Candidate Genes for Yellow Leaf Color in Common Wheat (Triticum aestivum L.) and Major Related Metabolic Pathways according to Transcriptome Profiling
Abstract
1. Introduction
2. Results
2.1. Sequence Analysis Using RNA-Seq
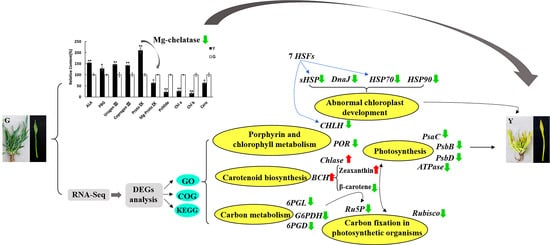
2.2. Identification and Functional Annotation of Differentially Expressed Genes (DEGs) in G and Y
2.3. Identification of DEGs Related to Chl Metabolism and Carotenoid Biosynthesis in Y and G Plants
2.4. Identification of DEGs Related to Photosynthesis
2.5. Analysis of TFs and Heat Shock Proteins (HSPs) in Y and G Plants
2.6. Quantitative Real-Time PCR (qRT-PCR) Analysis of Candidate Genes
2.7. Comparison of Chl Precursors in Y and G Plants
2.8. Analysis of the Carotenoid Composition in Y and G Plants by HPLC
3. Discussion
3.1. Yellow Leaf Phenotype is Closely Associated with Chl and Carotenoid Pigment Metabolism
3.2. Yellow Leaf Phenotype Affected by the Expression of Genes Related to Photosynthesis
3.3. HSFs, HSPs, and Chloroplasts
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Extraction, Library Construction, and RNA Sequencing
4.3. Sequence Alignment and Functional Annotation
4.4. DEG Analysis
4.5. qRT-PCR Analysis
4.6. Determination of Photosynthetic Pigments and Chl Precursors
4.7. Isolation and HPLC Analysis of Carotenoid Compounds
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Makino, A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 2011, 155, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Tanaka, R. Chlorophyll metabolism. Curr. Opin. Plant Biol. 2006, 9, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Nagata, N.; Tanaka, R.; Satoh, S.; Tanaka, A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 2005, 17, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tang, S.; Zhang, S.; Shan, J.; Tang, C.; Chen, Q.; Jia, G.; Han, Y.; Zhi, H.; Diao, X. Gene mapping and functional analysis of the novel leaf color gene SiYGL1 in foxtail millet [Setaria italica (L.) P. Beauv]. Physiol. Plant 2015, 157, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, X.; He, B.; Diao, L.; Sheng, S.; Wang, J.; Guo, X.; Su, N.; Wang, L.; Jiang, L.; et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 2007, 145, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Shuang, G.; Zhongwei, W.; Qing, D.; Yadi, X.; Tianquan, Z.; Wenqiang, S.; Xianchun, S.; Yinghua, L.; Guanghua, H. Map-based cloning and functional analysis of YGL8, which controls leaf colour in rice (Oryza sativa L.). BMC Plant Biol. 2016, 16, 134. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.-H.; Hur, J.; Ryu, C.-H.; Choi, Y.; Chung, Y.-Y.; Miyao, A.; Hirochika, H.; An, G. Characterization of a Rice Chlorophyll-Deficient Mutant Using the T-DNA Gene-Trap System. Plant Cell Physiol. 2003, 44, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Long, W.; Wang, Y.; Liu, L.; Wang, Y.; Niu, M.; Zheng, M.; Wang, D.; Wan, J. A rice White-stripe leaf3 (wsl3) mutant lacking an HD domain-containing protein affects chlorophyll biosynthesis and chloroplast development. J. Plant Biol. 2016, 59, 282–292. [Google Scholar] [CrossRef]
- Wang, Y.; He, Y.; Yang, M.; He, J.; Xu, P.; Shao, M.; Chu, P.; Guan, R. Fine mapping of a dominant gene conferring chlorophyll-deficiency in Brassica napus. Sci. Rep. 2016, 6, 31419. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Li, X.P.; Razeghifard, R.; Anderson, J.M.; Niyogi, K.K.; Pogson, B.J.; Chow, W.S. The multiple roles of light-harvesting chlorophyll a/b-protein complexes define structure and optimize function of Arabidopsis chloroplasts: A study using two chlorophyll b-less mutants. Biochim. Biophys. Acta 2009, 1787, 973–984. [Google Scholar] [CrossRef] [PubMed]
- Braumann, I.; Stein, N.; Hansson, M. Reduced chlorophyll biosynthesis in heterozygous barley magnesium chelatase mutants. Plant Physiol. Biochem. 2014, 78, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Chen, Z.; Sun, W.; Deng, T.; Chen, M. De novo sequencing of the Leaf transcriptome reveals complex light-responsive regulatory networks in Camellia sinensis cv. Baijiguan. Front. Plant Sci. 2016, 7, 332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, J.; Yoo, J.H.; Yoo, S.C.; Cho, S.H.; Koh, H.J.; Seo, H.S.; Paek, N.C. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol. Biol. 2006, 62, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Campoli, C.; Caffarri, S.; Svensson, J.T.; Bassi, R.; Stanca, A.M.; Cattivelli, L.; Crosatti, C. Parallel pigment and transcriptomic analysis of four barley Albina and Xantha mutants reveals the complex network of the chloroplast-dependent metabolism. Plant Mol. Biol. 2009, 71, 173–191. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.W.; Mani, D.; Curtin, S.J.; Slattery, R.A.; Michno, J.-M.; Ort, D.R.; Schaus, P.J.; Palmer, R.G.; Orf, J.H.; Stupar, R.M. Identical Substitutions in Magnesium Chelatase Paralogs Result in Chlorophyll-Deficient Soybean Mutants. G3 Genes Genomes Genet. 2015, 5, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Papenbrock, J.; GrMe, S.; Kruse, E.; Hanel, F.; Grimm, B. Mg-chelatase of tobacco: Identification of a Chl D cDNA sequence encoding a third subunit, analysis of the interaction of the three subunits with the yeast two-hybrid system, and reconstitution of the enzyme activity by co-expression of recombinant CHL D, CHL H and CHL I. Plant J. 1997, 12, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Gibson, L.C.; Willows, R.D.; Kannangara, C.G.; Wettstein, D.V.; Hunter, C.N. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: Reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 1941–1944. [Google Scholar] [CrossRef] [PubMed]
- Fodje, M.N.; Hansson, A.; Hansson, M.; Olsen, J.G.; Gough, S.; Willows, R.D.; Al-Karadaghi, S. Interplay Between an AAA Module and an Integrin I Domain May Regulate the Function of Magnesium Chelatase. J. Mol. Biol. 2001, 311, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Sirijovski, N.; Olsson, U.; Lundqvist, J.; Al-Karadaghi, S.; Willows, R.D.; Hansson, M. ATPase activity associated with the magnesium chelatase H-subunit of the chlorophyll biosynthetic pathway is an artefact. Biochem. J. 2006, 400, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki, N.; Brusslan, J.A.; Larkin, R.; Nagatani, A.; Chory, J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 2000, 98, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Larkin, R.M.; Alonso, J.M.; Ecker, J.R.; Chory, J. GUN4, a Regulator of Chlorophyll Synthesis and Intracellular Signaling. Science 2003, 299, 902–906. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Luo, S.; Araujo, W.L.; Schlicke, H.; Rothbart, M.; Yu, J.; Fan, T.; Fernie, A.R.; Grimm, B.; Luo, M. Virus-induced gene silencing of pea CHLI and CHLD affects tetrapyrrole biosynthesis, chloroplast development and the primary metabolic network. Plant Physiol. Biochem. 2013, 65, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Hansson, A.; Willows, R.D.; Roberts, T.H.; Hansson, M. Three semidominant barley mutants with single amino acid substitutions in the smallest magnesium chelatase subunit form defective AAA+ hexamers. Proc. Natl. Acad. Sci. USA 2002, 99, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Hansson, A.; Kannangara, C.G.; Wettstein, D.V.; Hansson, M. Molecular basis for semidominance of missense mutations in the XANTHA-H (42-kDa) subunit of magnesium chelatase. Proc. Natl. Acad. Sci. USA 1999, 96, 1744–1749. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, W.P.; Nguyen, L.V.; Wernsman, E.A.; Thompson, W.F.; Conkling, M.A. Transposon tagging of the sulfur gene of tobacco using engineered maize Ac/Ds elements. Genetics 1999, 153, 1919–1928. [Google Scholar] [PubMed]
- Sawers, R.J.; Viney, J.; Farmer, P.R.; Bussey, R.R.; Olsefski, G.; Anufrikova, K.; Hunter, C.N.; Brutnell, T.P. The maize Oil yellow1 (Oy1) gene encodes the I subunit of magnesium chelatase. Plant Mol. Biol. 2006, 60, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.D.; Joppa, L.R.; Duysen, M.E.; Freeman, T.P. Inheritance of Three Chlorophyll-Deficient Mutants of Common Wheat. Crop Sci. 1985, 25, 1023–1025. [Google Scholar] [CrossRef]
- Watanabe, N.; Koval, S.F. Mapping of chlorina mutant genes on the long arm of homoeologous group 7 chromosomes in common wheat with partial deletion lines. Euphytica 2003, 129, 259–265. [Google Scholar] [CrossRef]
- Ansari, M.J.; Al-Ghamdi, A.; Kumar, R.; Usmani, S.; Al-Attal, Y.; Nuru, A.; Mohamed, A.A.; Singh, K.; Dhaliwal, H.S. Characterization and gene mapping of a chlorophyll-deficient mutant clm1 of Triticum monococcum L. Biol. Plant. 2013, 57, 442–448. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, K.; Zhang, W.; Liu, H.Y.; Du, L.P.; Hu, H.R.; Ye, X.G. Comprehensive analysis of differently expressed genes and proteins in albino and green plantlets from a wheat anther culture. Biol. Plant. 2017, 61, 255–265. [Google Scholar] [CrossRef]
- Zhao, H.B.; Guo, H.J.; Zhao, L.S.; Gu, J.Y.; Zhao, S.R.; Li, J.H.; Liu, L.X. Agronomic Traits and Photosynthetic Characteristics of Chlorophyll-Deficient Wheat Mutant Induced by Spaceflight Environment. Acta Agron. Sin. 2011, 37, 119–126. [Google Scholar] [CrossRef]
- Kosuge, K.; Watanabe, N.; Kuboyama, T. Comparative genetic mapping of homoeologous genes for the chlorina phenotype in the genus Triticum. Euphytica 2011, 179, 257–263. [Google Scholar] [CrossRef]
- Brestic, M.; Zivcak, M.; Kunderlikova, K.; Sytar, O.; Shao, H.; Kalaji, H.M.; Allakhverdiev, S.I. Low PSI content limits the photoprotection of PSI and PSII in early growth stages of chlorophyll b-deficient wheat mutant lines. Photosynth. Res. 2015, 125, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Gu, J.; Guo, H.; Zhao, L.; Xie, Y.; Xiong, H.; Li, J.; Zhao, S.; Song, X.; Liu, L. Transcriptome and proteomic analyses reveal multiple differences associated with chloroplast development in the spaceflight-induced wheat albino mutant mta. PLoS ONE 2017, 12, e0177992. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.Y.; Xu, H.; Du, G.Y.; Lin, J.T.; Duan, M.; Guo, A.G. Proteome analysis of chloroplast proteins in stage albinism line of winter wheat (Triticum aestivum) FA85. BMB Rep. 2009, 42, 450–455. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.G.; Xu, H.; Zhang, J.Y.; Liang, G.W.; Liu, Y.T.; Guo, A.G. Effect of low temperature on chlorophyll biosynthesis in albinism line of wheat (Triticum aestivum) FA85. Physiol. Plant. 2012, 145, 384–394. [Google Scholar] [CrossRef] [PubMed]
- Falbel, T.G.; Staehelin, L.A. Characterization of a family of chlorophyll-deficient wheat (Triticum) and barley (Hordeum vulgare) mutants with defects in the magnesium-lnsertion step of chlorophyll biosynthesis. Plant Physiol. 1994, 104, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, H.J.; Zhao, B.; Sun, Q.Q.; Cao, Y.Y.; Li, R.; Wu, X.X.; Weeda, S.; Li, L.; Ren, S.; et al. The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J. Pineal Res. 2014, 56, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Bellieny-Rabelo, D.; De Oliveira, E.A.; Ribeiro, E.S.; Costa, E.P.; Oliveira, A.E.; Venancio, T.M. Transcriptome analysis uncovers key regulatory and metabolic aspects of soybean embryonic axes during germination. Sci. Rep. 2016, 6, 36009. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chen, X.; Xu, B.; Li, Y.; Ma, Y.; Wang, G. Phenotype and transcriptome analysis reveals chloroplast development and pigment biosynthesis together influenced the leaf color formation in mutants of Anthurium andraeanum ‘Sonate’. Front. Plant Sci. 2015, 6, 139. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Wang, P.; Wang, S.A.; Ma, L.; Li, L.; Yang, R.; Ma, Y.; Wang, Q. Comprehensive transcriptome analysis discovers novel candidate genes related to leaf color in a Lagerstroemia indica yellow leaf mutant. Genes Genom. 2017, 37, 851–863. [Google Scholar] [CrossRef]
- Zhang, L.L.; Liu, C.; An, X.Y.; Wu, H.Y.; Feng, Y.; Wang, H.; Sun, D.J. Identification and genetic mapping of a novel incompletely dominant yellow leaf color gene, Y1718, on chromosome 2BS in wheat. Euphytica 2017, 213, 141. [Google Scholar] [CrossRef]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. Fimo: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Willows, R.D.; Gibson, L.C.; Kanangara, C.G.; Hunter, C.N.; Wettstein, D. Three separate proteins constitute the magnesium chelatase of Rhodobacter Sphaeroides. Eur. J. Biochem. 1996, 235, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Takamiya, K. Novel insights into the enzymology, regulation and physiological functions of light-dependent protochlorophyllide oxidoreductase in angiosperms. Photosynth. Res. 2004, 81, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Su, Q.; Frick, G.; Armstrong, G.; Apel, K. POR C of Arabidopsis thaliana: A third light- and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol. Biol. 2001, 47, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Rahman, M.L.; Cho, S.H.; Kim, Y.S.; Koh, H.J.; Yoo, S.C.; Paek, N.C. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J. 2013, 74, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Xu, J.; Rao, Y.C.; Zeng, Y.J.; Liu, H.J.; Zheng, T.T.; Zhang, G.-H.; Hu, J.; Guo, L.B.; Qian, Q.; et al. Cloning and functional analysis of pale-green leaf (PGL10) in rice (Oryza sativa L.). Plant Growth Regul. 2016, 78, 69–77. [Google Scholar] [CrossRef]
- Takamiya, K.-I.; Tsuchiya, T.; Ohta, H. Degradation pathway(s) of chlorophyll: What has gene cloning revealed? Trends Plant Sci. 2000, 5, 426–431. [Google Scholar] [CrossRef]
- Harpaz-Saad, S.; Azoulay, T.; Arazi, T.; Ben-Yaakov, E.; Mett, A.; Shiboleth, Y.M.; Hortensteiner, S.; Gidoni, D.; Gal-On, A.; Goldschmidt, E.E.; et al. Chlorophyllase is a rate-limiting enzyme in chlorophyll catabolism and is posttranslationally regulated. Plant Cell 2007, 19, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Schenk, N.; Schelbert, S.; Kanwischer, M.; Goldschmidt, E.E.; Dormann, P.; Hortensteiner, S. The chlorophyllases ATCLH1 and ATCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett. 2007, 581, 5517–5525. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Makita, S.; Schelbert, S.; Sano, S.; Ochiai, M.; Tsuchiya, T.; Hasegawa, S.F.; Hortensteiner, S.; Tanaka, A.; Tanaka, R. Reexamination of chlorophyllase function implies its involvement in defense against chewing herbivores. Plant Physiol. 2015, 167, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Hortensteiner, S. Chlorophyll degradation during senescence. Annu. Rev. Plant Biol. 2006, 57, 55–77. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, C.E.; Arruda, P. Altering the expression of the chlorophyllase gene ATHCOR1 in transgenic Arabidopsis caused changes in the chlorophyll-to-chlorophyllide ratio. Plant Physiol. 2002, 128, 1255–1263. [Google Scholar] [CrossRef] [PubMed]
- Kariola, T.; Brader, G.; Li, J.; Palva, E.T. Chlorophyllase 1, a damage control enzyme, affects the balance between defense pathways in plants. Plant Cell 2005, 17, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Wilk, D.; Goldschmidt, D.H.E.E.; Riov, J.; Eyal, Y. Chlorophyll breakdown by chlorophyllase: Isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J. 1999, 20, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Azoulay Shemer, T.; Harpaz-Saad, S.; Belausov, E.; Lovat, N.; Krokhin, O.; Spicer, V.; Standing, K.G.; Goldschmidt, E.E.; Eyal, Y. Citrus chlorophyllase dynamics at ethylene-induced fruit color-break: A study of chlorophyllase expression, posttranslational processing kinetics, and in situ intracellular localization. Plant Physiol. 2008, 148, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Schelbert, S.; Aubry, S.; Burla, B.; Agne, B.; Kessler, F.; Krupinska, K.; Hortensteiner, S. Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 2009, 21, 767–785. [Google Scholar] [CrossRef] [PubMed]
- Demmig-Adams, B.; Adams, W.W., III. The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1996, 1, 21–26. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, G.; Kato, M.; Yamawaki, K.; Takagi, T.; Kiriiwa, Y.; Ikoma, Y.; Matsumoto, H.; Yoshioka, T.; Nesumi, H. Regulation of carotenoid accumulation and the expression of carotenoid metabolic genes in citrus juice sacs in vitro. J. Exp. Bot. 2012, 63, 871–886. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Smith, J.J.; Tian, L.; Dellapenna, D. The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol. 2009, 50, 463–479. [Google Scholar] [CrossRef] [PubMed]
- Diretto, G.; Welsch, R.; Tavazza, R.; Mourgues, F.; Pizzichini, D.; Beyer, P.; Giuliano, G. Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol. 2007, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Ahn, Y.O.; Ahn, M.J.; Lee, H.S.; Kwak, S.S. Down-regulation of beta-carotene hydroxylase increases beta-carotene and total carotenoids enhancing salt stress tolerance in transgenic cultured cells of sweetpotato. Phytochemistry 2012, 74, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.; Zorrilla-Lopez, U.; Sandmann, G.; Capell, T.; Christou, P.; Zhu, C. The silencing of carotenoid beta-hydroxylases by RNA interference in different maize genetic backgrounds increases the beta-carotene content of the endosperm. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Latowski, D.; Grzyb, J.; Strzałka, K. The xanthophyll cycle-molecular mechanism and physiological significance. Acta Physiol. Plant. 2004, 26, 197. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Adams, W.W. Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 599–626. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, G.; Ji, J.; Jin, C.; Wu, W.; Zhao, J. Over-expression of Arabidopsis thaliana β-carotene hydroxylase (chyB) gene enhances drought tolerance in transgenic tobacco. J. Plant Biochem. Biotechnol. 2014, 23, 190–198. [Google Scholar] [CrossRef]
- Wang, L.; Cao, H.; Chen, C.; Yue, C.; Hao, X.; Yang, Y.; Wang, X. Complementary transcriptomic and proteomic analyses of a chlorophyll-deficient tea plant cultivar reveal multiple metabolic pathway changes. J. Proteom. 2016, 130, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Li, C.F.; Xu, Y.X.; Ma, J.Q.; Jin, J.Q.; Huang, D.J.; Yao, M.Z.; Ma, C.L.; Chen, L. Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘Anji Baicha’ (Camellia sinensis). BMC Plant Biol. 2016, 16, 195. [Google Scholar] [CrossRef] [PubMed]
- Bashir, H.; Qureshi, M.I.; Ibrahim, M.M.; Iqbal, M. Chloroplast and photosystems: Impact of cadmium and iron deficiency. Photosynthetica 2015, 53, 321–335. [Google Scholar] [CrossRef]
- Allen, J.F.; de Paula, W.B.; Puthiyaveetil, S.; Nield, J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 2011, 16, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Albertsson, P. A quantitative model of the domain structure of the photosynthetic membrane. Trends Plant Sci. 2001, 6, 349–354. [Google Scholar] [CrossRef]
- Chu, P.; Yan, G.X.; Yang, Q.; Zhai, L.N.; Zhang, C.; Zhang, F.Q.; Guan, R.Z. iTRAQ-based quantitative proteomics analysis of Brassica napus leaves reveals pathways associated with chlorophyll deficiency. J. Proteom. 2015, 113, 244–259. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Cao, J.; Li, J.; Zhou, B.; Tang, J.; Miao, A. Phenotypic, histological and proteomic analyses reveal multiple differences associated with chloroplast development in yellow and variegated variants from Camellia sinensis. Sci. Rep. 2016, 6, 33369. [Google Scholar] [CrossRef] [PubMed]
- Hutin, C.; Nussaume, L.; Moise, N.; Moya, I.; Kloppstech, K.; Havaux, M. Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc. Natl. Acad. Sci. USA 2003, 100, 4921–4926. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.; Lohscheider, J.N.; Albert, S.; Andersson, U.; Mendgen, K.W.; Rojas-Stutz, M.C.; Adamska, I.; Funck, D. Small One-Helix Proteins Are Essential for Photosynthesis in Arabidopsis. Front. Plant Sci. 2017, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Adamska, I.; Kruse, E.; Kloppstech, K. Stable insertion of the early light-induced proteins into etioplast membranes requires chlorophyll A. J. Biol. Chem. 2001, 276, 8582–8587. [Google Scholar] [CrossRef] [PubMed]
- Heddad, M.; Noren, H.; Reiser, V.; Dunaeva, M.; Andersson, B.; Adamska, I. Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiol. 2006, 142, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Tzvetkova-Chevolleau, T.; Franck, F.; Alawady, A.E.; Dall’Osto, L.; Carrière, F.; Bassi, R.; Grimm, B.; Nussaume, L.; Havaux, M. The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J. 2007, 50, 795–809. [Google Scholar] [CrossRef] [PubMed]
- Parry, M.; Andralojc, P.J.; Scales, J.C.; Salvucci, M.E.; Carmo-Silva, A.E.; Alonso, H.; Whitney, S.M. Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 2013, 64, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Andersson, I.; Backlund, A. Structure and function of Rubisco. Plant Physiol. Biochem. 2008, 46, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Zhan, G.M.; Li, R.J.; Hu, Z.Y.; Liu, J.; Deng, L.B.; Lu, S.Y.; Hua, W. Cosuppression of RBCS3B in Arabidopsis leads to severe photoinhibition caused by ROS accumulation. Plant Cell Rep. 2014, 33, 1091–1108. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S. Nitrogen assimilation, Abiotic stress and glucose 6-phosphate dehydrogenase: The full circle of reductants. Plants 2016, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.; Tymoczko, J.; Stryer, L. 20.3 the pentose phosphate pathway generates NADPH and synthesizes five-carbon sugars. In Biochemistry, 5th ed.; W H Freeman: New York, NY, USA, 2002; ISBN-10 0-7167-3051-0. [Google Scholar]
- Waters, E.R.; Lee, G.J.; Vierling, E. Evolution, structure and function of the small heat shock proteins in plants. J. Exp. Bot. 1996, 47, 325–338. [Google Scholar] [CrossRef]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins (HSP). J. Proteom. 2008, 71, 391–411. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.P.; Drenth, J.; McIntyre, C.L. TaHsfA6f is a transcriptional activator that regulates a suite of heat stress protection genes in wheat (Triticum aestivum L.) including previously unknown Hsf targets. J. Exp. Bot. 2015, 66, 1025–1039. [Google Scholar] [CrossRef] [PubMed]
- Westerheidea, S.D.; Raynesa, R.; Powella, C.; Xueb, B.; Uversky, V.N. HSF transcription factor family, heat shock response, and protein intrinsic disorder. Curr. Protein Pept. Sci. 2012, 13, 86–103. [Google Scholar] [CrossRef]
- Nover, L.; Scharf, K.D. Heat stress proteins and transcription factors. Cell. Mol. Life Sci. 1997, 53, 80–103. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yang, Y.; Gong, F.; Zhang, D.; Zhang, L.; Wu, L.; Li, C.; Wang, W. Protein sHSP26 improves chloroplast performance under heat stress by interacting with specific chloroplast proteins in maize (Zea mays). J. Proteom. 2015, 115, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Alam, I.; Kim, Y.G.; Sharmin, S.A.; Lee, K.W.; Lee, S.H.; Lee, B.H. Overexpression of a chloroplast-localized small heat shock protein OsHSP26 confers enhanced tolerance against oxidative and heat stresses in tall fescue. Biotechnol. Lett. 2012, 34, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Zhou, W.; Wang, H.; Ding, S.; Lu, Q.; Wen, X.; Peng, L.; Zhang, L.; Lu, C. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 2013, 25, 2925–2943. [Google Scholar] [CrossRef] [PubMed]
- Latijnhouwers, M.; Xu, X.M.; Møller, S.G. Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta 2010, 232, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; An, G. Rice chloroplast-localized heat shock protein 70, OsHsp70CP1, is essential for chloroplast development under high-temperature conditions. J. Plant Physiol. 2013, 170, 854–863. [Google Scholar] [CrossRef] [PubMed]
- Schroda, M.; Vallon, O.; Wollman, F.A.; Beck, C.F. A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 1999, 11, 1165–1178. [Google Scholar] [CrossRef] [PubMed]
- Greene, M.K.; Maskos, K.; Landry, S.J. Role of the J-domain in the cooperation of HSP40 with HSP70. Proc. Natl. Acad. Sci. USA. 1998, 95, 6108–6113. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, M.E.; Caplan, A.J. Structure, function and evolution of DnaJ: Conservation and adaptation of chaperone function. Cell Stress Chaperones 1998, 3, 28–36. [Google Scholar] [CrossRef]
- Zhu, X.; Liang, S.; Yin, J.; Yuan, C.; Wang, J.; Li, W.; He, M.; Wang, J.; Chen, W.; Ma, B.; et al. The DnaJ OsDjA7/8 is essential for chloroplast development in rice (Oryza sativa). Gene 2015, 574, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Strand, A.; Asami, T.; Alonso, J.; Chory, J. Chloroplast to nucleus communication triggered by accumulation of Mg-ProtoporphyrinIX. Nature 2003, 421, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Chi, W.; Sun, X.; Zhang, L. Intracellular signaling from plastid to nucleus. Annu. Rev. Plant Biol. 2013, 64, 559–582. [Google Scholar] [CrossRef] [PubMed]
- Kropat, J.; Oster, U.; Rüdiger, W.; Beck, C.F. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc. Natl. Acad. Sci. USA 1997, 94, 14168–14172. [Google Scholar] [CrossRef] [PubMed]
- Kindgren, P.; Noren, L.; Lopez Jde, D.; Shaikhali, J.; Strand, A. Interplay between heat shock protein 90 and HY5 controls phANG expression in response to the GUN5 plastid signal. Mol. Plant 2012, 5, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.T.; He, N.Y.; Chen, J.H.; Guo, F.Q. Identification of core subunits of photosystem II as action sites of HSP21, which is activated by the GUN5-mediated retrograde pathway in Arabidopsis. Plant J. 2017, 89, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Mao, X.; Cai, T.; Luo, J.; Wei, L. Kobas server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.P.; Hou, W.Q.; Zhang, L.; Wu, H.Y.; Zhao, L.F.; Du, X.Y.; Ma, X.; Li, A.F.; Wang, H.W.; Kong, L.R. Functional analysis of a wheat pleiotropic drug resistance gene involved in Fusarium head blight resistance. J. Integr. Agric. 2016, 15, 2215–2227. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar] [CrossRef]
- Dei, M. Benzyladenine-induced stimulation of 5-aminolevulinic acid accumulation under various light intensities in levulinic acid-treated cotyledons of etiolated cucumber. Physiol. Plant. 1985, 64, 153–160. [Google Scholar] [CrossRef]
- Bogorad, L. Porphyrin synthesis. In Methods in Enzymology; Colowick, S.P., Kaplan, N.O., Eds.; Academic Press: New York, NY, USA, 1962; Volume 5, pp. 885–895. ISBN 0076-6879. [Google Scholar]
- Rebeiz, C.A.; Mattheis, J.R.; Smith, B.B.; Rebeiz, C.C.; Dayton, D.F. Chloroplast biogenesis: Biosynthesis and accumulation of protochlorophyll by isolated etioplasts and developing chloroplasts. Arch. Biochem. Biophys. 1975, 171, 549–567. [Google Scholar] [CrossRef]
- Norris, S.R.; Barrette, T.R.; DellaPenna, D. Genetic dissection of camtenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 1995, 7, 2139–2149. [Google Scholar] [CrossRef] [PubMed]














| Groups | Total Reads | Clean Reads | GC (%) | Q20 (%) | Total Mapped Reads | Ratio (%) |
|---|---|---|---|---|---|---|
| G-1 | 59,480,874 | 58,804,014 | 58.37 | 96.76 | 41,285,997 | 70.21 |
| G-2 | 61,434,418 | 60,676,246 | 58.68 | 96.54 | 42,662,324 | 70.31 |
| G-3 | 64,966,228 | 64,070,288 | 58.68 | 96.65 | 44935,906 | 70.14 |
| Y-1 | 67,083,278 | 66,278,074 | 59.25 | 96.87 | 46,615,892 | 70.33 |
| Y-2 | 58,156,566 | 57,490,204 | 57.87 | 96.95 | 40,650,136 | 70.71 |
| Y-3 | 81,071,740 | 80,112,586 | 57.29 | 96.61 | 55,817,063 | 69.67 |
| Total | 392,193,104 | 387,431,412 | 58.32 | 96.72 | 271,967,318 | 69.35 |
| Phenotype | Lutein | Zeaxanthin | β-Cryptoxanthin | α-Carotene | β-Carotene |
|---|---|---|---|---|---|
| G | 366.60 ± 23.26 | 3.89 ± 0.66 | 0.85 ± 0.24 | ND | 494.94 ± 45.00 |
| Y | 140.18 ± 9.43 * | 157.62 ± 15.63 ** | 0.56 ± 0.08 | ND | 105.73 ± 29.75 * |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Shi, N.; An, X.; Liu, C.; Fu, H.; Cao, L.; Feng, Y.; Sun, D.; Zhang, L. Candidate Genes for Yellow Leaf Color in Common Wheat (Triticum aestivum L.) and Major Related Metabolic Pathways according to Transcriptome Profiling. Int. J. Mol. Sci. 2018, 19, 1594. https://doi.org/10.3390/ijms19061594
Wu H, Shi N, An X, Liu C, Fu H, Cao L, Feng Y, Sun D, Zhang L. Candidate Genes for Yellow Leaf Color in Common Wheat (Triticum aestivum L.) and Major Related Metabolic Pathways according to Transcriptome Profiling. International Journal of Molecular Sciences. 2018; 19(6):1594. https://doi.org/10.3390/ijms19061594
Chicago/Turabian StyleWu, Huiyu, Narong Shi, Xuyao An, Cong Liu, Hongfei Fu, Li Cao, Yi Feng, Daojie Sun, and Lingli Zhang. 2018. "Candidate Genes for Yellow Leaf Color in Common Wheat (Triticum aestivum L.) and Major Related Metabolic Pathways according to Transcriptome Profiling" International Journal of Molecular Sciences 19, no. 6: 1594. https://doi.org/10.3390/ijms19061594
APA StyleWu, H., Shi, N., An, X., Liu, C., Fu, H., Cao, L., Feng, Y., Sun, D., & Zhang, L. (2018). Candidate Genes for Yellow Leaf Color in Common Wheat (Triticum aestivum L.) and Major Related Metabolic Pathways according to Transcriptome Profiling. International Journal of Molecular Sciences, 19(6), 1594. https://doi.org/10.3390/ijms19061594