Transcriptome Analysis Provides Insight into the Molecular Mechanisms Underlying gametophyte factor 2-Mediated Cross-Incompatibility in Maize
Abstract
:1. Introduction
2. Results
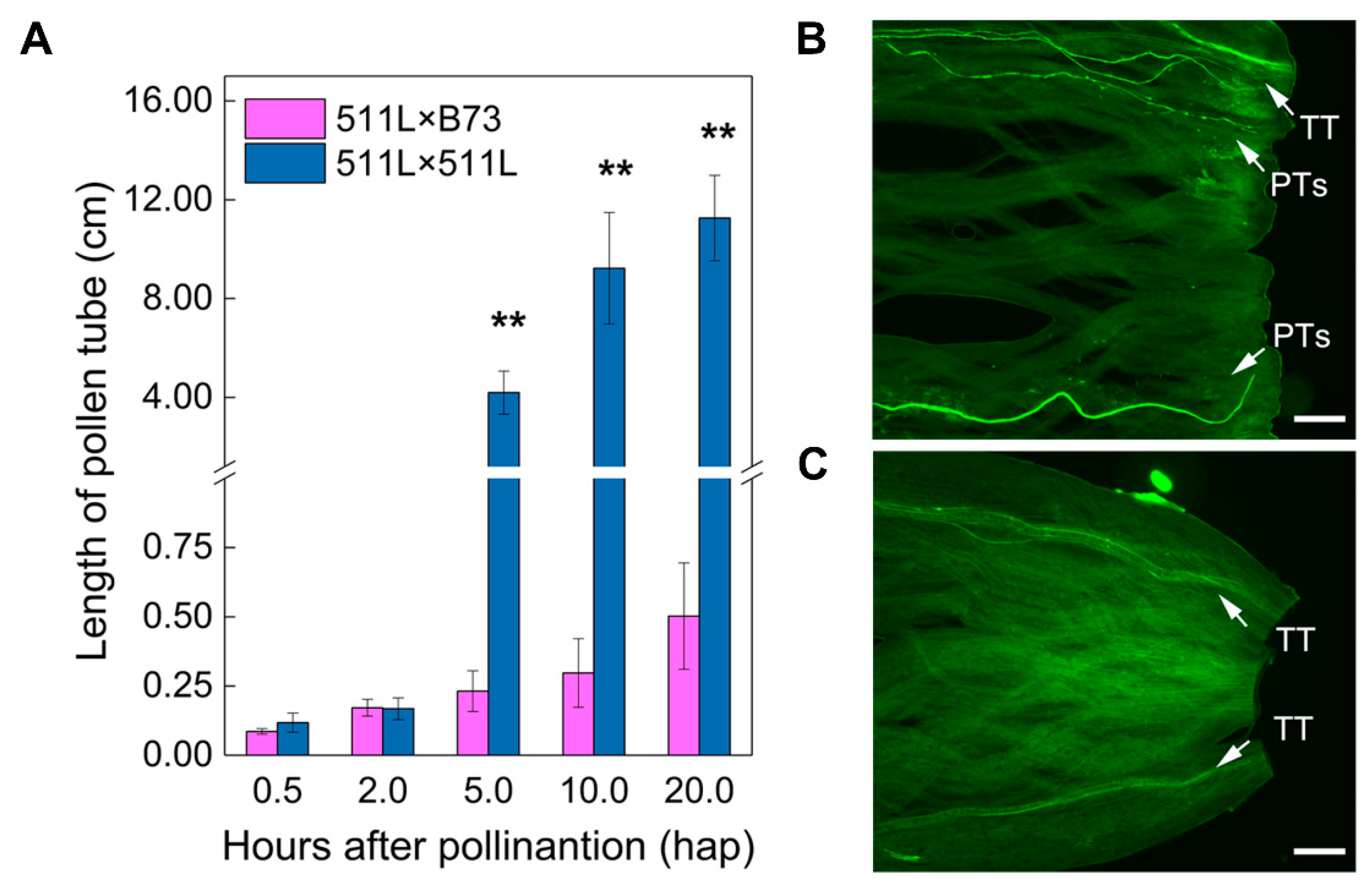
2.1. Phenotypic and Genotypic Analysis of Ga2-S
2.2. Transcriptome Profiling of Silks from Compatible and Incompatible Crosses
2.3. The Genes Involved in Signal Transduction Are Active in GB
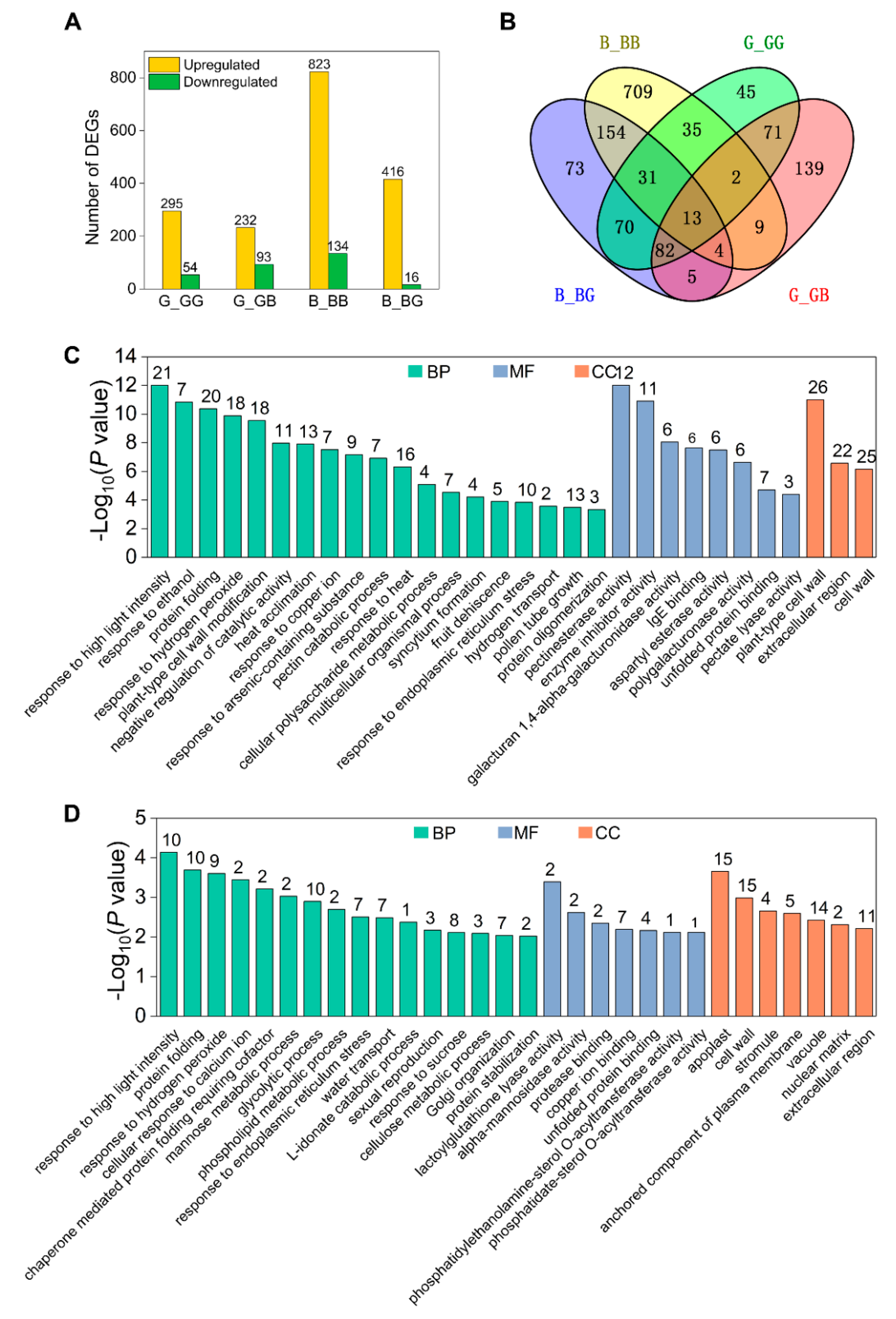
2.4. Analysis of DEGs Induced by Pollination
2.5. Cell Wall Metabolism Genes Are Upregulated by Pollination in 511L Silks
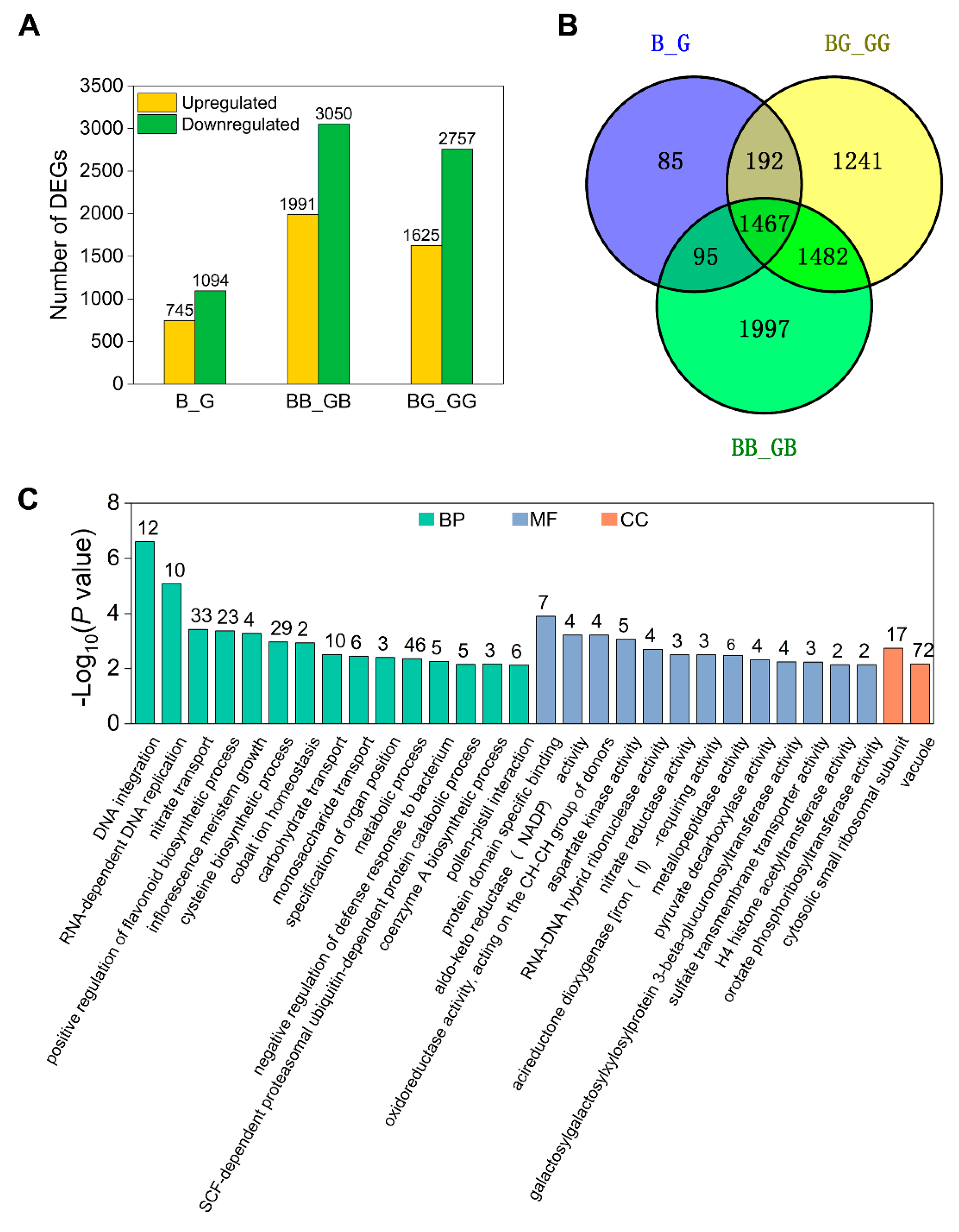
2.6. Cross-Incompatibility Pollination Involves Diverse Signaling Pathways in GB
2.7. Genetic Background-Dependent DEGs
2.8. Functional Network Analysis of Identified DEGs Potentially Involved in UCI
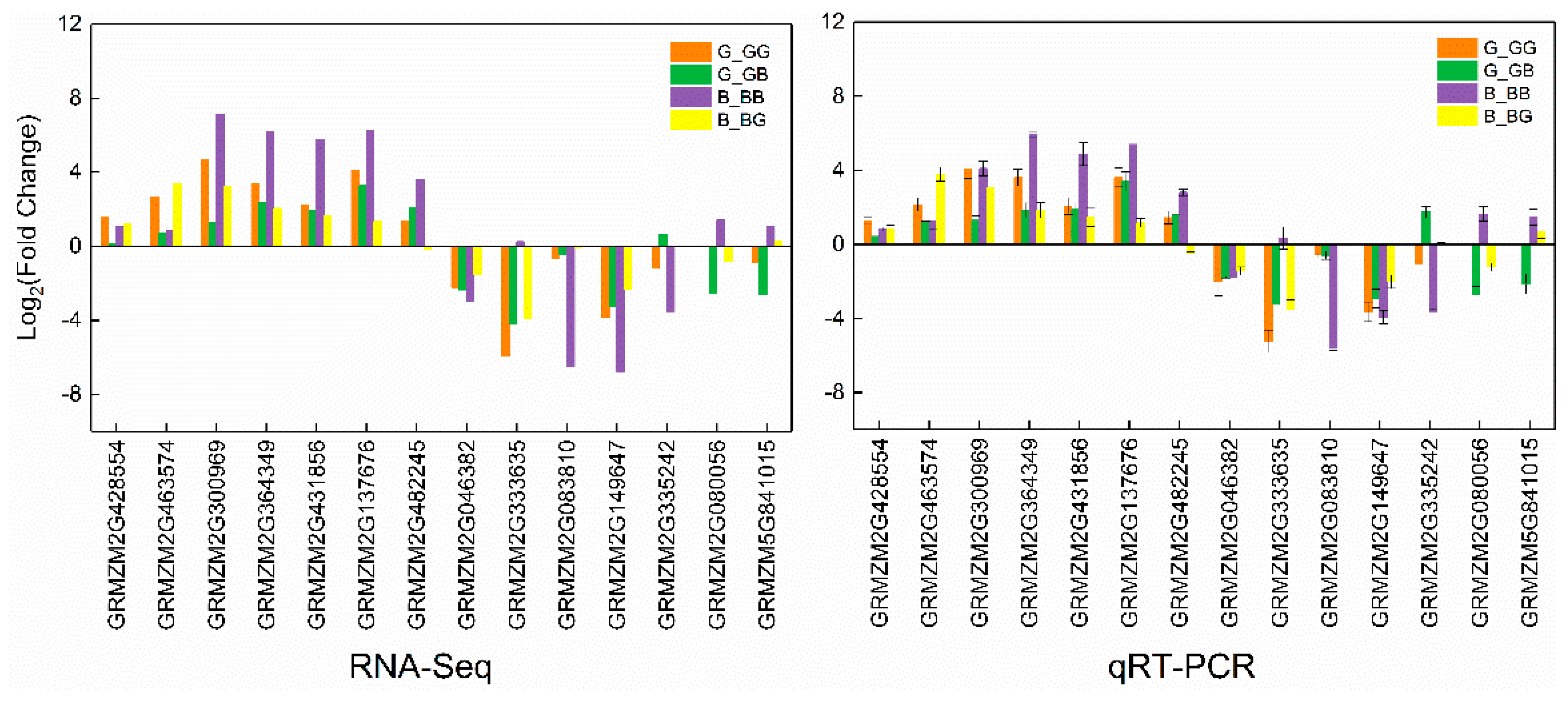
2.9. Validation of RNA-Seq Data by qRT-PCR
3. Discussion
3.1. Genes Involved in Signal Transduction and Defense Are Expressed in Ga2-S Silks
3.2. Cell Wall Metabolism Plays a Crucial Role in UCI
3.3. The Utilization of Ga2-S in Maize Breeding
4. Materials and Methods
4.1. Growth Conditions and Evaluation of Cross-Incompatibility in Maize
4.2. In Vivo Pollen Tube Staining and Growth Analysis
4.3. Tissue Harvest for RNA-Seq Analysis
4.4. RNA-Seq Library Construction and Sequencing
4.5. Sequencing Reads Analysis
4.6. DEGs and GO Analysis
4.7. qRT-PCR Validation of RNA-Seq Data
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Correns, C.E. Bastarde zwischen Maisrassen mit besonderer Berucksichtigung der Xenien. Bibl. Bot. 1901, 53, 1–161. [Google Scholar]
- Nelson, O.E., Jr. Non-reciprocal cross-sterility in maize. Genetics 1952, 37, 101–124. [Google Scholar] [PubMed]
- Schwartz, D. The analysis of a case of cross-sterility in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.F. Selective fertilization among the gametes from the same individuals. Proc. Natl. Acad. Sci. USA 1924, 10, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Correns, C.E. Scheinbare Ausnahmen von der Mendel’schen Spaltungsregel für Bastarde; Springer: Berlin/Heidelberg, Germany, 1924; pp. 1899–1924. [Google Scholar]
- Kermicle, J.L.; Evans, M.M. The Zea mays sexual compatibility gene ga2: Naturally occurring alleles, their distribution, and role in reproductive isolation. J. Hered. 2010, 101, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Burnham, C. Differential fertilization in the Bt Pr linkage group of maize. J. Am. Soc. Agron. 1936, 28, 968–975. [Google Scholar] [CrossRef]
- Evans, M.; Kermicle, J. Teosinte crossing barrier1, a locus governing hybridization of teosinte with maize. Theor. Appl. Genet. 2001, 103, 259–265. [Google Scholar] [CrossRef]
- Kermicle, J.; Allen, J. Cross-incompatibility between maize and teosinte. Maydica 1990, 35, 399–408. [Google Scholar]
- Jones, Z.G.; Goodman, M.M. Susceptibility of Dent-Sterile Popcorn to the Gametophyte Factor. Crop Sci. 2016, 56, 2594–2599. [Google Scholar] [CrossRef]
- Jones, Z.G.; Goodman, M.M.; Krakowsky, M.D. Identification of resistance to the Ga1-m gametophyte factor in maize. Euphytica 2015, 206, 785–791. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Zhang, Y.E.; Jiang, C.; Cui, D.; Liu, H.; Li, D.; Wang, L.; Chen, T.; Ning, L. Genetic analysis and fine mapping of the Ga1-S gene region conferring cross-incompatibility in maize. Theor. Appl. Genet. 2012, 124, 459–465. [Google Scholar] [CrossRef] [PubMed]
- González, M.D.; Pollak, L.M.; Goggi, A.S. Genotype × environment interactions in populations possessing Ga1-s and ga1 alleles for cross incompatibility in maize. Euphytica 2011, 185, 377–384. [Google Scholar] [CrossRef]
- Liu, X.; Sun, H.; Wu, P.; Tian, Y.; Cui, D.; Xu, C.; Li, S.; Li, P.; Zhang, H.; Chen, T.; et al. Fine Mapping of the Maize Cross-Incompatibility Locus Gametophytic Factor 1 (ga1) Using a Homogeneous Population. Crop Sci. 2014, 54, 873–881. [Google Scholar] [CrossRef]
- Bloom, J.C. Genomic localization of the maize cross-incompatibility gene, Gametophyte factor 1 (ga1). Maydica 2012, 56, 379–387. [Google Scholar]
- Mangelsdorf, P.C.; Jones, D.F. The Expression of Mendelian Factors in the Gametophyte of Maize. Genetics 1926, 11, 423–455. [Google Scholar] [PubMed]
- Nelson, O.E. The gametophyte factors of maize. In The Maize Handbook; Springer: Berlin/Heidelberg, Germany, 1994; pp. 496–503. [Google Scholar]
- Kermicle, J.L.; Evans, M.M.S. Pollen–pistil barriers to crossing in maize and teosinte result from incongruity rather than active rejection. Sex. Plant Reprod. 2005, 18, 187–194. [Google Scholar] [CrossRef]
- Johnson, M.A.; Preuss, D. Plotting a course: Multiple signals guide pollen tubes to their targets. Dev. Cell 2002, 2, 273–281. [Google Scholar] [CrossRef]
- Dresselhaus, T.; Lausser, A.; Marton, M.L. Using maize as a model to study pollen tube growth and guidance, cross-incompatibility and sperm delivery in grasses. Ann. Bot. 2011, 108, 727–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Kermicle, J.L.; Evans, M.M. Genetic and cellular analysis of cross-incompatibility in Zea mays. Plant Reprod. 2014, 27, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Moran Lauter, A.N.; Muszynski, M.G.; Huffman, R.D.; Scott, M.P. A Pectin Methylesterase ZmPme3 Is Expressed in Gametophyte factor1-s (Ga1-s) Silks and Maps to that Locus in Maize (Zea mays L.). Front. Plant Sci. 2017, 8, 1926. [Google Scholar] [CrossRef] [PubMed]
- Chebli, Y.; Kaneda, M.; Zerzour, R.; Geitmann, A. The cell wall of the Arabidopsis pollen tub—spatial distribution, recycling, and network formation of polysaccharides. Plant Physiol. 2012, 160, 1940–1955. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Guo, J.; Li, H.; Yang, Z. Signaling in pollen tube growth: Crosstalk, feedback, and missing links. Mol. Plant 2013, 6, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Rounds, C.M.; Bezanilla, M. Growth mechanisms in tip-growing plant cells. Annu. Rev. Plant Biol. 2013, 64, 243–265. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.F.; Gu, L.L.; Wang, H.Q.; Fei, C.F.; Fang, X.; Hussain, J.; Sun, S.J.; Dong, J.Y.; Liu, H.; Wang, Y.F. Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 3096–3101. [Google Scholar] [CrossRef] [PubMed]
- Hepler, P.K.; Kunkel, J.G.; Rounds, C.M.; Winship, L.J. Calcium entry into pollen tubes. Trends Plant Sci. 2012, 17, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Iwano, M.; Entani, T.; Shiba, H.; Kakita, M.; Nagai, T.; Mizuno, H.; Miyawaki, A.; Shoji, T.; Kubo, K.; Isogai, A.; et al. Fine-tuning of the cytoplasmic Ca2+ concentration is essential for pollen tube growth. Plant Physiol. 2009, 150, 1322–1334. [Google Scholar] [CrossRef] [PubMed]
- Iwano, M.; Shiba, H.; Miwa, T.; Che, F.S.; Takayama, S.; Nagai, T.; Miyawaki, A.; Isogai, A. Ca2+ dynamics in a pollen grain and papilla cell during pollination of Arabidopsis. Plant Physiol. 2004, 136, 3562–3571. [Google Scholar] [CrossRef] [PubMed]
- Holdaway-Clarke, T.L.; Feijó, J.A.; Hackett, G.R.; Kunkel, J.G.; Hepler, P.K. Pollen tube growth and the intracellular cytosolic calcium gradient oscillate in phase while extracellular calcium influx is delayed. Plant Cell Online 1997, 9, 1999–2010. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, C.; Teeri, T.; Siika-Aho, M.; Read, S.; Bacic, A. Location of cellulose and callose in pollen tubes and grains of Nicotiana tabacum. Planta 1998, 206, 452–460. [Google Scholar] [CrossRef]
- Moustacas, A.-M.; Nari, J.; Borel, M.; Noat, G.; Ricard, J. Pectin methylesterase, metal ions and plant cell-wall extension. The role of metal ions in plant cell-wall extension. Biochem. J. 1991, 279, 351–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosch, M.; Hepler, P.K. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 2005, 17, 3219–3226. [Google Scholar] [CrossRef] [PubMed]
- Bosch, M.; Cheung, A.Y.; Hepler, P.K. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 2005, 138, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Giovane, A.; Servillo, L.; Balestrieri, C.; Raiola, A.; D’avino, R.; Tamburrini, M.; Ciardiello, M.; Camardella, L. Pectin methylesterase inhibitor. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2004, 1696, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Jolie, R.P.; Duvetter, T.; Van Loey, A.M.; Hendrickx, M.E. Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr. Res. 2010, 345, 2583–2595. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Liu, Z.; Huang, S.; Li, C.; Ren, J.; Tang, X.; Liu, W.; Peng, S.; Feng, H. Pectin methylesterase inhibitor (PMEI) family can be related to male sterility in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genom. MGG 2017. [Google Scholar] [CrossRef] [PubMed]
- Woriedh, M.; Wolf, S.; Marton, M.L.; Hinze, A.; Gahrtz, M.; Becker, D.; Dresselhaus, T. External application of gametophyte-specific ZmPMEI1 induces pollen tube burst in maize. Plant Reprod. 2013, 26, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Dresselhaus, T.; Gu, H.; Qu, L.J. Active role of small peptides in Arabidopsis reproduction: Expression evidence. J. Integr. Plant Biol. 2015, 57, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Qu, L.J.; Li, L.; Lan, Z.; Dresselhaus, T. Peptide signalling during the pollen tube journey and double fertilization. J. Exp. Bot. 2015, 66, 5139–5150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. The male determinant of self-incompatibility in Brassica. Science 1999, 286, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, T.; Hatakeyama, K.; Suzuki, G.; Watanabe, M.; Isogai, A.; Hinata, K. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 2000, 403, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Mollet, J.C.; Dong, J.; Zhang, K.; Park, S.Y.; Lord, E.M. Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc. Natl. Acad. Sci. USA 2003, 100, 16125–16130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.-Y.; Jauh, G.-Y.; Mollet, J.-C.; Eckard, K.J.; Nothnagel, E.A.; Walling, L.L.; Lord, E.M. A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 2000, 12, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Zipfel, C. Complex regulation of plant sex by peptides. Science 2017, 358, 1544–1545. [Google Scholar] [CrossRef] [PubMed]
- Mecchia, M.A.; Santos-Fernandez, G.; Duss, N.N.; Somoza, S.C.; Boisson-Dernier, A.; Gagliardini, V.; Martínez-Bernardini, A.; Fabrice, T.N.; Ringli, C.; Muschietti, J.P. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science 2017, 358, 1600–1603. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Bergonci, T.; Zhao, Y.; Zou, Y.; Du, S.; Liu, M.-C.; Luo, X.; Ruan, H.; García-Valencia, L.E.; Zhong, S. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 2017, 358, 1596–1600. [Google Scholar] [CrossRef] [PubMed]
- De Graaf, B.H.; Rudd, J.J.; Wheeler, M.J.; Perry, R.M.; Bell, E.M.; Osman, K.; Franklin, F.C.; Franklin-Tong, V.E. Self-incompatibility in Papaver targets soluble inorganic pyrophosphatases in pollen. Nature 2006, 444, 490–493. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.G.; Franklin-Tong, V.E. Self-incompatibility triggers programmed cell death in Papaver pollen. Nature 2004, 429, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Sijacic, P.; Wang, X.; Skirpan, A.L.; Wang, Y.; Dowd, P.E.; McCubbin, A.G.; Huang, S.; Kao, T.-H. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 2004, 429, 302–305. [Google Scholar] [CrossRef] [PubMed]
- McClure, B.A.; Haring, V.; Ebert, P.R.; Anderson, M.A.; Simpson, R.J.; Sakiyama, F.; Clarke, A.E. Style self-incompatibility gene products of Nicotlana alata are ribonucleases. Nature 1989, 342, 955–957. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chetelat, R.T. Unilateral incompatibility gene ui1.1 encodes an S-locus F-box protein expressed in pollen of Solanum species. Proc. Natl. Acad. Sci. USA 2015, 112, 4417–4422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Chetelat, R.T. A pollen factor linking inter-and intraspecific pollen rejection in tomato. Science 2010, 330, 1827–1830. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, T.; Barker, D.; Birney, E.; Cameron, G.; Chen, Y.; Clark, L.; Cox, T.; Cuff, J.; Curwen, V.; Down, T.; et al. The Ensembl genome database project. Nucleic Acids Res. 2002, 30, 38–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, N.; Xu, X.W.; Wang, R.R.; Peng, W.L.; Cai, L.; Song, J.M.; Li, W.; Luo, X.; Niu, L.; Wang, Y.; et al. Contributions of Zea mays subspecies mexicana haplotypes to modern maize. Nat. Commun. 2017, 8, 1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kost, B.; Lemichez, E.; Spielhofer, P.; Hong, Y.; Tolias, K.; Carpenter, C.; Chua, N.-H. Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 1999, 145, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Dowd, P.E.; Coursol, S.; Skirpan, A.L.; Kao, T.-H.; Gilroy, S. Petunia phospholipase C1 is involved in pollen tube growth. Plant Cell 2006, 18, 1438–1453. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 2005, 6, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Mouille, G.; Pelloux, J. Homogalacturonan methyl-esterification and plant development. Mol. Plant 2009, 2, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Crawford, B.C.; Yanofsky, M.F. The formation and function of the female reproductive tract in flowering plants. Curr. Biol. 2008, 18, R972–R978. [Google Scholar] [CrossRef] [PubMed]
- Lausser, A.; Kliwer, I.; Srilunchang, K.-O.; Dresselhaus, T. Sporophytic control of pollen tube growth and guidance in maize. J. Exp. Bot. 2010, 61, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Lord, R.G.; Brown, D.J. Leadership Processes and Follower Self-Identity; Psychology Press: London, UK, 2003. [Google Scholar]
- Jones, Z.G.; Goodman, M.M.; Krakowsky, M.D. Identification of maize-derived dominant gametophyte factors. Euphytica 2016, 209, 63–69. [Google Scholar] [CrossRef]
- Boavida, L.C.; Borges, F.; Becker, J.D.; Feijó, J.A. Whole genome analysis of gene expression reveals coordinated activation of signaling and metabolic pathways during pollen-pistil interactions in Arabidopsis. Plant Physiol. 2011, 155, 2066–2080. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, T.; Matsushima, M.; Nabemoto, M.; Osaka, M.; Sakazono, S.; Masuko-Suzuki, H.; Takahashi, H.; Nakazono, M.; Iwano, M.; Takayama, S. Transcriptional characteristics and differences in Arabidopsis stigmatic papilla cells pre-and post-pollination. Plant Cell Physiol. 2014, 56, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.H.; Wang, F.; Chen, H.; Sun, W.; Zhang, X.S. Transcript profile analyses of maize silks reveal effective activation of genes involved in microtubule-based movement, ubiquitin-dependent protein degradation, and transport in the pollination process. PLoS ONE 2013, 8, e53545. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, S.; Murata, T.; Sakurai-Ozato, N.; Kubo, M.; Demura, T.; Fukuda, H.; Hasebe, M. ANXUR1 and 2, sister genes to FERONIA/SIRENE, are male factors for coordinated fertilization. Curr. Biol. 2009, 19, 1327–1331. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liang, L.; Xue, Y.; Jia, P.F.; Chen, W.; Zhang, M.X.; Wang, Y.C.; Li, H.J.; Yang, W.C. A receptor heteromer mediates the male perception of female attractants in plants. Nature 2016, 531, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H.; Higashiyama, T. Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature 2016, 531, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Cadena, D.L.; Gill, G.N. Receptor tyrosine kinases. FASEB J. 1992, 6, 2332–2337. [Google Scholar] [CrossRef] [PubMed]
- Goring, D.R.; Rothstein, S.J. The S-locus receptor kinase gene in a self-incompatible Brassica napus line encodes a functional serine/threonine kinase. Plant Cell 1992, 4, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Van der Biezen, E.A.; Jones, J.D. The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 1998, 8, R226–R228. [Google Scholar] [CrossRef]
- O’Neill, M.; Albersheim, P.; Darvill, A. 12-The Pectic Polysaccharides of Primary Cell Walls. Carbohydrates 1990, 2, 415–441. [Google Scholar]
- Staehelin, L.A.; Moore, I. The plant Golgi apparatus: Structure, functional organization and trafficking mechanisms. Annu. Rev. Plant Biol. 1995, 46, 261–288. [Google Scholar] [CrossRef]
- Sterling, J.D.; Quigley, H.F.; Orellana, A.; Mohnen, D. The catalytic site of the pectin biosynthetic enzyme α-1, 4-galacturonosyltransferase is located in the lumen of the Golgi. Plant Physiol. 2001, 127, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Micheli, F. Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef]
- Peaucelle, A.; Braybrook, S.; Hofte, H. Cell wall mechanics and growth control in plants: The role of pectins revisited. Front. Plant Sci. 2012, 3, 121. [Google Scholar] [CrossRef] [PubMed]
- Hocq, L.; Senechal, F.; Lefebvre, V.; Lehner, A.; Domon, J.M.; Mollet, J.C.; Dehors, J.; Pageau, K.; Marcelo, P.; Guerineau, F.; et al. Combined Experimental and Computational Approaches Reveal Distinct pH Dependence of Pectin Methylesterase Inhibitors. Plant Physiol. 2017, 173, 1075–1093. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yang, S.L.; Xie, L.F.; Puah, C.S.; Zhang, X.Q.; Yang, W.C.; Sundaresan, V.; Ye, D. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 2005, 17, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Leroux, C.; Bouton, S.; Kiefer-Meyer, M.C.; Fabrice, T.N.; Mareck, A.; Guenin, S.; Fournet, F.; Ringli, C.; Pelloux, J.; Driouich, A.; et al. PECTIN METHYLESTERASE48 is involved in Arabidopsis pollen grain germination. Plant Physiol. 2015, 167, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Francis, K.E.; Lam, S.Y.; Copenhaver, G.P. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol. 2006, 142, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Valášková, V.; Baldrian, P. Degradation of cellulose and hemicelluloses by the brown rot fungus Piptoporus betulinus—Production of extracellular enzymes and characterization of the major cellulases. Microbiology 2006, 152, 3613–3622. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Li, Y.; Lim, E.-K.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, reviews3004-1. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Wang, Y.; Qiu, S.; Kang, Z.; Che, S.; Wang, G.; Huang, J. Overexpression of OsEXPA8, a root-specific gene, improves rice growth and root system architecture by facilitating cell extension. PLoS ONE 2013, 8, e75997. [Google Scholar] [CrossRef] [PubMed]
- Pien, S.; Wyrzykowska, J.; McQueen-Mason, S.; Smart, C.; Fleming, A. Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc. Natl. Acad. Sci. USA 2001, 98, 11812–11817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, A.E.; Westgate, M.E. Relationship between desiccation and viability of maize pollen. Field Crops Res. 2005, 94, 114–125. [Google Scholar] [CrossRef]
- Della Porta, G.; Ederle, D.; Bucchini, L.; Prandi, M.; Verderio, A.; Pozzi, C. Maize pollen mediated gene flow in the Po valley (Italy): Source–recipient distance and effect of flowering time. Eur. J. Agron. 2008, 28, 255–265. [Google Scholar] [CrossRef]
- Kermicle, J.L. A Selfish Gene Governing Pollen-Pistil Compatibility Confers Reproductive Isolation Between Maize Relatives. Genetics 2006, 172, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Kermicle, J.; Taba, S.; Evans, M.M.S. The gametophyte-1 locus and reproductive isolation among Zea mays subspecies. Maydica 2006, 51, 219–225. [Google Scholar]
- MaizeGDB Stock Center. Available online: https://www.maizegdb.org/data_center/stock?id=9020818 (accessed on 15 January 2012).
- Hummon, A.B.; Lim, S.R.; Difilippantonio, M.J.; Ried, T. Isolation and solubilization of proteins after TRIzol® extraction of RNA and DNA from patient material following prolonged storage. Biotechniques 2007, 42, 467. [Google Scholar] [CrossRef] [PubMed]
- B73 RenfGen_v3. Available online: http://www.MaizeSequence.org (accessed on 1 September 2017).
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- BMK Cloud Platform. Available online: https://www.biocloud.net/ (accessed on 2 October 2017).
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Thakare, D.; Ma, C.; Lloyd, A.; Nixon, N.M.; Arakaki, A.M.; Burnett, W.J.; Logan, K.O.; Wang, D.; Wang, X.; et al. RNA Sequencing of Laser-Capture Microdissected Compartments of the Maize Kernel Identifies Regulatory Modules Associated with Endosperm Cell Differentiation. Plant Cell 2015, 27, 513–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhang, Z.; Fu, J.; Wang, G.; Wang, J.; Liu, Y. Transcriptome Analysis of Maize Immature Embryos Reveals the Roles of Cysteine in Improving Agrobacterium Infection Efficiency. Front. Plant Sci. 2017, 8, 1778. [Google Scholar] [CrossRef] [PubMed]
- Spotfire. Available online: http://spotfire.tibco.com/ (accessed on 7 November 2017).
- AgriGO. Available online: http://bioinfo.cau.edu.cn/agriGO/ (accessed on 15 November 2017).
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| B | BB | BG | G | GG | GB | |
|---|---|---|---|---|---|---|
| Total reads | 30,497,892 | 31,920,376 | 30,669,706 | 31,936,897 | 31,246,441 | 27,568,918 |
| Total mapped (%) | 28,814,261 94.47% | 29,381,957 92.05% | 28,992,358 94.53% | 24,751,691 77.44% | 24,027,406 76.94% | 21,441,071 77.78% |
| Unique mapped (%) | 26,205,774 85.95% | 26,984,829 84.53% | 27,122,861 88.43% | 22,377,204 69.99% | 21,930,700 70.20% | 19,252,301 69.84% |
| Gene numbers | 21,847 | 22,329 | 22,210 | 21,453 | 21,911 | 21,864 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Chen, Z.; Zhang, H.; Chen, H.; Gao, X. Transcriptome Analysis Provides Insight into the Molecular Mechanisms Underlying gametophyte factor 2-Mediated Cross-Incompatibility in Maize. Int. J. Mol. Sci. 2018, 19, 1757. https://doi.org/10.3390/ijms19061757
Wang M, Chen Z, Zhang H, Chen H, Gao X. Transcriptome Analysis Provides Insight into the Molecular Mechanisms Underlying gametophyte factor 2-Mediated Cross-Incompatibility in Maize. International Journal of Molecular Sciences. 2018; 19(6):1757. https://doi.org/10.3390/ijms19061757
Chicago/Turabian StyleWang, Man, Zhibin Chen, Huairen Zhang, Huabang Chen, and Xiquan Gao. 2018. "Transcriptome Analysis Provides Insight into the Molecular Mechanisms Underlying gametophyte factor 2-Mediated Cross-Incompatibility in Maize" International Journal of Molecular Sciences 19, no. 6: 1757. https://doi.org/10.3390/ijms19061757





