. Plant. 2009, 136, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pérez, F.; Pomar, F.; Pedreño, M.A.; Novo-Uzal, E. Suppression of Arabidopsis peroxidase 72 alters cell wall and phenylpropanoid metabolism. Plant Sci. 2015, 239, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kim, K.W.; Cho, M.H.; Franceschi, V.R.; Davin, L.B.; Lewis, N.G. Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: Lessons for database annotations? Phytochemistry 2007, 68, 1957–1974. [Google Scholar] [CrossRef] [PubMed]
- Liszkay, A.; Kenk, B.; Schopfer, P. Evidence for the involvement of cell wall peroxidase in the generation of hydroxyl radicals mediating extension growth. Planta 2003, 217, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Podgórska, A.; Burian, M.; Szal, B. Extra-cellular but extra-ordinarily important for cells: Apoplastic reactive oxygen species metabolism. Front. Plant Sci. 2017b, 8, 1353. [Google Scholar] [CrossRef] [PubMed]
- Seifert, G.J.; Blaukopf, C. Irritable walls: The plant extracellular matrix and signaling. Plant Physiol. 2010, 153, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Tenhaken, R. Cell wall remodeling under abiotic stress. Front. Plant Sci. 2014, 5, 771. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Höfte, H. Growth control: A saga of cell walls, ROS, and peptide receptors. Plant Cell 2014, 26, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, H.-M.; Cheung, A.Y. FERONIA and her pals: Functions and mechanisms. Plant Physiol. 2016, 171, 2379–2392. [Google Scholar] [CrossRef] [PubMed]
- Nissen, K.S.; Willats, W.G.T.; Malinovsky, F.G. Understanding CrRLK1L function: Cell walls and growth control. Trends Plant Sci. 2016, 21, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Vanderhaeghen, R.; Cools, T.; Wang, Y.; De Clercq, I.; Leroux, O.; Nguyen, L.; Belt, K.; Millar, A.H.; Audenaert, D.; et al. Mitochondrial defects confer tolerance against cellulose deficiency. Plant Cell 2016, 28, 2276–2290. [Google Scholar] [CrossRef] [PubMed]
- Harrington, G.N.; Bush, D.R. The bifunctional role of hexokinase in metabolism and glucose signaling. Plant Cell 2003, 15, 2493–2496. [Google Scholar] [CrossRef] [PubMed]
- Sheen, J. Master regulators in plant glucose signaling networks. J. Plant Biol. 2014, 57, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.H.; Yoo, S.D.; Sheen, J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 2006, 127, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.C.; León, P.; Zhou, L.; Sheen, J. Hexokinase as a sugar sensor in higher plants. Plant Cell 1997, 9, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.; Zhou, L.; Rolland, F.; Hall, Q.; Cheng, W.H.; Liu, Y.X.; Hwang, I.; Jones, T.; Sheen, J. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 2003, 300, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Heinzel, N.; Giese, J.; Koeber, J.; Melzer, M.; Rutten, T.; Von Wirén, N.; Sonnewald, U.; Hajirezaei, M. Dual function of hexokinases in tobacco plants. Plant Cell Environ. 2013, 36, 1311–1327. [Google Scholar] [CrossRef] [PubMed]
- Simon, N.M.L.; Kusakina, J.; Fernández-López, Á.; Chembath, A.; Belbin, F.E.; Dodd, A.N. The energy-signaling hub SnRK1 is important for sucrose-induced hypocotyl elongation. Plant Physiol. 2018, 176, 1299–1310. [Google Scholar] [CrossRef] [PubMed]
- Wurzinger, B.; Nukarinen, E.; Nägele, T.; Weckwerth, W.; Teige, M. The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiol. 2018, 176, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–942. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Su, Z.-Z.; Huang, L.; Xia, F.-N.; Qi, H.; Xie, L.-J.; Xiao, S.; Chen, Q.-F. The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front. Plant Sci. 2017, 8, 1201. [Google Scholar] [CrossRef] [PubMed]
- Pedrotti, L.; Weiste, C.; Nägele, T.; Wolf, E.; Lorenzin, F.; Dietrich, K.; Mair, A.; Weckwerth, W.; Teige, M.; Baena-González, E.; et al. Snf1-RELATED KINASE1-controlled C/S1-bZIP signaling activates alternative mitochondrial metabolic pathways to ensure plant survival in extended darkness. Plant Cell 2018, 30, 495–509. [Google Scholar] [CrossRef] [PubMed]
- Ghillebert, R.; Swinnen, E.; Wen, J.; Vandesteene, L.; Ramon, M.; Norga, K.; Rolland, F.; Winderickx, J. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: Structure, function and regulation. FEBS J. 2011, 278, 3978–3990. [Google Scholar] [CrossRef] [PubMed]
- Wurzinger, B.; Mair, A.; Fischer-Schrader, K.; Nukarinen, E.; Roustan, V.; Weckwerth, W.; Teige, M. Redox state-dependent modulation of plant SnRK1 kinase activity differs from AMPK regulation in animals. FEBS Lett. 2017, 591, 3625–3636. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. Arabidopsis Bax inhibitor-1 functions as an attenuator of biotic and abiotic types of cell death. Plant J. 2006, 45, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Lam, E. BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem. 2008, 283, 3200–3210. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, W.G.; Beers, E.P.; Dangl, J.L.; Franklin-Tong, V.E.; Gallois, P.; Hara-Nishimura, I.; Jones, A.M.; Kawai-Yamada, M.; Lam, E.; Mundy, J.; et al. Morphological classification of plant cell deaths. Cell Death Differ. 2011, 18, 1241. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.I.; Cho, H.J.; Jung, J.H.; Yoshimoto, K.; Park, O.K. The RabGTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 2010, 64, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Lasa, B.; Frechilla, S.; Apricio-Tejo, P.M.; Lamsfus, C. Alternative pathway respiration is associated with ammonium ion sensitivity in spinach and pea plants. Plant Growth Regul. 2002, 37, 49–55. [Google Scholar] [CrossRef]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Görlach, J. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 2002, 13, 1499–1510. [Google Scholar] [CrossRef]
- Pogány, M.; von Rad, U.; Grün, S.; Dongó, A.; Pintye, A.; Simoneau, P.; Bahnweg, G.; Kiss, L.; Barna, B.; Durner, J. Dual roles of reactive oxygen species and NADPH oxidase RBOHD in an Arabidopsis-Alternaria pathosystem. Plant Physiol. 2009, 151, 1459–1475. [Google Scholar] [CrossRef] [PubMed]
- Weremczuk, A.; Ruszczyńska, A.; Bulska, E.; Antosiewicz, D.M. NO-Dependent programmed cell death is involved in the formation of Zn-related lesions in tobacco leaves. Metallomics 2017, 19, 924–935. [Google Scholar] [CrossRef] [PubMed]
- Solecka, D.; Zebrowski, J.; Kacperska, A. Are pectins involved in cold acclimation and de-acclimation of winter oil-seed rape plants? Ann. Bot. 2008, 101, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Updegraff, D. Semimicro determination of cellulose in biological materials. Anal. Biochem. 1969, 32, 420–424. [Google Scholar] [CrossRef]
- Johnson, D.B.; Moore, W.E.; Zank, L.C. The spectrophotometric determination of lignin in small wood samples. Tappi 1961, 44, 793–798. [Google Scholar] [CrossRef]
- Hatfield, R.D.; Grabber, J.; Ralph, J.; Brei, K. Using the acetylbromide assay to determine lignin concentrations in herbaceous plants: Some cautionarynotes. J. Agric. Food Chem. 1999, 47, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Forrest, G.I.; Bendall, D.S. The distribution of polyphenols in the tea plant (Camellia sinensis L.). Biochem. J. 1969, 113, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Solecka, D.; Boudet, A.M.; Kacperska, A. Phenylpropanoid and anthocyanin changes in low-temperature treated winter ilseed rape leaves. Plant Physiol. Biochem. 1999, 37, 491–496. [Google Scholar] [CrossRef]
- Szal, B.; Jastrzębska, A.; Kulka, M.; Leśniak, K.; Podgórska, A.; Pärnik, T.; Ivanova, H.; Keerberg, O.; Gardeström, P.; Rychter, A.M. Influence of mitochondrial genome rearrangement on cucumber leaf carbon and nitrogen metabolism. Planta 2010, 232, 1371–1382. [Google Scholar] [CrossRef] [PubMed]
- Kunst, A.; Draeger, B.; Ziegenhorn, J. Colorimertic methods with glucose oxidase and peroxidase. In Methods in Enzymatic Analysis, Metabolites: Carbohydrates; Bergmeyer, H.U., Bergmeyer, J., Grassl, M., Eds.; Verlag Chemie: Weinheim, Germany, 1985; pp. 178–185. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Huber, S.C.; Akazawa, T. A nove1 sucrose synthase pathway for sucrose degradation in cultured cells. Plant Physiol. 1986, 81, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Escobar, M.A.; Franklin, K.A.; Svensson, A.S.; Salter, M.G.; Whitelam, G.C.; Rasmusson, A.G. Light regulation of the Arabidopsis respiratory chain. Multiple discrete photoreceptor responses contribute to induction of type II NAD(P)H dehydrogenase genes. Plant Physiol. 2004, 136, 2710–2721. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, MK.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Podgórska, A.; Ostaszewska-Bugajska, M.; Tarnowska, A.; Burian, M.; Borysiuk, K.; Gardeström, P.; Szal, B. Nitrogen Source Dependent Changes in Central Sugar Metabolism Maintain Cell Wall Assembly in Mitochondrial Complex I-Defective frostbite1 and Secondarily Affect Programmed Cell Death. Int. J. Mol. Sci. 2018, 19, 2206. https://doi.org/10.3390/ijms19082206
Podgórska A, Ostaszewska-Bugajska M, Tarnowska A, Burian M, Borysiuk K, Gardeström P, Szal B. Nitrogen Source Dependent Changes in Central Sugar Metabolism Maintain Cell Wall Assembly in Mitochondrial Complex I-Defective frostbite1 and Secondarily Affect Programmed Cell Death. International Journal of Molecular Sciences. 2018; 19(8):2206. https://doi.org/10.3390/ijms19082206
Chicago/Turabian StylePodgórska, Anna, Monika Ostaszewska-Bugajska, Agata Tarnowska, Maria Burian, Klaudia Borysiuk, Per Gardeström, and Bożena Szal. 2018. "Nitrogen Source Dependent Changes in Central Sugar Metabolism Maintain Cell Wall Assembly in Mitochondrial Complex I-Defective frostbite1 and Secondarily Affect Programmed Cell Death" International Journal of Molecular Sciences 19, no. 8: 2206. https://doi.org/10.3390/ijms19082206
APA StylePodgórska, A., Ostaszewska-Bugajska, M., Tarnowska, A., Burian, M., Borysiuk, K., Gardeström, P., & Szal, B. (2018). Nitrogen Source Dependent Changes in Central Sugar Metabolism Maintain Cell Wall Assembly in Mitochondrial Complex I-Defective frostbite1 and Secondarily Affect Programmed Cell Death. International Journal of Molecular Sciences, 19(8), 2206. https://doi.org/10.3390/ijms19082206