Combined Transcriptomics, Proteomics and Bioinformatics Identify Drug Targets in Spinal Cord Injury
Abstract
:1. Introduction
2. Results
2.1. Transcriptomics and Proteomics Analysis 7 Days and 8 Weeks after Rat SCI
2.2. Integration of Transcriptomics and Proteomics Datasets to Identify Persistently Differentially Regulated Molecules
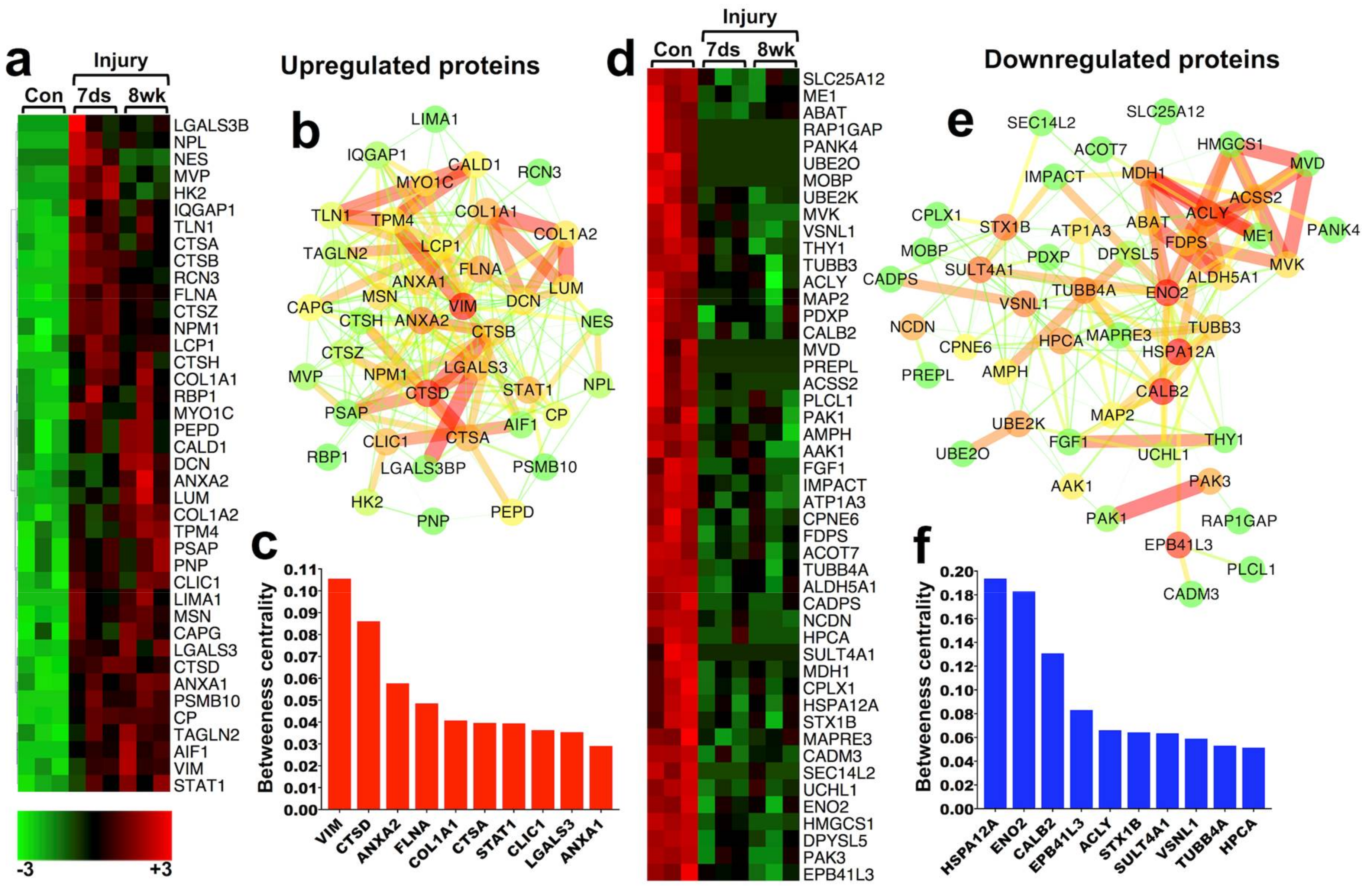
2.2.1. 40 Persistently Upregulated Proteins
2.2.2. 48 Persistently Downregulated Proteins
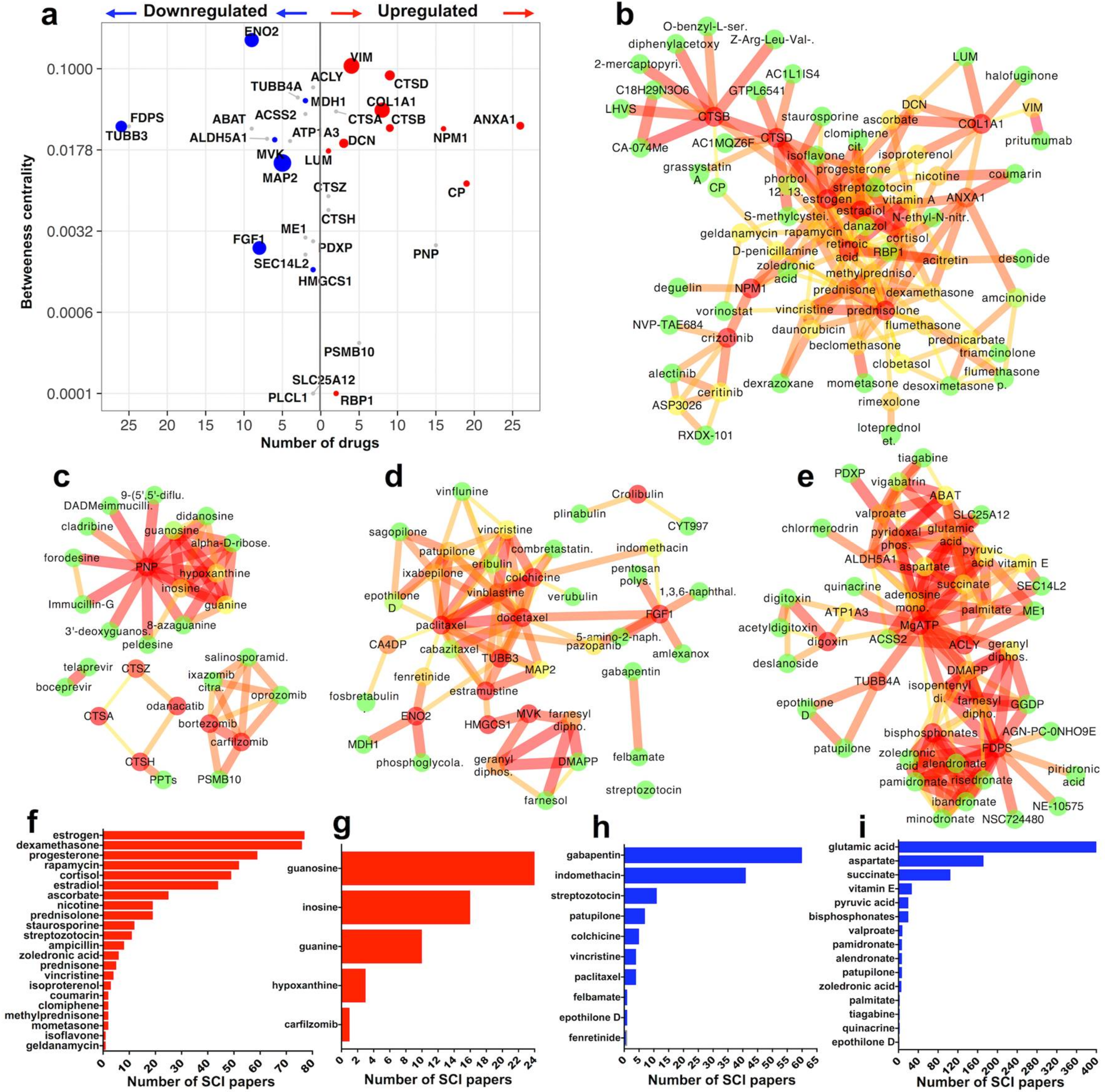
2.3. Identification of Druggable Proteins in SCI
2.3.1. Upregulated Druggable Proteins with SCI Citations (Figure 2b,f)
2.3.2. Upregulated Druggable Proteins without SCI Citations (Figure 2c,g)
2.3.3. Downregulated Druggable Proteins with SCI Citations (Figure 2d,h)
2.3.4. Downregulated Druggable Proteins without SCI Citations (Figure 2e,i)
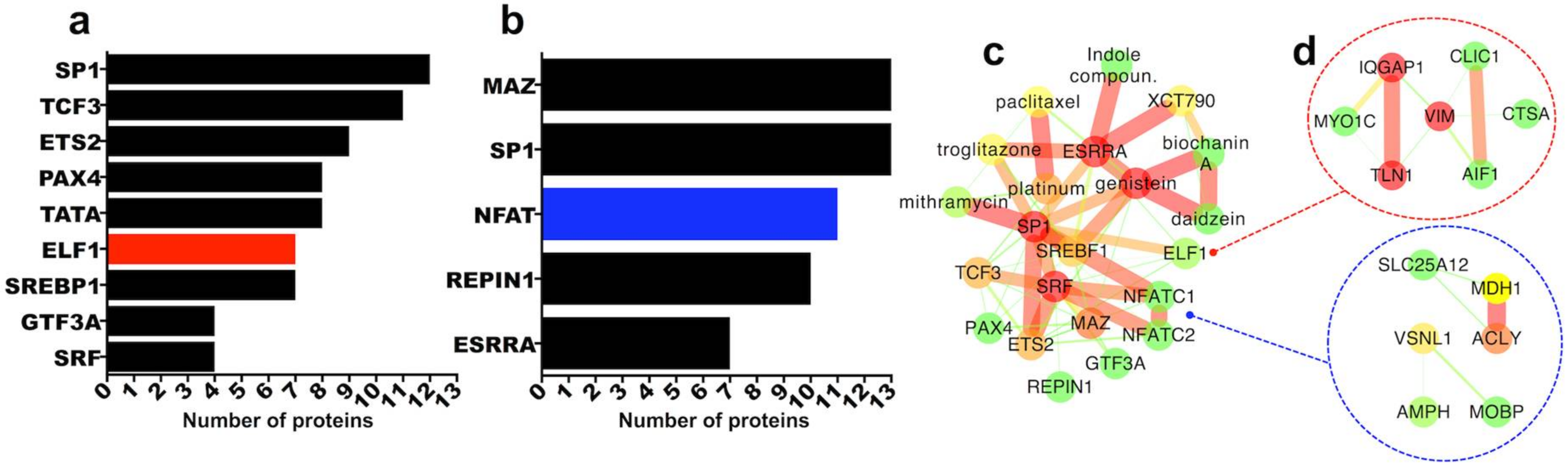
2.3.5. Transcription Factor Regulation of Persistently Differentially Regulated Proteins
3. Discussion
4. Materials and Methods
4.1. Rat Transcriptomics—Microarray Analysis of Rat SCI
4.2. Spinal Cord Injury Model in Rats for Proteomics Analysis
4.3. Shotgun LC-MS/MS Proteomics—Proteomics Analysis of Rat SCI
4.4. Computational and Bioinformatics Analysis of High-Throughput Data
4.5. Gene and Drug Java Text-Mining Tool
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| SCI | Spinal cord injury |
| LC-MS/MS | Liquid-chromatography and tandem mass-spectrometry |
References
- Cregg, J.M.; DePaul, M.A.; Filous, A.R.; Lang, B.T.; Tran, A.; Silver, J. Functional regeneration beyond the glial scar. Exp. Neurol. 2014, 253, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Ramer, L.M.; Ramer, M.S.; Bradbury, E.J. Restoring function after spinal cord injury: Towards clinical translation of experimental strategies. Lancet Neurol. 2014, 13, 1241–1256. [Google Scholar] [CrossRef]
- Fitch, M.T.; Silver, J. CNS injury, glial scars and inflammation: Inhibitory extracellular matrices and regeneration failure. Exp. Neurol. 2008, 209, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, J.W.; Schwab, M.E.; Montani, L.; Brazda, N.; Muller, H.W. Defeating inhibition of regeneration by scar and myelin components. Handb. Clin. Neurol. 2012, 109, 503–522. [Google Scholar] [PubMed]
- Hellal, F.; Hurtado, A.; Ruschel, J.; Flynn, K.C.; Laskowski, C.J.; Umlauf, M.; Kapitein, L.C.; Strikis, D.; Lemmon, V.; Bixby, J.; et al. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science 2011, 331, 928–931. [Google Scholar] [CrossRef] [PubMed]
- Ruschel, J.; Hellal, F.; Flynn, K.C.; Dupraz, S.; Elliott, D.A.; Tedeschi, A.; Bates, M.; Sliwinski, C.; Brook, G.; Dobrindt, K.; et al. Axonal regeneration. Systemic administration of epothilone B promotes axon regeneration after spinal cord injury. Science 2015, 348, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Bartus, K.; James, N.D.; Didangelos, A.; Bosch, K.D.; Verhaagen, J.; Yanez-Munoz, R.J.; Rogers, J.H.; Schneider, B.L.; Muir, E.M.; Bradbury, E.J. Large-scale chondroitin sulfate proteoglycan digestion with chondroitinase gene therapy leads to reduced pathology and modulates macrophage phenotype following spinal cord contusion injury. J. Neurosci. 2014, 34, 4822–4836. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, E.J.; Moon, L.D.; Popat, R.J.; King, V.R.; Bennett, G.S.; Patel, P.N.; Fawcett, J.W.; McMahon, S.B. Chondroitinase ABC promotes functional recovery after spinal cord injury. Nature 2002, 416, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Didangelos, A.; Iberl, M.; Vinsland, E.; Bartus, K.; Bradbury, E.J. Regulation of IL-10 by chondroitinase ABC promotes a distinct immune response following spinal cord injury. J. Neurosci. 2014, 34, 16424–16432. [Google Scholar] [CrossRef] [PubMed]
- Didangelos, A.; Puglia, M.; Iberl, M.; Sanchez-Bellot, C.; Roschitzki, B.; Bradbury, E.J. High-throughput proteomics reveal alarmins as amplifiers of tissue pathology and inflammation after spinal cord injury. Sci. Rep. 2016, 6, 21607. [Google Scholar] [CrossRef] [PubMed]
- Chamankhah, M.; Eftekharpour, E.; Karimi-Abdolrezaee, S.; Boutros, P.C.; San-Marina, S.; Fehlings, M.G. Genome-wide gene expression profiling of stress response in a spinal cord clip compression injury model. BMC Genom. 2013, 14, 583. [Google Scholar] [CrossRef] [PubMed]
- Didangelos, A.; Yin, X.; Mandal, K.; Baumert, M.; Jahangiri, M.; Mayr, M. Proteomics characterization of extracellular space components in the human aorta. Mol. Cell. Proteom. 2010, 9, 2048–2062. [Google Scholar] [CrossRef] [PubMed]
- Didangelos, A.; Yin, X.; Mandal, K.; Saje, A.; Smith, A.; Xu, Q.; Jahangiri, M.; Mayr, M. Extracellular matrix composition and remodelling in human abdominal aortic aneurysms: A proteomics approach. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Arike, L.; Peil, L. Spectral counting label-free proteomics. Methods Mol. Biol. 2014, 1156, 213–222. [Google Scholar] [PubMed]
- Koschutzki, D.; Schreiber, F. Centrality analysis methods for biological networks and their application to gene regulatory networks. Gene Regul. Syst. Biol. 2008, 2, 193–201. [Google Scholar] [CrossRef]
- Shree, T.; Olson, O.C.; Elie, B.T.; Kester, J.C.; Garfall, A.L.; Simpson, K.; Bell-McGuinn, K.M.; Zabor, E.C.; Brogi, E.; Joyce, J.A. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011, 25, 2465–2479. [Google Scholar] [CrossRef] [PubMed]
- Saher, G.; Brugger, B.; Lappe-Siefke, C.; Mobius, W.; Tozawa, R.; Wehr, M.C.; Wieland, F.; Ishibashi, S.; Nave, K.A. High cholesterol level is essential for myelin membrane growth. Nat. Neurosci. 2005, 8, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Goodrum, J.F. Cholesterol synthesis is down-regulated during regeneration of peripheral nerve. J. Neurochem. 1990, 54, 1709–1715. [Google Scholar] [CrossRef] [PubMed]
- Griffith, M.; Griffith, O.L.; Coffman, A.C.; Weible, J.V.; McMichael, J.F.; Spies, N.C.; Koval, J.; Das, I.; Callaway, M.B.; Eldred, J.M.; et al. DGIdb: Mining the druggable genome. Nat. Methods 2013, 10, 1209–1210. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef] [PubMed]
- Perretti, M.; D’Acquisto, F. Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat. Rev. Immunol. 2009, 9, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Nesathurai, S. Steroids and spinal cord injury: Revisiting the NASCIS 2 and NASCIS 3 trials. J. Trauma 1998, 45, 1088–1093. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, L.H.; Fehlings, M.G. Epidemiology, demographics and pathophysiology of acute spinal cord injury. Spine 2001, 26, S2–S12. [Google Scholar] [CrossRef] [PubMed]
- Elkabes, S.; Nicot, A.B. Sex steroids and neuroprotection in spinal cord injury: A review of preclinical investigations. Exp. Neurol. 2014, 259, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Banik, N.L.; Hogan, E.L.; Powers, J.M.; Smith, K.P. Proteolytic enzymes in experimental spinal cord injury. J. Neurol. Sci. 1986, 73, 245–256. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, S.; Wang, P.; Zhang, H.; Wang, F.; Bing, L.; Gao, J.; Yang, J.; Hao, A. Granulocyte colony-stimulating factor improves neuron survival in experimental spinal cord injury by regulating nucleophosmin-1 expression. J. Neurosci. Res. 2014, 92, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Babic, I.; Nurmemmedov, E.; Yenugonda, V.M.; Juarez, T.; Nomura, N.; Pingle, S.C.; Glassy, M.C.; Kesari, S. Pritumumab, the first therapeutic antibody for glioma patients. Hum. Antib. 2018, 26, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Menet, V.; Prieto, M.; Privat, A.; Gimenez y Ribotta, M. Axonal plasticity and functional recovery after spinal cord injury in mice deficient in both glial fibrillary acidic protein and vimentin genes. Proc. Natl. Acad. Sci. USA 2003, 100, 8999–9004. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Xie, X.; Luo, D.; Wang, Y.; Gao, Y. The role of halofuginone in fibrosis: More to be explored? J. Leukoc. Biol. 2017, 102, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Jalil, J.E.; Doering, C.W.; Janicki, J.S.; Pick, R.; Shroff, S.G.; Weber, K.T. Fibrillar collagen and myocardial stiffness in the intact hypertrophied rat left ventricle. Circ. Res. 1989, 64, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Ciccarelli, R.; Ballerini, P.; Sabatino, G.; Rathbone, M.P.; D’Onofrio, M.; Caciagli, F.; Di Iorio, P. Involvement of astrocytes in purine-mediated reparative processes in the brain. Int. J. Dev. Neurosci. 2001, 19, 395–414. [Google Scholar] [CrossRef]
- Kicska, G.A.; Long, L.; Horig, H.; Fairchild, C.; Tyler, P.C.; Furneaux, R.H.; Schramm, V.L.; Kaufman, H.L. Immucillin H, a powerful transition-state analog inhibitor of purine nucleoside phosphorylase, selectively inhibits human T lymphocytes. Proc. Natl. Acad. Sci. USA 2001, 98, 4593–4598. [Google Scholar] [CrossRef] [PubMed]
- Miles, R.W.; Tyler, P.C.; Furneaux, R.H.; Bagdassarian, C.K.; Schramm, V.L. One-third-the-sites transition-state inhibitors for purine nucleoside phosphorylase. Biochemistry 1998, 37, 8615–8621. [Google Scholar] [CrossRef] [PubMed]
- Rathbone, M.; Pilutti, L.; Caciagli, F.; Jiang, S. Neurotrophic effects of extracellular guanosine. Nucleosides Nucleotides Nucleic Acids 2008, 27, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Gong, B.; Radulovic, M.; Figueiredo-Pereira, M.E.; Cardozo, C. The Ubiquitin-Proteasome System: Potential Therapeutic Targets for Alzheimer’s Disease and Spinal Cord Injury. Front. Mol. Neurosci. 2016, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.S.; Muresanu, D.F.; Lafuente, J.V.; Sjoquist, P.O.; Patnaik, R.; Sharma, A. Nanoparticles Exacerbate Both Ubiquitin and Heat Shock Protein Expressions in Spinal Cord Injury: Neuroprotective Effects of the Proteasome Inhibitor Carfilzomib and the Antioxidant Compound H-290/51. Mol. Neurobiol. 2015, 52, 882–898. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Arcuino, G.; Takano, T.; Lin, J.; Peng, W.G.; Wan, P.; Li, P.; Xu, Q.; Liu, Q.S.; Goldman, S.A.; et al. P2X7 receptor inhibition improves recovery after spinal cord injury. Nat. Med. 2004, 10, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.D.; Mocchetti, I.; Wrathall, J.R. Basic and acidic fibroblast growth factors protect spinal motor neurones in vivo after experimental spinal cord injury. Eur. J. Neurosci. 1998, 10, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Ray, S.K.; Cox, A.; Banik, N.L. Neuron specific enolase: A promising therapeutic target in acute spinal cord injury. Metab. Brain Dis. 2015, 31, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Vales, R.; Redensek, A.; Skinner, T.A.; Rathore, K.I.; Ghasemlou, N.; Wojewodka, G.; DeSanctis, J.; Radzioch, D.; David, S. Fenretinide promotes functional recovery and tissue protection after spinal cord contusion injury in mice. J. Neurosci. 2010, 30, 3220–3226. [Google Scholar] [CrossRef] [PubMed]
- Eftekharpour, E.; Nagakannan, P.; Iqbal, M.A.; Chen, Q.M. Mevalonate Cascade and Small Rho GTPase in Spinal Cord Injury. Curr. Mol. Pharmacol. 2017, 10, 141–151. [Google Scholar] [PubMed]
- Abate, M.; Laezza, C.; Pisanti, S.; Torelli, G.; Seneca, V.; Catapano, G.; Montella, F.; Ranieri, R.; Notarnicola, M.; Gazzerro, P.; et al. Deregulated expression and activity of Farnesyl Diphosphate Synthase (FDPS) in Glioblastoma. Sci. Rep. 2017, 7, 14123. [Google Scholar] [CrossRef] [PubMed]
- Nance, P.W.; Schryvers, O.; Leslie, W.; Ludwig, S.; Krahn, J.; Uebelhart, D. Intravenous pamidronate attenuates bone density loss after acute spinal cord injury. Arch. Phys. Med. Rehabil. 1999, 80, 243–251. [Google Scholar] [CrossRef]
- Beigneux, A.P.; Kosinski, C.; Gavino, B.; Horton, J.D.; Skarnes, W.C.; Young, S.G. ATP-citrate lyase deficiency in the mouse. J. Biol. Chem. 2004, 279, 9557–9564. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, M.L.; Wang, Z.; Silver, M.; Boardman, J.P.; Ball, G.; Counsell, S.J.; Walley, A.J.; Montana, G.; Edwards, A.D. Possible relationship between common genetic variation and white matter development in a pilot study of preterm infants. Brain Behav. 2016, 6, e00434. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Parada, I.; Prince, D.A. Temporal and topographic alterations in expression of the alpha3 isoform of Na+, K(+)-ATPase in the rat freeze lesion model of microgyria and epileptogenesis. Neuroscience 2009, 162, 339–348. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho Aguiar, P.; Sweadner, K.J.; Penniston, J.T.; Zaremba, J.; Liu, L.; Caton, M.; Linazasoro, G.; Borg, M.; Tijssen, M.A.; Bressman, S.B.; et al. Mutations in the Na+/K+-ATPase alpha3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron 2004, 43, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Besse, A.; Wu, P.; Bruni, F.; Donti, T.; Graham, B.H.; Craigen, W.J.; McFarland, R.; Moretti, P.; Lalani, S.; Scott, K.L.; et al. The GABA transaminase, ABAT, is essential for mitochondrial nucleoside metabolism. Cell Metab. 2015, 21, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Sauer, S.W.; Kolker, S.; Hoffmann, G.F.; Ten Brink, H.J.; Jakobs, C.; Gibson, K.M.; Okun, J.G. Enzymatic and metabolic evidence for a region specific mitochondrial dysfunction in brains of murine succinic semialdehyde dehydrogenase deficiency (Aldh5a1−/− mice). Neurochem. Int. 2007, 50, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Drewes, A.M.; Andreasen, A.; Poulsen, L.H. Valproate for treatment of chronic central pain after spinal cord injury. A double-blind cross-over study. Paraplegia 1994, 32, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2016, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Durbin, J.E.; Hackenmiller, R.; Simon, M.C.; Levy, D.E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 1996, 84, 443–450. [Google Scholar] [CrossRef]
- Meraz, M.A.; White, J.M.; Sheehan, K.C.; Bach, E.A.; Rodig, S.J.; Dighe, A.S.; Kaplan, D.H.; Riley, J.K.; Greenlund, A.C.; Campbell, D.; et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 1996, 84, 431–442. [Google Scholar] [CrossRef]
- Profyris, C.; Cheema, S.S.; Zang, D.; Azari, M.F.; Boyle, K.; Petratos, S. Degenerative and regenerative mechanisms governing spinal cord injury. Neurobiol. Dis. 2004, 15, 415–436. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, L.; Mei, X.; Yu, Y. Selective inhibition of STAT1 reduces spinal cord injury in mice. Neurosci. Lett. 2014, 580, 7–11. [Google Scholar] [CrossRef] [PubMed]
- James, N.D.; Bartus, K.; Grist, J.; Bennett, D.L.; McMahon, S.B.; Bradbury, E.J. Conduction failure following spinal cord injury: Functional and anatomical changes from acute to chronic stages. J. Neurosci. 2011, 31, 18543–18555. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D.; Hains, B.C.; Johnson, K.M.; Hulsebosch, C.E. Strain and model differences in behavioral outcomes after spinal cord injury in rat. J. Neurotrauma 2001, 18, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Kjell, J.; Olson, L. Rat models of spinal cord injury: From pathology to potential therapies. Dis. Model. Mech. 2016, 9, 1125–1137. [Google Scholar] [CrossRef] [PubMed]
- Cheriyan, T.; Ryan, D.J.; Weinreb, J.H.; Cheriyan, J.; Paul, J.C.; Lafage, V.; Kirsch, T.; Errico, T.J. Spinal cord injury models: A review. Spinal Cord 2014, 52, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Sharif-Alhoseini, M.; Khormali, M.; Rezaei, M.; Safdarian, M.; Hajighadery, A.; Khalatbari, M.M.; Meknatkhah, S.; Rezvan, M.; Chalangari, M.; Derakhshan, P.; et al. Animal models of spinal cord injury: A systematic review. Spinal Cord 2017, 55, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Pinzon, A.; Marcillo, A.; Pabon, D.; Bramlett, H.M.; Bunge, M.B.; Dietrich, W.D. A re-assessment of erythropoietin as a neuroprotective agent following rat spinal cord compression or contusion injury. Exp. Neurol. 2008, 213, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Geremia, N.M.; Hryciw, T.; Bao, F.; Streijger, F.; Okon, E.; Lee, J.H.T.; Weaver, L.C.; Dekaban, G.A.; Kwon, B.K.; Brown, A. The effectiveness of the anti-CD11d treatment is reduced in rat models of spinal cord injury that produce significant levels of intraspinal hemorrhage. Exp. Neurol. 2017, 295, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Kjell, J.; Sandor, K.; Josephson, A.; Svensson, C.I.; Abrams, M.B. Rat substrains differ in the magnitude of spontaneous locomotor recovery and in the development of mechanical hypersensitivity after experimental spinal cord injury. J. Neurotrauma 2013, 30, 1805–1811. [Google Scholar] [CrossRef] [PubMed]
- Popovich, P.G.; Wei, P.; Stokes, B.T. Cellular inflammatory response after spinal cord injury in Sprague-Dawley and Lewis rats. J. Comp. Neurol. 1997, 377, 443–464. [Google Scholar] [CrossRef]
- Schmitt, C.; Miranpuri, G.S.; Dhodda, V.K.; Isaacson, J.; Vemuganti, R.; Resnick, D.K. Changes in spinal cord injury-induced gene expression in rat are strain-dependent. Spine J. 2006, 6, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Bantia, S.; Ananth, S.L.; Parker, C.D.; Horn, L.L.; Upshaw, R. Mechanism of inhibition of T-acute lymphoblastic leukemia cells by PNP inhibitor--BCX-1777. Int. Immunopharmacol. 2003, 3, 879–887. [Google Scholar] [CrossRef]
- Somech, R.; Lev, A.; Grisaru-Soen, G.; Shiran, S.I.; Simon, A.J.; Grunebaum, E. Purine nucleoside phosphorylase deficiency presenting as severe combined immune deficiency. Immunol. Res. 2013, 56, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Hiraiwa, M. Cathepsin A/protective protein: An unusual lysosomal multifunctional protein. Cell. Mol. Life Sci. 1999, 56, 894–907. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Z.; Mao, Y.; Zheng, Z.; Chen, Y.; Khor, S.; Shi, K.; He, Z.; Li, J.; Gong, F.; et al. Liraglutide activates autophagy via GLP-1R to improve functional recovery after spinal cord injury. Oncotarget 2017, 8, 85949–85968. [Google Scholar] [CrossRef] [PubMed]
- Calhan, O.Y.; Seyrantepe, V. Mice with Catalytically Inactive Cathepsin A Display Neurobehavioral Alterations. Behav. Neurol. 2017, 2017, 4261873. [Google Scholar] [CrossRef] [PubMed]
- Carecchio, M.; Zorzi, G.; Ragona, F.; Zibordi, F.; Nardocci, N. ATP1A3-related disorders: An update. Eur. J. Paediatr. Neurol. 2018, 22, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Searle, B.C. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 2010, 10, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.; Thiagarajan, M.; White, J.A.; Quackenbush, J. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2010, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tica, J.; Bradbury, E.J.; Didangelos, A. Combined Transcriptomics, Proteomics and Bioinformatics Identify Drug Targets in Spinal Cord Injury. Int. J. Mol. Sci. 2018, 19, 1461. https://doi.org/10.3390/ijms19051461
Tica J, Bradbury EJ, Didangelos A. Combined Transcriptomics, Proteomics and Bioinformatics Identify Drug Targets in Spinal Cord Injury. International Journal of Molecular Sciences. 2018; 19(5):1461. https://doi.org/10.3390/ijms19051461
Chicago/Turabian StyleTica, Jure, Elizabeth J. Bradbury, and Athanasios Didangelos. 2018. "Combined Transcriptomics, Proteomics and Bioinformatics Identify Drug Targets in Spinal Cord Injury" International Journal of Molecular Sciences 19, no. 5: 1461. https://doi.org/10.3390/ijms19051461




