At the Crossroads of Clinical and Preclinical Research for Muscular Dystrophy—Are We Closer to Effective Treatment for Patients?
Abstract
:1. Introduction
2. Muscular Dystrophy
2.1. General Characterization
2.2. Classification and Frequency
2.3. Genetics
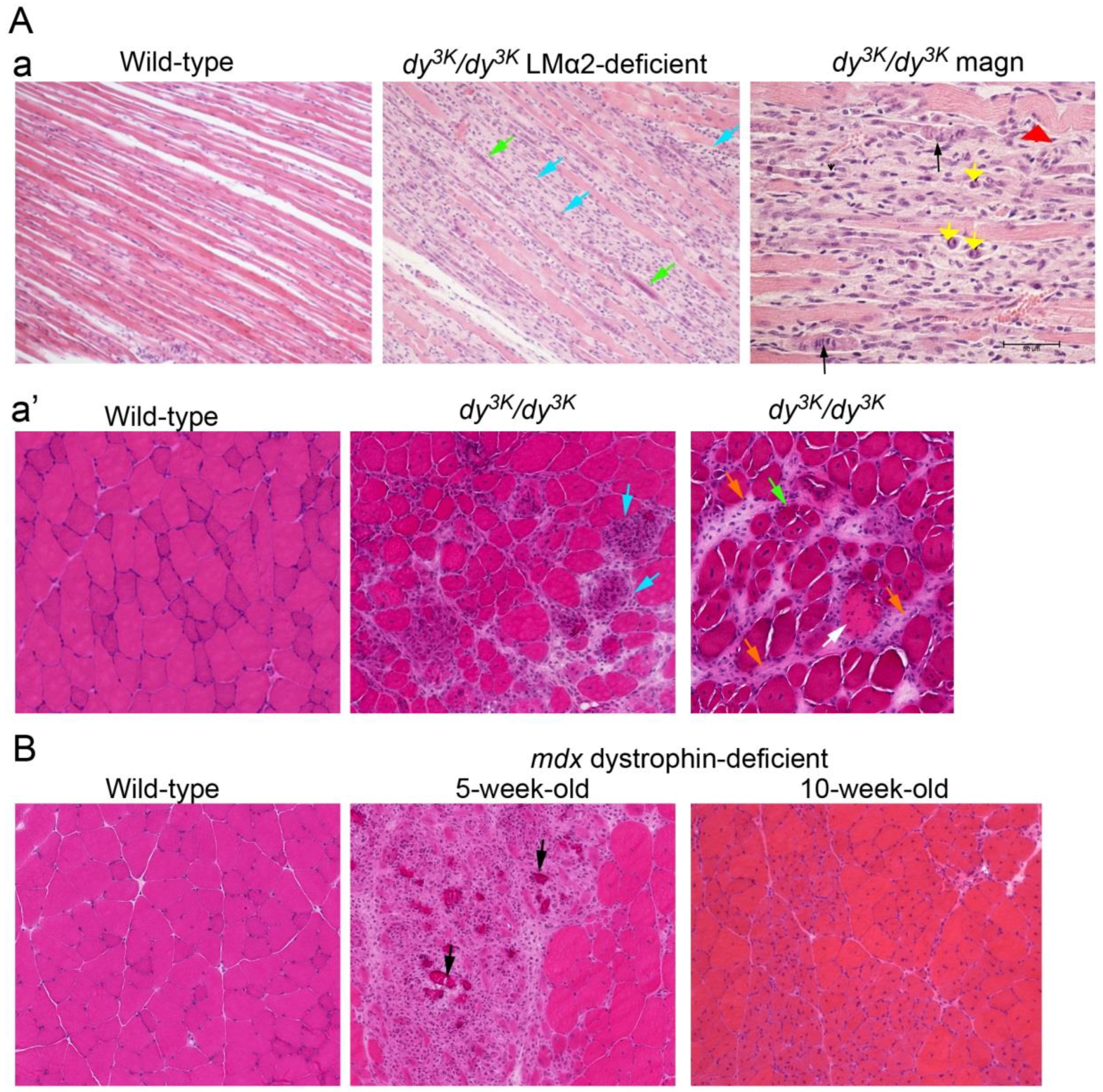
2.4. Dystrophic Pattern of Muscle Biopsy
2.5. Diagnosis
2.6. Current Management
2.7. Animal Models
3. Preclinical Studies: Strategies for Treatment
3.1. Rescue of the Primary Genetic Defect: Classical Concept
3.1.1. Virus Delivery: From Proof of Concept to Implementation
3.1.2. Cell Therapy
3.1.3. Protein Therapy
3.1.4. Endogenous Up-Regulation of Paralogous Genes
3.2. Targeting Primary Genetic Defects: Mutation Repair
3.2.1. Exon Skipping
3.2.2. CRISPR/Cas9 Genome Editing
3.2.3. Alternative Use of Antisense Methods
3.2.4. Suppression of Stop Codons
3.3. Targeting Secondary Defects of Muscular Dystrophy
Genetic Modifiers
4. Clinical Trials
Animal Models Versus Clinical Trials—Alternative Preclinical Research
5. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Engel, A.G.; Franzini-Armstrong, C. Myology, 3rd ed.; McGraw-Hill: New York, NY, USA, 2004; Volume 2. [Google Scholar]
- Norwood, F.L.; Harling, C.; Chinnery, P.F.; Eagle, M.; Bushby, K.; Straub, V. Prevalence of Genetic Muscle Disease in Northern England: In-Depth Analysis of a Muscle Clinic Population. Brain 2009, 132, 3175–3186. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, N.E.; Seto, J.T.; Hall, J.K.; Chamberlain, J.S.; Odom, G.L. Progress and Prospects of Gene Therapy Clinical Trials for the Muscular Dystrophies. Hum. Mol. Genet. 2016, 25, R9–R17. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E.; Muntoni, F. Muscular Dystrophies. Lancet 2013, 381, 845–860. [Google Scholar] [CrossRef]
- Adam, M.P.; Ardinger, H.H.; Pagon, R.A.; Wallace, S.E. Gene Reviews [Internet]; University of Washington: Seattle, WA, USA, 1993–2018. [Google Scholar]
- Amberger, J.; Bocchini, C.; Hamosh, A. A New Face and New Challenges for Online Mendelian Inheritance in Man (Omim(R)). Hum. Mutat. 2011, 32, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, C.; Clement, E.; Mein, R.; Brockington, M.; Smith, J.; Talim, B.; Straub, V.; Robb, S.; Quinlivan, R.; Feng, L.; et al. Refining Genotype Phenotype Correlations in Muscular Dystrophies with Defective Glycosylation of Dystroglycan. Brain 2007, 130, 2725–2735. [Google Scholar] [CrossRef] [PubMed]
- Bushby, K.; Finkel, R.; Birnkrant, D.J.; Case, L.E.; Clemens, P.R.; Cripe, L.; Kaul, A.; Kinnett, K.; McDonald, C.; Pandya, S.; et al. Diagnosis and Management of Duchenne Muscular Dystrophy, Part 1: Diagnosis, and Pharmacological and Psychosocial Management. Lancet Neurol. 2010, 9, 77–93. [Google Scholar] [CrossRef]
- Romitti, P.A.; Zhu, Y.; Puzhankara, S.; James, K.A.; Nabukera, S.K.; Zamba, G.K.; Ciafaloni, E.; Cunniff, C.; Druschel, C.M.; Mathews, K.D.; et al. Prevalence of Duchenne and Becker Muscular Dystrophies in the United States. Pediatrics 2015, 135, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Shilling, C.; Leslie, N.D.; Flanigan, K.M.; al-Dahhak, R.; Gastier-Foster, J.; Kneile, K.; Dunn, D.M.; Duval, B.; Aoyagi, A.; et al. Evidence-Based Path to Newborn Screening for Duchenne Muscular Dystrophy. Ann. Neurol. 2012, 71, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Emery, A.E. Population Frequencies of Inherited Neuromuscular Diseases—A World Survey. Neuromuscul. Disord. 1991, 1, 19–29. [Google Scholar] [CrossRef]
- Deenen, J.C.; Arnts, H.; van der Maarel, S.M.; Padberg, G.W.; Verschuuren, J.J.; Bakker, E.; Weinreich, S.S.; Verbeek, A.L.; van Engelen, B.G. Population-Based Incidence and Prevalence of Facioscapulohumeral Dystrophy. Neurology 2014, 83, 1056–1059. [Google Scholar] [CrossRef] [PubMed]
- Sposito, R.; Pasquali, L.; Galluzzi, F.; Rocchi, A.; Solito, B.; Soragna, D.; Tupler, R.; Siciliano, G. Facioscapulohumeral Muscular Dystrophy Type 1a in Northwestern Tuscany: A Molecular Genetics-Based Epidemiological and Genotype-Phenotype Study. Genet. Test. 2005, 9, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Clement, E.M.; Feng, L.; Mein, R.; Sewry, C.A.; Robb, S.A.; Manzur, A.Y.; Mercuri, E.; Godfrey, C.; Cullup, T.; Abbs, S.; et al. Relative Frequency of Congenital Muscular Dystrophy Subtypes: Analysis of the Uk Diagnostic Service 2001–2008. Neuromuscul. Disord. 2012, 22, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Sframeli, M.; Sarkozy, A.; Bertoli, M.; Astrea, G.; Hudson, J.; Scoto, M.; Mein, R.; Yau, M.; Phadke, R.; Feng, L.; et al. Congenital Muscular Dystrophies in the UK Population: Clinical and Molecular Spectrum of a Large Cohort Diagnosed over a 12-Year Period. Neuromuscul. Disord. 2017, 27, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Monaco, A.P.; Bertelson, C.J.; Liechti-Gallati, S.; Moser, H.; Kunkel, L.M. An Explanation for the Phenotypic Differences between Patients Bearing Partial Deletions of the Dmd Locus. Genomics 1988, 2, 90–95. [Google Scholar] [CrossRef]
- Ciafaloni, E.; Fox, D.J.; Pandya, S.; Westfield, C.P.; Puzhankara, S.; Romitti, P.A.; Mathews, K.D.; Miller, T.M.; Matthews, D.J.; Miller, L.A.; et al. Delayed Diagnosis in Duchenne Muscular Dystrophy: Data from the Muscular Dystrophy Surveillance, Tracking, and Research Network (Md Starnet). J. Pediatr. 2009, 155, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.A.; Hejtmancik, J.F.; Witkowski, J.A.; Baumbach, L.L.; Gunnell, S.; Speer, J.; Hawley, P.; Tantravahi, U.; Caskey, C.T. Prenatal Diagnosis of Duchenne Muscular Dystrophy: Prospective Linkage Analysis and Retrospective Dystrophin Cdna Analysis. Am. J. Hum. Genet. 1989, 44, 270–281. [Google Scholar] [PubMed]
- Vainzof, M.; Richard, P.; Herrmann, R.; Jimenez-Mallebrera, C.; Talim, B.; Yamamoto, L.U.; Ledeuil, C.; Mein, R.; Abbs, S.; Brockington, M.; et al. Prenatal Diagnosis in Laminin Alpha2 Chain (Merosin)-Deficient Congenital Muscular Dystrophy: A Collective Experience of Five International Centers. Neuromuscul. Disord. 2005, 15, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Lovering, R.M.; Porter, N.C.; Bloch, R.J. The Muscular Dystrophies: From Genes to Therapies. Phys. Ther. 2005, 85, 1372–1388. [Google Scholar] [PubMed]
- Bushby, K.; Finkel, R.; Birnkrant, D.J.; Case, L.E.; Clemens, P.R.; Cripe, L.; Kaul, A.; Kinnett, K.; McDonald, C.; Pandya, S.; et al. Diagnosis and Management of Duchenne Muscular Dystrophy, Part 2: Implementation of Multidisciplinary Care. Lancet Neurol. 2010, 9, 177–189. [Google Scholar] [CrossRef]
- Biggar, W.D.; Gingras, M.; Fehlings, D.L.; Harris, V.A.; Steele, C.A. Deflazacort Treatment of Duchenne Muscular Dystrophy. J. Pediatr. 2001, 138, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Durbeej, M.; Campbell, K.P. Muscular Dystrophies Involving the Dystrophin-Glycoprotein Complex: An Overview of Current Mouse Models. Curr. Opin. Genet. Dev. 2002, 12, 349–361. [Google Scholar] [CrossRef]
- Ng, R.; Banks, G.B.; Hall, J.K.; Muir, L.A.; Ramos, J.N.; Wicki, J.; Odom, G.L.; Konieczny, P.; Seto, J.; Chamberlain, J.R.; et al. Animal Models of Muscular Dystrophy. Prog. Mol. Biol. Transl. Sci. 2012, 105, 83–111. [Google Scholar] [PubMed]
- Wang, Z.; Chamberlain, J.S.; Tapscott, S.J.; Storb, R. Gene Therapy in Large Animal Models of Muscular Dystrophy. ILAR J. 2009, 50, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Lieschke, G.J.; Currie, P.D. Animal Models of Human Disease: Zebrafish Swim into View. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Bulfield, G.; Siller, W.G.; Wight, P.A.; Moore, K.J. X Chromosome-Linked Muscular Dystrophy (Mdx) in the Mouse. Proc. Natl. Acad. Sci. USA 1984, 81, 1189–1192. [Google Scholar] [CrossRef] [PubMed]
- Sicinski, P.; Geng, Y.; Ryder-Cook, A.S.; Barnard, E.A.; Darlison, M.G.; Barnard, P.J. The Molecular Basis of Muscular Dystrophy in the Mdx Mouse: A Point Mutation. Science 1989, 244, 1578–1580. [Google Scholar] [CrossRef] [PubMed]
- Kornegay, J.N.; Spurney, C.F.; Nghiem, P.P.; Brinkmeyer-Langford, C.L.; Hoffman, E.P.; Nagaraju, K. Pharmacologic Management of Duchenne Muscular Dystrophy: Target Identification and Preclinical Trials. ILAR J. 2014, 55, 119–149. [Google Scholar] [CrossRef] [PubMed]
- Deconinck, A.E.; Rafael, J.A.; Skinner, J.A.; Brown, S.C.; Potter, A.C.; Metzinger, L.; Watt, D.J.; Dickson, J.G.; Tinsley, J.M.; Davies, K.E. Utrophin-Dystrophin-Deficient Mice as a Model for Duchenne Muscular Dystrophy. Cell 1997, 90, 717–727. [Google Scholar] [CrossRef]
- Grady, R.M.; Teng, H.; Nichol, M.C.; Cunningham, J.C.; Wilkinson, R.S.; Sanes, J.R. Skeletal and Cardiac Myopathies in Mice Lacking Utrophin and Dystrophin: A Model for Duchenne Muscular Dystrophy. Cell 1997, 90, 729–738. [Google Scholar] [CrossRef]
- Coley, W.D.; Bogdanik, L.; Vila, M.C.; Yu, Q.; van der Meulen, J.H.; Rayavarapu, S.; Novak, J.S.; Nearing, M.; Quinn, J.L.; Saunders, A.; et al. Effect of Genetic Background on the Dystrophic Phenotype in Mdx Mice. Hum. Mol. Genet. 2016, 25, 130–145. [Google Scholar] [CrossRef] [PubMed]
- Gawlik, K.I.; Durbeej, M. Skeletal Muscle Laminin and Mdc1a: Pathogenesis and Treatment Strategies. Skelet. Muscle 2011, 1, 9. [Google Scholar] [CrossRef] [PubMed]
- Yurchenco, P.D.; McKee, K.K.; Reinhard, J.R.; Ruegg, M.A. Laminin-Deficient Muscular Dystrophy: Molecular Pathogenesis and Structural Repair Strategies. Matrix Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Cordova, G.; Negroni, E.; Cabello-Verrugio, C.; Mouly, V.; Trollet, C. Combined Therapies for Duchenne Muscular Dystrophy to Optimize Treatment Efficacy. Front. Genet. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Meinen, S.; Lin, S.; Thurnherr, R.; Erb, M.; Meier, T.; Ruegg, M.A. Apoptosis Inhibitors and Mini-Agrin Have Additive Benefits in Congenital Muscular Dystrophy Mice. EMBO Mol. Med. 2011, 3, 465–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, G.A.; Cole, N.M.; Matsumura, K.; Phelps, S.F.; Hauschka, S.D.; Campbell, K.P.; Faulkner, J.A.; Chamberlain, J.S. Overexpression of Dystrophin in Transgenic Mdx Mice Eliminates Dystrophic Symptoms without Toxicity. Nature 1993, 364, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Phelps, S.F.; Hauser, M.A.; Cole, N.M.; Rafael, J.A.; Hinkle, R.T.; Faulkner, J.A.; Chamberlain, J.S. Expression of Full-Length and Truncated Dystrophin Mini-Genes in Transgenic Mdx Mice. Hum. Mol. Genet. 1995, 4, 1251–1258. [Google Scholar] [CrossRef] [PubMed]
- Harper, S.Q.; Hauser, M.A.; DelloRusso, C.; Duan, D.; Crawford, R.W.; Phelps, S.F.; Harper, H.A.; Robinson, A.S.; Engelhardt, J.F.; Brooks, S.V.; et al. Modular Flexibility of Dystrophin: Implications for Gene Therapy of Duchenne Muscular Dystrophy. Nat. Med. 2002, 8, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Tinsley, J.; Deconinck, N.; Fisher, R.; Kahn, D.; Phelps, S.; Gillis, J.M.; Davies, K. Expression of Full-Length Utrophin Prevents Muscular Dystrophy in Mdx Mice. Nat. Med. 1998, 4, 1441–1444. [Google Scholar] [CrossRef] [PubMed]
- Tinsley, J.M.; Potter, A.C.; Phelps, S.R.; Fisher, R.; Trickett, J.I.; Davies, K.E. Amelioration of the Dystrophic Phenotype of Mdx Mice Using a Truncated Utrophin Transgene. Nature 1996, 384, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Gawlik, K.; Miyagoe-Suzuki, Y.; Ekblom, P.; Takeda, S.; Durbeej, M. Laminin Alpha1 Chain Reduces Muscular Dystrophy in Laminin Alpha2 Chain Deficient Mice. Hum. Mol. Genet. 2004, 13, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Gawlik, K.I.; Harandi, V.M.; Cheong, R.Y.; Petersen, A.; Durbeej, M. Laminin Alpha1 Reduces Muscular Dystrophy in Dy(2j) Mice. Matrix Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Barresi, R.; Michele, D.E.; Kanagawa, M.; Harper, H.A.; Dovico, S.A.; Satz, J.S.; Moore, S.A.; Zhang, W.; Schachter, H.; Dumanski, J.P.; et al. Large Can Functionally Bypass Alpha-Dystroglycan Glycosylation Defects in Distinct Congenital Muscular Dystrophies. Nat. Med. 2004, 10, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; He, Y.; Wang, K.; Zhang, P.; Zhang, S.; Hu, H. Adeno-Associated Viral-Mediated Large Gene Therapy Rescues the Muscular Dystrophic Phenotype in Mouse Models of Dystroglycanopathy. Hum. Gene Ther. 2013, 24, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Moll, J.; Barzaghi, P.; Lin, S.; Bezakova, G.; Lochmuller, H.; Engvall, E.; Muller, U.; Ruegg, M.A. An Agrin Minigene Rescues Dystrophic Symptoms in a Mouse Model for Congenital Muscular Dystrophy. Nature 2001, 413, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Shaw, K.; Phillips, A.; Cooper, D.N. The Human Gene Mutation Database: Building a Comprehensive Mutation Repository for Clinical and Molecular Genetics, Diagnostic Testing and Personalized Genomic Medicine. Hum. Genet. 2014, 133, 1–9. [Google Scholar] [CrossRef] [PubMed]
- McKee, K.K.; Crosson, S.C.; Meinen, S.; Reinhard, J.R.; Ruegg, M.A.; Yurchenco, P.D. Chimeric Protein Repair of Laminin Polymerization Ameliorates Muscular Dystrophy Phenotype. J. Clin. Investig. 2017, 127, 1075–1089. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, J.R.; Lin, S.; McKee, K.K.; Meinen, S.; Crosson, S.C.; Sury, M.; Hobbs, S.; Maier, G.; Yurchenco, P.D.; Ruegg, M.A. Linker Proteins Restore Basement Membrane and Correct Lama2-Related Muscular Dystrophy in Mice. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Daya, S.; Berns, K.I. Gene Therapy Using Adeno-Associated Virus Vectors. Clin. Microbiol. Rev. 2008, 21, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Gregorevic, P.; Blankinship, M.J.; Allen, J.M.; Crawford, R.W.; Meuse, L.; Miller, D.G.; Russell, D.W.; Chamberlain, J.S. Systemic Delivery of Genes to Striated Muscles Using Adeno-Associated Viral Vectors. Nat. Med. 2004, 10, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Gregorevic, P.; Blankinship, M.J.; Chamberlain, J.S. Viral Vectors for Gene Transfer to Striated Muscle. Curr. Opin. Mol. Ther. 2004, 6, 491–498. [Google Scholar] [PubMed]
- Durbeej, M.; Sawatzki, S.M.; Barresi, R.; Schmainda, K.M.; Allamand, V.; Michele, D.E.; Campbell, K.P. Gene Transfer Establishes Primacy of Striated Vs. Smooth Muscle Sarcoglycan Complex in Limb-Girdle Muscular Dystrophy. Proc. Natl. Acad. Sci. USA 2003, 100, 8910–8915. [Google Scholar] [CrossRef] [PubMed]
- Allamand, V.; Donahue, K.M.; Straub, V.; Davisson, R.L.; Davidson, B.L.; Campbell, K.P. Early Adenovirus-Mediated Gene Transfer Effectively Prevents Muscular Dystrophy in Alpha-Sarcoglycan-Deficient Mice. Gene Ther. 2000, 7, 1385–1391. [Google Scholar] [CrossRef] [PubMed]
- Cordier, L.; Hack, A.A.; Scott, M.O.; Barton-Davis, E.R.; Gao, G.; Wilson, J.M.; McNally, E.M.; Sweeney, H.L. Rescue of Skeletal Muscles of Gamma-Sarcoglycan-Deficient Mice with Adeno-Associated Virus-Mediated Gene Transfer. Mol. Ther. 2000, 1, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Gicquel, E.; Maizonnier, N.; Foltz, S.J.; Martin, W.J.; Bourg, N.; Svinartchouk, F.; Charton, K.; Beedle, A.M.; Richard, I. Aav-Mediated Transfer of Fkrp Shows Therapeutic Efficacy in a Murine Model but Requires Control of Gene Expression. Hum. Mol. Genet. 2017, 26, 1952–1965. [Google Scholar] [CrossRef] [PubMed]
- Qiao, C.; Li, J.; Zhu, T.; Draviam, R.; Watkins, S.; Ye, X.; Chen, C.; Li, J.; Xiao, X. Amelioration of Laminin-Alpha2-Deficient Congenital Muscular Dystrophy by Somatic Gene Transfer of Miniagrin. Proc. Natl. Acad. Sci. USA 2005, 102, 11999–12004. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Storb, R.; Halbert, C.L.; Banks, G.B.; Butts, T.M.; Finn, E.E.; Allen, J.M.; Miller, A.D.; Chamberlain, J.S.; Tapscott, S.J. Successful Regional Delivery and Long-Term Expression of a Dystrophin Gene in Canine Muscular Dystrophy: A Preclinical Model for Human Therapies. Mol. Ther. 2012, 20, 1501–1507. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Pan, X.; Hakim, C.H.; Yang, H.T.; Yue, Y.; Zhang, K.; Terjung, R.L.; Duan, D. Microdystrophin Ameliorates Muscular Dystrophy in the Canine Model of Duchenne Muscular Dystrophy. Mol. Ther. 2013, 21, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Kornegay, J.N.; Li, J.; Bogan, J.R.; Bogan, D.J.; Chen, C.; Zheng, H.; Wang, B.; Qiao, C.; Howard, J.F., Jr.; Xiao, X. Widespread Muscle Expression of an Aav9 Human Mini-Dystrophin Vector after Intravenous Injection in Neonatal Dystrophin-Deficient Dogs. Mol. Ther. 2010, 18, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, J.R.; Chamberlain, J.S. Progress toward Gene Therapy for Duchenne Muscular Dystrophy. Mol. Ther. 2017, 25, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
- Crist, C. Emerging New Tools to Study and Treat Muscle Pathologies: Genetics and Molecular Mechanisms Underlying Skeletal Muscle Development, Regeneration, and Disease. J. Pathol. 2017, 241, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, S.; Hoshiya, H.; Tedesco, F.S. Repair or Replace? Exploiting Novel Gene and Cell Therapy Strategies for Muscular Dystrophies. FEBS J. 2013, 280, 4263–4280. [Google Scholar] [CrossRef] [PubMed]
- Sampaolesi, M.; Blot, S.; D’Antona, G.; Granger, N.; Tonlorenzi, R.; Innocenzi, A.; Mognol, P.; Thibaud, J.L.; Galvez, B.G.; Barthelemy, I.; et al. Mesoangioblast Stem Cells Ameliorate Muscle Function in Dystrophic Dogs. Nature 2006, 444, 574–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sampaolesi, M.; Torrente, Y.; Innocenzi, A.; Tonlorenzi, R.; D’Antona, G.; Pellegrino, M.A.; Barresi, R.; Bresolin, N.; de Angelis, M.G.; Campbell, K.P.; et al. Cell Therapy of Alpha-Sarcoglycan Null Dystrophic Mice through Intra-Arterial Delivery of Mesoangioblasts. Science 2003, 301, 487–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domi, T.; Porrello, E.; Velardo, D.; Capotondo, A.; Biffi, A.; Tonlorenzi, R.; Amadio, S.; Ambrosi, A.; Miyagoe-Suzuki, Y.; Takeda, S.; et al. Mesoangioblast Delivery of Miniagrin Ameliorates Murine Model of Merosin-Deficient Congenital Muscular Dystrophy Type 1a. Skelet. Muscle 2015, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Sienkiewicz, D.; Kulak, W.; Okurowska-Zawada, B.; Paszko-Patej, G.; Kawnik, K. Duchenne Muscular Dystrophy: Current Cell Therapies. Ther. Adv. Neurol. Disord. 2015, 8, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Cossu, G.; Previtali, S.C.; Napolitano, S.; Cicalese, M.P.; Tedesco, F.S.; Nicastro, F.; Noviello, M.; Roostalu, U.; Sora, M.G.N.; Scarlato, M.; et al. Intra-Arterial Transplantation of Hla-Matched Donor Mesoangioblasts in Duchenne Muscular Dystrophy. EMBO Mol. Med. 2015, 7, 1513–1528. [Google Scholar] [CrossRef] [PubMed]
- Rooney, J.E.; Knapp, J.R.; Hodges, B.L.; Wuebbles, R.D.; Burkin, D.J. Laminin-111 Protein Therapy Reduces Muscle Pathology and Improves Viability of a Mouse Model of Merosin-Deficient Congenital Muscular Dystrophy. Am. J. Pathol. 2012, 180, 1593–1602. [Google Scholar] [CrossRef] [PubMed]
- Sonnemann, K.J.; Heun-Johnson, H.; Turner, A.J.; Baltgalvis, K.A.; Lowe, D.A.; Ervasti, J.M. Functional Substitution by Tat-Utrophin in Dystrophin-Deficient Mice. PLoS Med. 2009, 6, e1000083. [Google Scholar] [CrossRef] [PubMed]
- Rooney, J.E.; Gurpur, P.B.; Burkin, D.J. Laminin-111 Protein Therapy Prevents Muscle Disease in the Mdx Mouse Model for Duchenne Muscular Dystrophy. Proc. Natl. Acad. Sci. USA 2009, 106, 7991–7996. [Google Scholar] [CrossRef] [PubMed]
- Amenta, A.R.; Yilmaz, A.; Bogdanovich, S.; McKechnie, B.A.; Abedi, M.; Khurana, T.S.; Fallon, J.R. Biglycan Recruits Utrophin to the Sarcolemma and Counters Dystrophic Pathology in Mdx Mice. Proc. Natl. Acad. Sci. USA 2011, 108, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Von Maltzahn, J.; Renaud, J.M.; Parise, G.; Rudnicki, M.A. Wnt7a Treatment Ameliorates Muscular Dystrophy. Proc. Natl. Acad. Sci. USA 2012, 109, 20614–20619. [Google Scholar] [CrossRef] [PubMed]
- Van Ry, P.M.; Wuebbles, R.D.; Key, M.; Burkin, D.J. Galectin-1 Protein Therapy Prevents Pathology and Improves Muscle Function in the Mdx Mouse Model of Duchenne Muscular Dystrophy. Mol. Ther. 2015, 23, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Gawlik, K.I.; Durbeej, M. Transgenic Overexpression of Laminin Alpha1 Chain in Laminin Alpha2 Chain-Deficient Mice Rescues the Disease Throughout the Lifespan. Muscle Nerve 2010, 42, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Moorwood, C.; Lozynska, O.; Suri, N.; Napper, A.D.; Diamond, S.L.; Khurana, T.S. Drug Discovery for Duchenne Muscular Dystrophy Via Utrophin Promoter Activation Screening. PLoS ONE 2011, 6, e26169. [Google Scholar] [CrossRef] [PubMed]
- Miura, P.; Chakkalakal, J.V.; Boudreault, L.; Belanger, G.; Hebert, R.L.; Renaud, J.M.; Jasmin, B.J. Pharmacological Activation of Pparbeta/Delta Stimulates Utrophin a Expression in Skeletal Muscle Fibers and Restores Sarcolemmal Integrity in Mature Mdx Mice. Hum. Mol. Genet. 2009, 18, 4640–4649. [Google Scholar] [CrossRef] [PubMed]
- Di Certo, M.G.; Corbi, N.; Strimpakos, G.; Onori, A.; Luvisetto, S.; Severini, C.; Guglielmotti, A.; Batassa, E.M.; Pisani, C.; Floridi, A.; et al. The Artificial Gene Jazz, a Transcriptional Regulator of Utrophin, Corrects the Dystrophic Pathology in Mdx Mice. Hum. Mol. Genet. 2010, 19, 752–760. [Google Scholar] [CrossRef] [PubMed]
- Miura, P.; Thompson, J.; Chakkalakal, J.V.; Holcik, M.; Jasmin, B.J. The Utrophin a 5’-Untranslated Region Confers Internal Ribosome Entry Site-Mediated Translational Control during Regeneration of Skeletal Muscle Fibers. J. Biol. Chem. 2005, 280, 32997–33005. [Google Scholar] [CrossRef] [PubMed]
- Perrin, A.; Rousseau, J.; Tremblay, J.P. Increased Expression of Laminin Subunit Alpha 1 Chain by Dcas9-Vp160. Mol. Ther. Nucleic Acids 2017, 6, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Tanganyika-de Winter, C.L.; Heemskerk, H.; Karnaoukh, T.G.; van Putten, M.; de Kimpe, S.J.; van Deutekom, J.; Aartsma-Rus, A. Long-Term Exon Skipping Studies with 2’-O-Methyl Phosphorothioate Antisense Oligonucleotides in Dystrophic Mouse Models. Mol. Ther. Nucleic Acids 2012, 1, e44. [Google Scholar] [CrossRef] [PubMed]
- Arnett, A.L.; Chamberlain, J.R.; Chamberlain, J.S. Therapy for Neuromuscular Disorders. Curr. Opin. Genet. Dev. 2009, 19, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Lu, Q.; Wood, M. Effective Exon Skipping and Restoration of Dystrophin Expression by Peptide Nucleic Acid Antisense Oligonucleotides in Mdx Mice. Mol. Ther. 2008, 16, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Goemans, N.M.; Tulinius, M.; van den Akker, J.T.; Burm, B.E.; Ekhart, P.F.; Heuvelmans, N.; Holling, T.; Janson, A.A.; Platenburg, G.J.; Sipkens, J.A.; et al. Systemic Administration of Pro051 in Duchenne’s Muscular Dystrophy. N. Engl. J. Med. 2011, 364, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Cirak, S.; Arechavala-Gomeza, V.; Guglieri, M.; Feng, L.; Torelli, S.; Anthony, K.; Abbs, S.; Garralda, M.E.; Bourke, J.; Wells, D.J.; et al. Exon Skipping and Dystrophin Restoration in Patients with Duchenne Muscular Dystrophy after Systemic Phosphorodiamidate Morpholino Oligomer Treatment: An Open-Label, Phase 2, dose-escalation study. Lancet 2011, 2, 595–605. [Google Scholar] [CrossRef]
- Mendell, J.R.; Rodino-Klapac, L.R.; Sahenk, Z.; Roush, K.; Bird, L.; Lowes, L.P.; Alfano, L.; Gomez, A.M.; Lewis, S.; Kota, J.; et al. Eteplirsen for the Treatment of Duchenne Muscular Dystrophy. Ann. Neurol. 2013, 74, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Goemans, N.; Lowes, L.P.; Alfano, L.N.; Berry, K.; Shao, J.; Kaye, E.M.; Mercuri, E.; Eteplirsen Study Group and Telethon Foundation DMD Italian Network. Longitudinal Effect of Eteplirsen versus Historical Control on Ambulation in Duchenne Muscular Dystrophy. Ann. Neurol. 2016, 79, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Kole, R.; Krieg, A.M. Exon Skipping Therapy for Duchenne Muscular Dystrophy. Adv. Drug Deliv. Rev. 2015, 87, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.L.; Cirak, S.; Partridge, T. What Can We Learn from Clinical Trials of Exon Skipping for Dmd? Mol. Ther. Nucleic Acids 2014, 3, e152. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.R.; Maruyama, R.; Yokota, T. Eteplirsen in the Treatment of Duchenne Muscular Dystrophy. Drug Des. Dev. Ther. 2017, 11, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Le Guiner, C.; Montus, M.; Servais, L.; Cherel, Y.; Francois, V.; Thibaud, J.L.; Wary, C.; Matot, B.; Larcher, T.; Guigand, L.; et al. Forelimb Treatment in a Large Cohort of Dystrophic Dogs Supports Delivery of a Recombinant Aav for Exon Skipping in Duchenne Patients. Mol. Ther. 2014, 22, 1923–1935. [Google Scholar] [CrossRef] [PubMed]
- Goyenvalle, A.; Griffith, G.; Babbs, A.; el Andaloussi, S.; Ezzat, K.; Avril, A.; Dugovic, B.; Chaussenot, R.; Ferry, A.; Voit, T.; et al. Functional Correction in Mouse Models of Muscular Dystrophy Using Exon-Skipping Tricyclo-DNA Oligomers. Nat. Med. 2015, 21, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using Crispr/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; McAnally, J.R.; Shelton, J.M.; Mireault, A.A.; Bassel-Duby, R.; Olson, E.N. Prevention of Muscular Dystrophy in Mice by Crispr/Cas9-Mediated Editing of Germline DNA. Science 2014, 345, 1184–1188. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; Amoasii, L.; Mireault, A.A.; McAnally, J.R.; Li, H.; Sanchez-Ortiz, E.; Bhattacharyya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Postnatal Genome Editing Partially Restores Dystrophin Expression in a Mouse Model of Muscular Dystrophy. Science 2016, 351, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Rivera, R.M.C.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In Vivo Genome Editing Improves Muscle Function in a Mouse Model of Duchenne Muscular Dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In Vivo Gene Editing in Dystrophic Mouse Muscle and Muscle Stem Cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, N.E.; Hall, J.K.; Odom, G.L.; Phelps, M.P.; Andrus, C.R.; Hawkins, R.D.; Hauschka, S.D.; Chamberlain, J.R.; Chamberlain, J.S. Muscle-Specific Crispr/Cas9 Dystrophin Gene Editing Ameliorates Pathophysiology in a Mouse Model for Duchenne Muscular Dystrophy. Nat. Commun. 2017, 8, 14454. [Google Scholar] [CrossRef] [PubMed]
- Kemaladewi, D.U.; Maino, E.; Hyatt, E.; Hou, H.; Ding, M.; Place, K.M.; Zhu, X.; Bassi, P.; Baghestani, Z.; Deshwar, A.G.; et al. Correction of a Splicing Defect in a Mouse Model of Congenital Muscular Dystrophy Type 1a Using a Homology-Directed-Repair-Independent Mechanism. Nat. Med. 2017, 23, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.M.; Sobczak, K.; Lueck, J.D.; Osborne, R.J.; Lin, X.; Dirksen, R.T.; Thornton, C.A. Reversal of RNA Dominance by Displacement of Protein Sequestered on Triplet Repeat RNA. Science 2009, 325, 336–339. [Google Scholar] [CrossRef] [PubMed]
- Dent, K.M.; Dunn, D.M.; von Niederhausern, A.C.; Aoyagi, A.T.; Kerr, L.; Bromberg, M.B.; Hart, K.J.; Tuohy, T.; White, S.; den Dunnen, J.T.; et al. Improved Molecular Diagnosis of Dystrophinopathies in an Unselected Clinical Cohort. Am. J. Med. Genet. A 2005, 134, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Welch, E.M.; Barton, E.R.; Zhuo, J.; Tomizawa, Y.; Friesen, W.J.; Trifillis, P.; Paushkin, S.; Patel, M.; Trotta, C.R.; Hwang, S.; et al. Ptc124 Targets Genetic Disorders Caused by Nonsense Mutations. Nature 2007, 447, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Hirawat, S.; Welch, E.M.; Elfring, G.L.; Northcutt, V.J.; Paushkin, S.; Hwang, S.; Leonard, E.M.; Almstead, N.G.; Ju, W.; Peltz, S.W.; et al. Safety, Tolerability, and Pharmacokinetics of Ptc124, a Nonaminoglycoside Nonsense Mutation Suppressor, Following Single- and Multiple-Dose Administration to Healthy Male and Female Adult Volunteers. J. Clin. Pharmacol. 2007, 47, 430–444. [Google Scholar] [CrossRef] [PubMed]
- Finkel, R.S.; Flanigan, K.M.; Wong, B.; Bonnemann, C.; Sampson, J.; Sweeney, H.L.; Reha, A.; Northcutt, V.J.; Elfring, G.; Barth, J.; et al. Phase 2a Study of Ataluren-Mediated Dystrophin Production in Patients with Nonsense Mutation Duchenne Muscular Dystrophy. PLoS ONE 2013, 8, e81302. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.P.; Connor, E.M. Orphan Drug Development in Muscular Dystrophy: Update on Two Large Clinical Trials of Dystrophin Rescue Therapies. Discov. Med. 2013, 16, 233–239. [Google Scholar] [PubMed]
- Bushby, K.; Finkel, R.; Wong, B.; Barohn, R.; Campbell, C.; Comi, G.P.; Connolly, A.M.; Day, J.W.; Flanigan, K.M.; Goemans, N.; et al. Ataluren Treatment of Patients with Nonsense Mutation Dystrophinopathy. Muscle Nerve 2014, 50, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Haas, M.; Vlcek, V.; Balabanov, P.; Salmonson, T.; Bakchine, S.; Markey, G.; Weise, M.; Schlosser-Weber, G.; Brohmann, H.; Yerro, C.P.; et al. European Medicines Agency Review of Ataluren for the Treatment of Ambulant Patients Aged 5 Years and Older with Duchenne Muscular Dystrophy Resulting from a Nonsense Mutation in the Dystrophin Gene. Neuromuscul. Disord. 2015, 25, 5–13. [Google Scholar] [CrossRef] [PubMed]
- McDonald, C.M.; Henricson, E.K.; Abresch, R.T.; Florence, J.; Eagle, M.; Gappmaier, E.; Glanzman, A.M.; Spiegel, R.; Barth, J.; Elfring, G.; et al. The 6-Minute Walk Test and Other Clinical Endpoints in Duchenne Muscular Dystrophy: Reliability, Concurrent Validity, and Minimal Clinically Important Differences from a Multicenter Study. Muscle Nerve 2013, 48, 357–368. [Google Scholar] [CrossRef] [PubMed]
- McDonald, C.M.; Henricson, E.K.; Abresch, R.T.; Florence, J.M.; Eagle, M.; Gappmaier, E.; Glanzman, A.M.; Spiegel, R.; Barth, J.; Elfring, G.; et al. The 6-Minute Walk Test and Other Endpoints in Duchenne Muscular Dystrophy: Longitudinal Natural History Observations over 48 Weeks from a Multicenter Study. Muscle Nerve 2013, 48, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Peltz, S.W.; Morsy, M.; Welch, E.M.; Jacobson, A. Ataluren as an Agent for Therapeutic Nonsense Suppression. Annu. Rev. Med. 2013, 64, 407–425. [Google Scholar] [CrossRef] [PubMed]
- McDonald, C.M.; Campbell, C.; Torricelli, R.E.; Finkel, R.S.; Flanigan, K.M.; Goemans, N.; Heydemann, P.; Kaminska, A.; Kirschner, J.; Muntoni, F.; et al. Ataluren in Patients with Nonsense Mutation Duchenne Muscular Dystrophy (Act Dmd): A Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2017, 390, 1489–1498. [Google Scholar] [CrossRef]
- Ryan, N.J. Ataluren: First Global Approval. Drugs 2014, 74, 1709–1714. [Google Scholar] [CrossRef] [PubMed]
- Millay, D.P.; Sargent, M.A.; Osinska, H.; Baines, C.P.; Barton, E.R.; Vuagniaux, G.; Sweeney, H.L.; Robbins, J.; Molkentin, J.D. Genetic and Pharmacologic Inhibition of Mitochondrial-Dependent Necrosis Attenuates Muscular Dystrophy. Nat. Med. 2008, 14, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Percival, J.M.; Whitehead, N.P.; Adams, M.E.; Adamo, C.M.; Beavo, J.A.; Froehner, S.C. Sildenafil Reduces Respiratory Muscle Weakness and Fibrosis in the Mdx Mouse Model of Duchenne Muscular Dystrophy. J. Pathol. 2012, 228, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Jahnke, V.E.; van der Meulen, J.H.; Johnston, H.K.; Ghimbovschi, S.; Partridge, T.; Hoffman, E.P.; Nagaraju, K. Metabolic Remodeling Agents Show Beneficial Effects in the Dystrophin-Deficient Mdx Mouse Model. Skelet. Muscle 2012, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Cozzoli, A.; Nico, B.; Sblendorio, V.T.; Capogrosso, R.F.; Dinardo, M.M.; Longo, V.; Gagliardi, S.; Montagnani, M.; de Luca, A. Enalapril Treatment Discloses an Early Role of Angiotensin Ii in Inflammation- and Oxidative Stress-Related Muscle Damage in Dystrophic Mdx Mice. Pharmacol. Res. 2011, 64, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Buyse, G.M.; van der Mieren, G.; Erb, M.; D’Hooge, J.; Herijgers, P.; Verbeken, E.; Jara, A.; van den Bergh, A.; Mertens, L.; Courdier-Fruh, I.; et al. Long-Term Blinded Placebo-Controlled Study of Snt-Mc17/Idebenone in the Dystrophin Deficient Mdx Mouse: Cardiac Protection and Improved Exercise Performance. Eur. Heart J. 2009, 30, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Girgenrath, M.; Beermann, M.L.; Vishnudas, V.K.; Homma, S.; Miller, J.B. Pathology Is Alleviated by Doxycycline in a Laminin-Alpha2-Null Model of Congenital Muscular Dystrophy. Ann. Neurol. 2009, 65, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Erb, M.; Meinen, S.; Barzaghi, P.; Sumanovski, L.T.; Courdier-Fruh, I.; Ruegg, M.A.; Meier, T. Omigapil Ameliorates the Pathology of Muscle Dystrophy Caused by Laminin-Alpha2 Deficiency. J. Pharmacol. Exp. Ther. 2009, 331, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Irwin, W.A.; Bergamin, N.; Sabatelli, P.; Reggiani, C.; Megighian, A.; Merlini, L.; Braghetta, P.; Columbaro, M.; Volpin, D.; Bressan, G.M.; et al. Mitochondrial Dysfunction and Apoptosis in Myopathic Mice with Collagen Vi Deficiency. Nat. Genet. 2003, 35, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Tiepolo, T.; Angelin, A.; Palma, E.; Sabatelli, P.; Merlini, L.; Nicolosi, L.; Finetti, F.; Braghetta, P.; Vuagniaux, G.; Dumont, J.M.; et al. The Cyclophilin Inhibitor Debio 025 Normalizes Mitochondrial Function, Muscle Apoptosis and Ultrastructural Defects in Col6a1−/− Myopathic Mice. Br. J. Pharmacol. 2009, 157, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Palma, E.; Tiepolo, T.; Angelin, A.; Sabatelli, P.; Maraldi, N.M.; Basso, E.; Forte, M.A.; Bernardi, P.; Bonaldo, P. Genetic Ablation of Cyclophilin D Rescues Mitochondrial Defects and Prevents Muscle Apoptosis in Collagen Vi Myopathic Mice. Hum. Mol. Genet. 2009, 18, 2024–2031. [Google Scholar] [CrossRef] [PubMed]
- Cohn, R.D.; van Erp, C.; Habashi, J.P.; Soleimani, A.A.; Klein, E.C.; Lisi, M.T.; Gamradt, M.; Rhys, C.M.A.; Holm, T.M.; Loeys, B.L.; et al. Angiotensin Ii Type 1 Receptor Blockade Attenuates Tgf-Beta-Induced Failure of Muscle Regeneration in Multiple Myopathic States. Nat. Med. 2007, 13, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Turgeman, T.; Hagai, Y.; Huebner, K.; Jassal, D.S.; Anderson, J.E.; Genin, O.; Nagler, A.; Halevy, O.; Pines, M. Prevention of Muscle Fibrosis and Improvement in Muscle Performance in the Mdx Mouse by Halofuginone. Neuromuscul. Disord. 2008, 18, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Nevo, Y.; Halevy, O.; Genin, O.; Moshe, I.; Turgeman, T.; Harel, M.; Biton, E.; Reif, S.; Pines, M. Fibrosis Inhibition and Muscle Histopathology Improvement in Laminin-Alpha2-Deficient Mice. Muscle Nerve 2010, 42, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Pines, M.; Halevy, O. Halofuginone and Muscular Dystrophy. Histol. Histopathol. 2011, 26, 135–146. [Google Scholar] [PubMed]
- Accorsi, A.; Kumar, A.; Rhee, Y.; Miller, A.; Girgenrath, M. Igf-1/Gh Axis Enhances Losartan Treatment in Lama2-Related Muscular Dystrophy. Hum. Mol. Genet. 2016, 25, 4624–4634. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, J.; Kumar, A.; Duarte, L.; Mehuron, T.; Girgenrath, M. Triggering Regeneration and Tackling Apoptosis: A Combinatorial Approach to Treating Congenital Muscular Dystrophy Type 1 A. Hum. Mol. Genet. 2013, 22, 4306–4317. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.D.; Flanigan, K.M.; Thrush, P.T.; Dvorchik, I.; Yin, H.; Canter, C.; Connolly, A.M.; Parrish, M.; McDonald, C.M.; Braunlin, E.; et al. A Randomized, Double-Blind Trial of Lisinopril and Losartan for the Treatment of Cardiomyopathy in Duchenne Muscular Dystrophy. PLoS Curr. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Elbaz, M.; Yanay, N.; Aga-Mizrachi, S.; Brunschwig, Z.; Kassis, I.; Ettinger, K.; Barak, V.; Nevo, Y. Losartan, a Therapeutic Candidate in Congenital Muscular Dystrophy: Studies in the Dy(2j) /Dy(2j) Mouse. Ann. Neurol. 2012, 71, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Lemos, D.R.; Babaeijandaghi, F.; Low, M.; Chang, C.K.; Lee, S.T.; Fiore, D.; Zhang, R.H.; Natarajan, A.; Nedospasov, S.A.; Rossi, F.M. Nilotinib Reduces Muscle Fibrosis in Chronic Muscle Injury by Promoting Tnf-Mediated Apoptosis of Fibro/Adipogenic Progenitors. Nat. Med. 2015, 21, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Sinadinos, A.; Young, C.N.; Al-Khalidi, R.; Teti, A.; Kalinski, P.; Mohamad, S.; Floriot, L.; Henry, T.; Tozzi, G.; Jiang, T.; et al. P2rx7 Purinoceptor: A Therapeutic Target for Ameliorating the Symptoms of Duchenne Muscular Dystrophy. PLoS Med. 2015, 12, e1001888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Khalidi, R.; Panicucci, C.; Cox, P.; Chira, N.; Rog, J.; Young, C.N.J.; McGeehan, R.E.; Ambati, K.; Ambati, J.; Zablocki, K.; et al. Zidovudine Ameliorates Pathology in the Mouse Model of Duchenne Muscular Dystrophy Via P2rx7 Purinoceptor Antagonism. Acta Neuropathol. Commun. 2018, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Kharraz, Y.; Guerra, J.; Mann, C.J.; Serrano, A.L.; Munoz-Canoves, P. Macrophage Plasticity and the Role of Inflammation in Skeletal Muscle Repair. Mediat. Inflamm. 2013, 2013, 491497. [Google Scholar] [CrossRef] [PubMed]
- Capote, J.; Kramerova, I.; Martinez, L.; Vetrone, S.; Barton, E.R.; Sweeney, H.L.; Miceli, M.C.; Spencer, M.J. Osteopontin Ablation Ameliorates Muscular Dystrophy by Shifting Macrophages to a Pro-Regenerative Phenotype. J. Cell Biol. 2016, 213, 275–288. [Google Scholar] [CrossRef] [PubMed]
- Carmignac, V.; Quere, R.; Durbeej, M. Proteasome Inhibition Improves the Muscle of Laminin Alpha2 Chain-Deficient Mice. Hum. Mol. Genet. 2011, 20, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, T.; Fujimoto, S.; Ito, T.; Horinouchi, H.; Ueyama, H.; Tsuda, T. Proteasome Expression in the Skeletal Muscles of Patients with Muscular Dystrophy. Acta Neuropathol. 2000, 100, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Korner, Z.; Fontes-Oliveira, C.C.; Holmberg, J.; Carmignac, V.; Durbeej, M. Bortezomib Partially Improves Laminin Alpha2 Chain-Deficient Muscular Dystrophy. Am. J. Pathol. 2014, 184, 1518–1528. [Google Scholar] [CrossRef] [PubMed]
- Gazzerro, E.; Assereto, S.; Bonetto, A.; Sotgia, F.; Scarfi, S.; Pistorio, A.; Bonuccelli, G.; Cilli, M.; Bruno, C.; Zara, F.; et al. Therapeutic Potential of Proteasome Inhibition in Duchenne and Becker Muscular Dystrophies. Am. J. Pathol. 2010, 176, 1863–1877. [Google Scholar] [CrossRef] [PubMed]
- Carmignac, V.; Svensson, M.; Korner, Z.; Elowsson, L.; Matsumura, C.; Gawlik, K.I.; Allamand, V.; Durbeej, M. Autophagy Is Increased in Laminin Alpha2 Chain-Deficient Muscle and Its Inhibition Improves Muscle Morphology in a Mouse Model of Mdc1a. Hum. Mol. Genet. 2011, 20, 4891–4902. [Google Scholar] [CrossRef] [PubMed]
- Grumati, P.; Coletto, L.; Sabatelli, P.; Cescon, M.; Angelin, A.; Bertaggia, E.; Blaauw, B.; Urciuolo, A.; Tiepolo, T.; Merlini, L.; et al. Autophagy Is Defective in Collagen Vi Muscular Dystrophies, and Its Reactivation Rescues Myofiber Degeneration. Nat. Med. 2010, 16, 1313–1320. [Google Scholar] [CrossRef] [PubMed]
- De Palma, C.; Morisi, F.; Cheli, S.; Pambianco, S.; Cappello, V.; Vezzoli, M.; Rovere-Querini, P.; Moggio, M.; Ripolone, M.; Francolini, M.; et al. Autophagy as a New Therapeutic Target in Duchenne Muscular Dystrophy. Cell Death Dis. 2012, 3, e418. [Google Scholar] [CrossRef] [PubMed]
- Ramos, F.J.; Chen, S.C.; Garelick, M.G.; Dai, D.F.; Liao, C.Y.; Schreiber, K.H.; MacKay, V.L.; An, E.H.; Strong, R.; Ladiges, W.C.; et al. Rapamycin Reverses Elevated Mtorc1 Signaling in Lamin a/C-Deficient Mice, Rescues Cardiac and Skeletal Muscle Function, and Extends Survival. Sci. Transl. Med. 2012, 4, 144ra03. [Google Scholar] [CrossRef] [PubMed]
- Castagnaro, S.; Pellegrini, C.; Pellegrini, M.; Chrisam, M.; Sabatelli, P.; Toni, S.; Grumati, P.; Ripamonti, C.; Pratelli, L.; Maraldi, N.M.; et al. Autophagy Activation in Col6 Myopathic Patients by a Low-Protein-Diet Pilot Trial. Autophagy 2016, 12, 2484–2495. [Google Scholar] [CrossRef] [PubMed]
- Guridi, M.; Kupr, B.; Romanino, K.; Lin, S.; Falcetta, D.; Tintignac, L.; Ruegg, M.A. Alterations to Mtorc1 Signaling in the Skeletal Muscle Differentially Affect Whole-Body Metabolism. Skelet. Muscle 2016, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Fontes-Oliveira, C.C.; Steinz, M.; Schneiderat, P.; Mulder, H.; Durbeej, M. Bioenergetic Impairment in Congenital Muscular Dystrophy Type 1a and Leigh Syndrome Muscle Cells. Sci. Rep. 2017, 7, 45272. [Google Scholar] [CrossRef] [PubMed]
- Cruz Guzman Odel, R.; Garcia, A.L.C.; Rodriguez-Cruz, M. Muscular Dystrophies at Different Ages: Metabolic and Endocrine Alterations. Int. J. Endocrinol. 2012, 2012, 485376. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, B.M.; Matsumura, C.Y.; Fontes-Oliveira, C.C.; Gawlik, K.I.; Acosta, H.; Wernhoff, P.; Durbeej, M. Quantitative Proteomic Analysis Reveals Metabolic Alterations, Calcium Dysregulation, and Increased Expression of Extracellular Matrix Proteins in Laminin Alpha2 Chain-Deficient Muscle. Mol. Cell. Proteom. 2014, 13, 3001–3013. [Google Scholar] [CrossRef] [PubMed]
- Barry-Hamilton, V.; Spangler, R.; Marshall, D.; McCauley, S.; Rodriguez, H.M.; Oyasu, M.; Mikels, A.; Vaysberg, M.; Ghermazien, H.; Wai, C.; et al. Allosteric Inhibition of Lysyl Oxidase-Like-2 Impedes the Development of a Pathologic Microenvironment. Nat. Med. 2010, 16, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.G.; Gutierrez, J.; Cabello-Verrugio, C.; Cabrera, D.; Lipson, K.E.; Goldschmeding, R.; Brandan, E. Reducing Ctgf/Ccn2 Slows Down Mdx Muscle Dystrophy and Improves Cell Therapy. Hum. Mol. Genet. 2013, 22, 4938–4951. [Google Scholar] [CrossRef] [PubMed]
- Rozo, M.; Li, L.; Fan, C.M. Targeting Beta1-Integrin Signaling Enhances Regeneration in Aged and Dystrophic Muscle in Mice. Nat. Med. 2016, 22, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Quattrocelli, M.; Spencer, M.J.; McNally, E.M. Outside In: The Matrix as a Modifier of Muscular Dystrophy. Biochim. Biophys. Acta 2016, 1864, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Flanigan, K.M.; Ceco, E.; Lamar, K.M.; Kaminoh, Y.; Dunn, D.M.; Mendell, J.R.; King, W.M.; Pestronk, A.; Florence, J.M.; Mathews, K.D.; et al. Ltbp4 Genotype Predicts Age of Ambulatory Loss in Duchenne Muscular Dystrophy. Ann. Neurol. 2013, 73, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Pegoraro, E.; Hoffman, E.P.; Piva, L.; Gavassini, B.F.; Cagnin, S.; Ermani, M.; Bello, L.; Soraru, G.; Pacchioni, B.; Bonifati, M.D.; et al. Spp1 Genotype Is a Determinant of Disease Severity in Duchenne Muscular Dystrophy. Neurology 2011, 76, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Vieira, N.M.; Elvers, I.; Alexander, M.S.; Moreira, Y.B.; Eran, A.; Gomes, J.P.; Marshall, J.L.; Karlsson, E.K.; Verjovski-Almeida, S.; Lindblad-Toh, K.; et al. Jagged 1 Rescues the Duchenne Muscular Dystrophy Phenotype. Cell 2015, 163, 1204–1213. [Google Scholar] [CrossRef] [PubMed]
- Vetrone, S.A.; Montecino-Rodriguez, E.; Kudryashova, E.; Kramerova, I.; Hoffman, E.P.; Liu, S.D.; Miceli, M.C.; Spencer, M.J. Osteopontin Promotes Fibrosis in Dystrophic Mouse Muscle by Modulating Immune Cell Subsets and Intramuscular Tgf-Beta. J. Clin. Investig. 2009, 119, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Kemaladewi, D.U.; Benjamin, J.S.; Hyatt, E.; Ivakine, E.A.; Cohn, R.D. Increased Polyamines as Protective Disease Modifiers in Congenital Muscular Dystrophy. Hum. Mol. Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Vanhoutte, D.; Schips, T.G.; Kwong, J.Q.; Davis, J.; Tjondrokoesoemo, A.; Brody, M.J.; Sargent, M.A.; Kanisicak, O.; Yi, H.; Gao, Q.Q.; et al. Thrombospondin Expression in Myofibers Stabilizes Muscle Membranes. Elife 2016, 5, e17589. [Google Scholar] [CrossRef] [PubMed]
- Perrin, S. Preclinical Research: Make Mouse Studies Work. Nature 2014, 507, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, N. Poor Quality Animal Studies cause Clinical Trials to Follow False Leads. BMJ 2015, 351, h5453. [Google Scholar] [CrossRef] [PubMed]
- Hematti, P. Why Most Clinical Trials Fail. ALS Worldwide. 29 January 2015. Available online: http://alsworldwide.org/whats-new/article/why-most-clinical-trials-fail (accessed on 21 March 2018).
- Mak, I.W.; Evaniew, N.; Ghert, M. Lost in Translation: Animal Models and Clinical Trials in Cancer Treatment. Am. J. Transl. Res. 2014, 6, 114–118. [Google Scholar] [PubMed]
- Wood, C. Is Animal Testing About to Become Obsolete? Casey Research. 11 April 2012. Available online: https://www.caseyresearch.com/animal-testing-about-become-obsolete/ (accessed on 21 March 2018).
- Hayden, C.E. Misleading Mouse Studies Waste Medical Resources. Nature. 26 March 2014. Available online: https://www.scientificamerican.com/article/misleading-mouse-studies-waste-medical-resources/ (accessed on 21 March 2018).









© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gawlik, K.I. At the Crossroads of Clinical and Preclinical Research for Muscular Dystrophy—Are We Closer to Effective Treatment for Patients? Int. J. Mol. Sci. 2018, 19, 1490. https://doi.org/10.3390/ijms19051490
Gawlik KI. At the Crossroads of Clinical and Preclinical Research for Muscular Dystrophy—Are We Closer to Effective Treatment for Patients? International Journal of Molecular Sciences. 2018; 19(5):1490. https://doi.org/10.3390/ijms19051490
Chicago/Turabian StyleGawlik, Kinga I. 2018. "At the Crossroads of Clinical and Preclinical Research for Muscular Dystrophy—Are We Closer to Effective Treatment for Patients?" International Journal of Molecular Sciences 19, no. 5: 1490. https://doi.org/10.3390/ijms19051490




