Biochemical Basis of E. coli Topoisomerase I Relaxation Activity Reduction by Nonenzymatic Lysine Acetylation
Abstract
:1. Introduction
2. Results
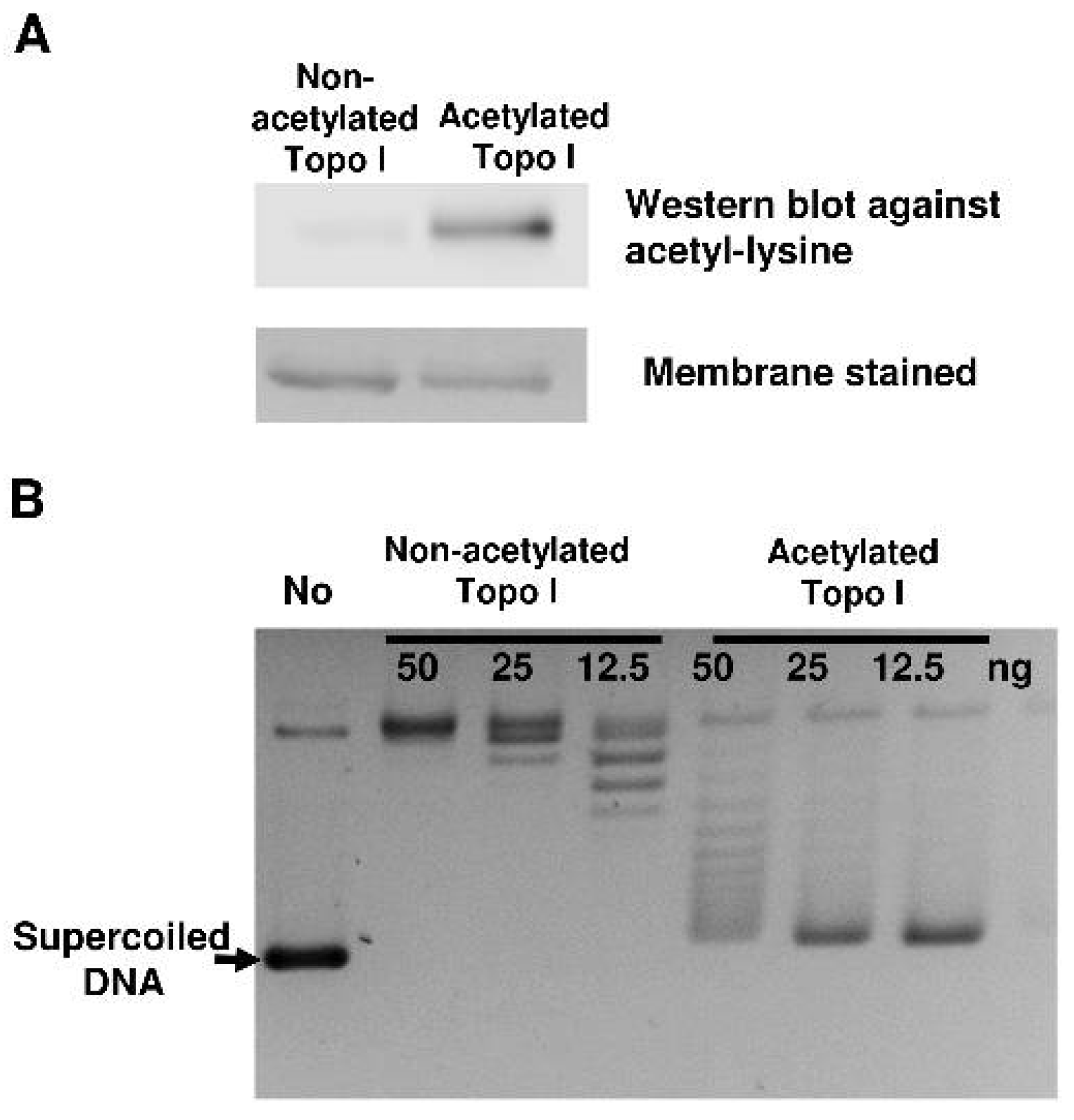
2.1. Reduction of Topoisomerase I Relaxation Activity Following Nonenzymatic Acetylation
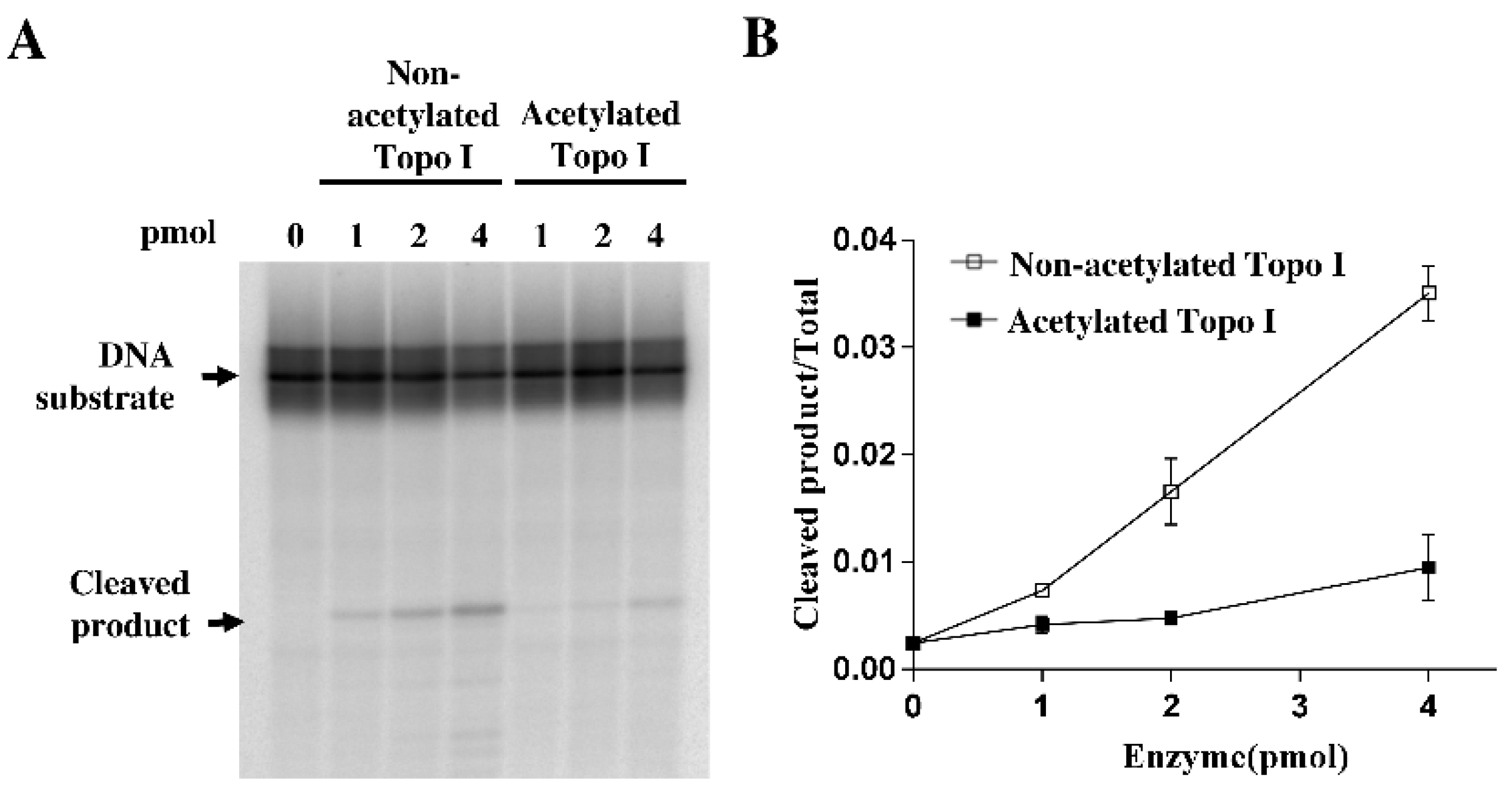
2.2. Decreased DNA Cleavage by Acetylated Topoisomerase I
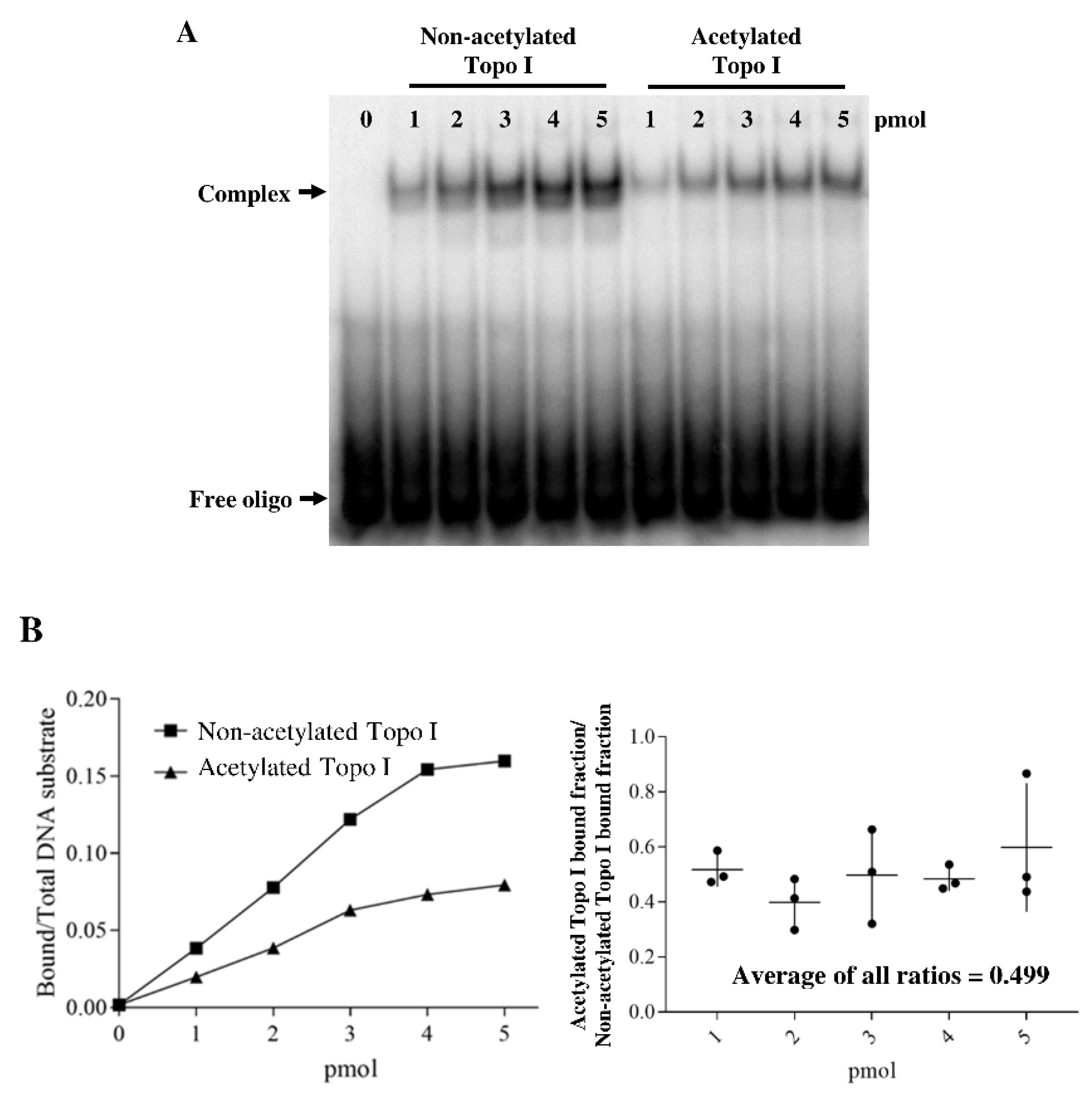
2.3. Acetylation of Topoisomerase I Reduced Binding Affinity of the DNA Substrate
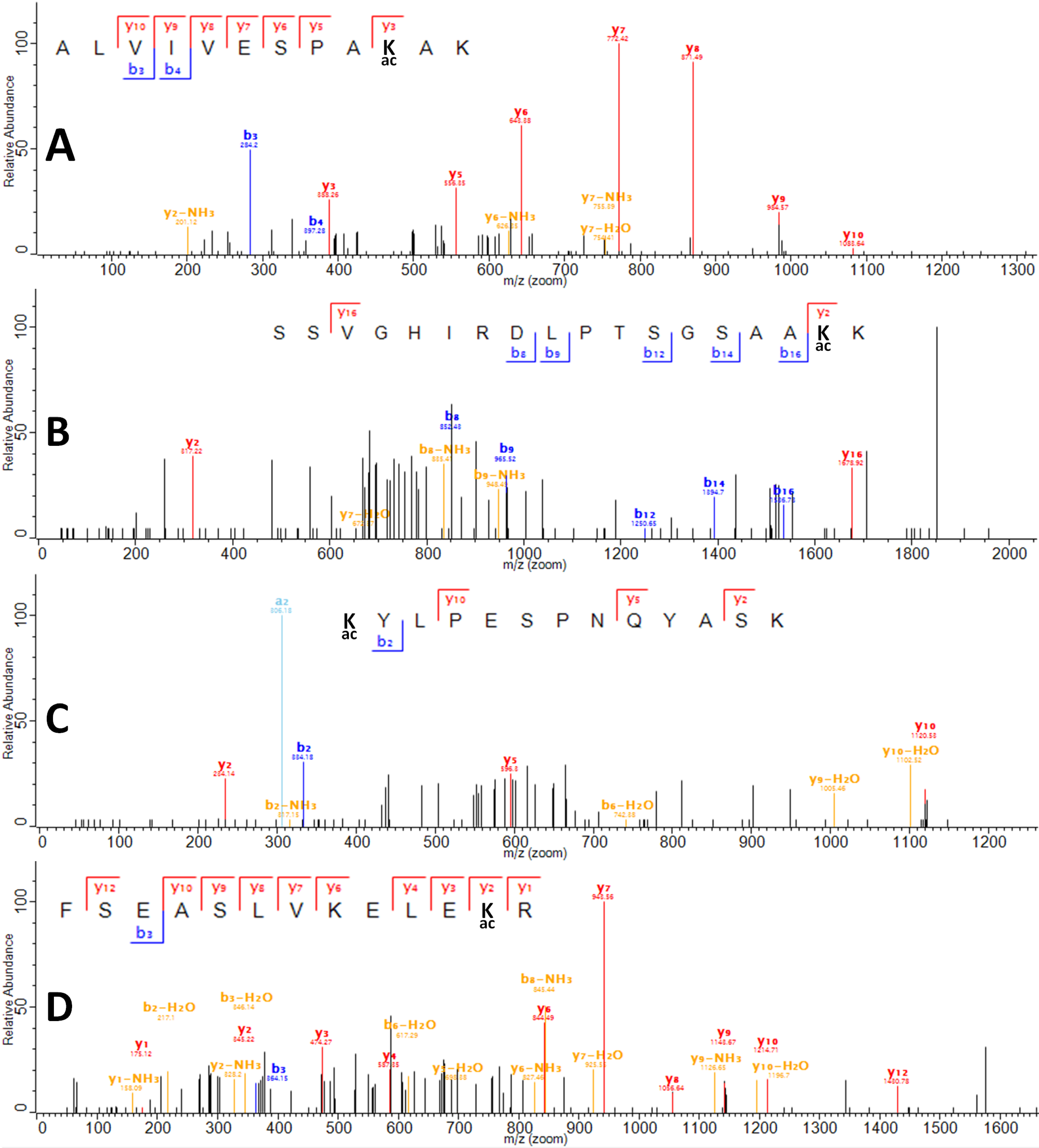
2.4. Identification of Lysines in Topoisomerase I Modified by Nonenzymatic Acetylation
3. Discussion
4. Materials and Methods
4.1. Acetyl Phosphate Treatment of E. coli Topoisomerase I
4.2. Topoisomerase I Relaxation Activity Assay
4.3. DNA Cleavage Assay
4.4. DNA Binding Assay
4.5. In-Gel Tryptic Digestion
4.6. Bottom-Up Proteomics Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| Topo I | Topoisomerase I |
| TCEP | Tris[2-carboxyethyl]phosphine |
| MS | Mass Spectrometry |
| EIC | Extracted Ion Chromatograms |
| Q-TOF | Quadrupole Time of Flight |
| CID | Collision Induced Dissociation |
References
- Sobetzko, P. Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of Bacterial genes. Nucleic Acids Res. 2016, 44, 1514–1524. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.M.; Lewis, D.E.; Lee, H.J.; Liu, M.; Adhya, S. Effect of varying the supercoiling of DNA on transcription and its regulation. Biochemistry 2003, 42, 10718–10725. [Google Scholar] [CrossRef] [PubMed]
- Usongo, V.; Drolet, M. Roles of type 1A topoisomerases in genome maintenance in Escherichia coli. PLoS Genet. 2014, 10, e1004543. [Google Scholar] [CrossRef] [PubMed]
- Drlica, K. Control of bacterial DNA supercoiling. Mol. Microbiol. 1992, 6, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Zechiedrich, E.L.; Khodursky, A.B.; Bachellier, S.; Schneider, R.; Chen, D.; Lilley, D.M.; Cozzarelli, N.R. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000, 275, 8103–8113. [Google Scholar] [CrossRef] [PubMed]
- Drolet, M. Growth inhibition mediated by excess negative supercoiling: The interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 2006, 59, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Usongo, V.; Martel, M.; Balleydier, A.; Drolet, M. Mutations reducing replication from R-loops suppress the defects of growth, chromosome segregation and DNA supercoiling in cells lacking topoisomerase I and RNase HI activity. DNA Repair 2016, 40, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.S.; Hall, A.N.; Merrikh, C.N.; Ragheb, M.; Tabakh, H.; Pollock, A.J.; Woodward, J.J.; Dreifus, J.E.; Merrikh, H. Replication-transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell 2017, 170, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Garnier, F.; Debat, H.; Nadal, M. Type IA DNA topoisomerases: A universal core and multiple activities. Methods Mol. Biol. 2018, 1703, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P.; Gadelle, D. Phylogenomics of DNA topoisomerases: Their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009, 37, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Wang, J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 1987, 84, 7024–7027. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.Y.; Shyy, S.H.; Wang, J.C.; Liu, L.F. Transcription generates positively and negatively supercoiled domains in the template. Cell 1988, 53, 433–440. [Google Scholar] [CrossRef]
- Masse, E.; Drolet, M. Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1999, 274, 16654–16658. [Google Scholar] [CrossRef] [PubMed]
- Drolet, M.; Broccoli, S.; Rallu, F.; Hraiky, C.; Fortin, C.; Masse, E.; Baaklini, I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front. Biosci. 2003, 8, d210–d221. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Menzel, R.; Tse-Dinh, Y.C. Increased thermosensitivity associated with topoisomerase I deletion and promoter mutations in Escherichia coli. FEMS Microbiol. Lett. 1999, 178, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Rui, S.; Tse-Dinh, Y.C. Topoisomerase function during bacterial responses to environmental challenge. Front. Biosci. 2003, 8, d256–263. [Google Scholar] [CrossRef] [PubMed]
- Weinstein-Fischer, D.; Elgrably-Weiss, M.; Altuvia, S. Escherichia coli response to hydrogen peroxide: A role for DNA supercoiling, topoisomerase I and Fis. Mol. Microbiol. 2000, 35, 1413–1420. [Google Scholar] [CrossRef] [PubMed]
- Stewart, N.; Feng, J.; Liu, X.; Chaudhuri, D.; Foster, J.W.; Drolet, M.; Tse-Dinh, Y.C. Loss of topoisomerase I function affects the RpoS-dependent and GAD systems of acid resistance in Escherichia coli. Microbiology 2005, 151, 2783–2791. [Google Scholar] [CrossRef] [PubMed]
- Tse-Dinh, Y.C. Increased sensitivity to oxidative challenges associated with topA deletion in Escherichia coli. J. Bacteriol. 2000, 182, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Viard, T.; de la Tour, C.B. Type IA topoisomerases: A simple puzzle? Biochimie 2007, 89, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhou, Y.N.; Jin, D.J.; Tse-Dinh, Y.C. Deacetylation of topoisomerase I is an important physiological function of E. coli CobB. Nucleic Acids Res. 2017, 45, 5349–5358. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.J.; Kim, J.A.; Moon, J.H.; Ryu, S.E.; Pan, J.G. The diversity of lysine-acetylated proteins in Escherichia coli. J. Microbiol. Biotechnol. 2008, 18, 1529–1536. [Google Scholar] [PubMed]
- Zhang, K.; Zheng, S.; Yang, J.S.; Chen, Y.; Cheng, Z. Comprehensive profiling of protein lysine acetylation in Escherichia coli. J. Proteome Res. 2013, 12, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Weinert, B.T.; Iesmantavicius, V.; Wagner, S.A.; Scholz, C.; Gummesson, B.; Beli, P.; Nystrom, T.; Choudhary, C. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell 2013, 51, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Castano-Cerezo, S.; Bernal, V.; Post, H.; Fuhrer, T.; Cappadona, S.; Sanchez-Diaz, N.C.; Sauer, U.; Heck, A.J.; Altelaar, A.F.; Canovas, M. Protein acetylation affects acetate metabolism, motility and acid stress response in Escherichia coli. Mol. Syst. Biol. 2014, 10, 762. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Wu, F.L.; Jiang, H.W.; He, X.; Guo, S.J.; Tao, S.C. Global identification of CobB interactors by an Escherichia coli proteome microarray. Acta Biochim. Biophys. Sin. 2014, 46, 548–555. [Google Scholar] [CrossRef] [PubMed]
- AbouElfetouh, A.; Kuhn, M.L.; Hu, L.I.; Scholle, M.D.; Sorensen, D.J.; Sahu, A.K.; Becher, D.; Antelmann, H.; Mrksich, M.; Anderson, W.F.; et al. The E. coli Sirtuin CobB shows no preference for enzymatic and nonenzymatic lysine acetylation substrate sites. MicrobiologyOpen 2015, 4, 66–83. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.H.; Shulla, A.; Reimann, S.A.; Keating, D.H.; Wolfe, A.J. The intracellular concentration of acetyl phosphate in Escherichia coli is sufficient for direct phosphorylation of two-component response regulators. J. Bacteriol. 2007, 189, 5574–5581. [Google Scholar] [CrossRef] [PubMed]
- Strahs, D.; Zhu, C.X.; Cheng, B.; Chen, J.; Tse-Dinh, Y.C. Experimental and computational investigations of Ser10 and Lys13 in the binding and cleavage of DNA substrates by Escherichia coli DNA topoisomerase I. Nucleic Acids Res. 2006, 34, 1785–1797. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, A.J. Bacterial protein acetylation: New discoveries unanswered questions. Curr. Genet. 2016, 62, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.M.; Rajan, R.; Mondragon, A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009, 37, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cheng, B.; Tse-Dinh, Y.C. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 2011, 108, 6939–6944. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Zhou, Q.; Cheng, B.; Zhang, Z.; Joachimiak, A.; Tse-Dinh, Y.C. Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I. Nucleic Acids Res. 2015, 43, 11031–11046. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.I.; Chi, B.K.; Kuhn, M.L.; Filippova, E.V.; Walker-Peddakotla, A.J.; Bäsell, K.; Becher, D.; Anderson, W.F.; Antelmann, H.; Wolfe, A.J. Acetylation of the response regulator RcsB controls transcription from a small RNA promoter. J. Bacteriol. 2013, 195, 4174–4186. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, E.P.; Cheng, B.; Rathi, S.; Aedo, S.J.; Abrenica, M.V.; Tse-Dinh, Y.C. Inhibition of Mg2+ binding and DNA religation by bacterial topoisomerase I via introduction of an additional positive charge into the active site region. Nucleic Acids Res. 2008, 36, 4788–4796. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.L.; Zemaitaitis, B.; Hu, L.I.; Sahu, A.; Sorensen, D.; Minasov, G.; Lima, B.P.; Scholle, M.; Mrksich, M.; Anderson, W.F.; et al. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS ONE 2014, 9, e94816. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.; Annamalai, T.; Sandhaus, S.; Bansod, P.; Tse-Dinh, Y.C. Inhibition of Zn(II) binding type IA topoisomerases by organomercury compounds and Hg(II). PLoS ONE 2015, 10, e0120022. [Google Scholar] [CrossRef] [PubMed]
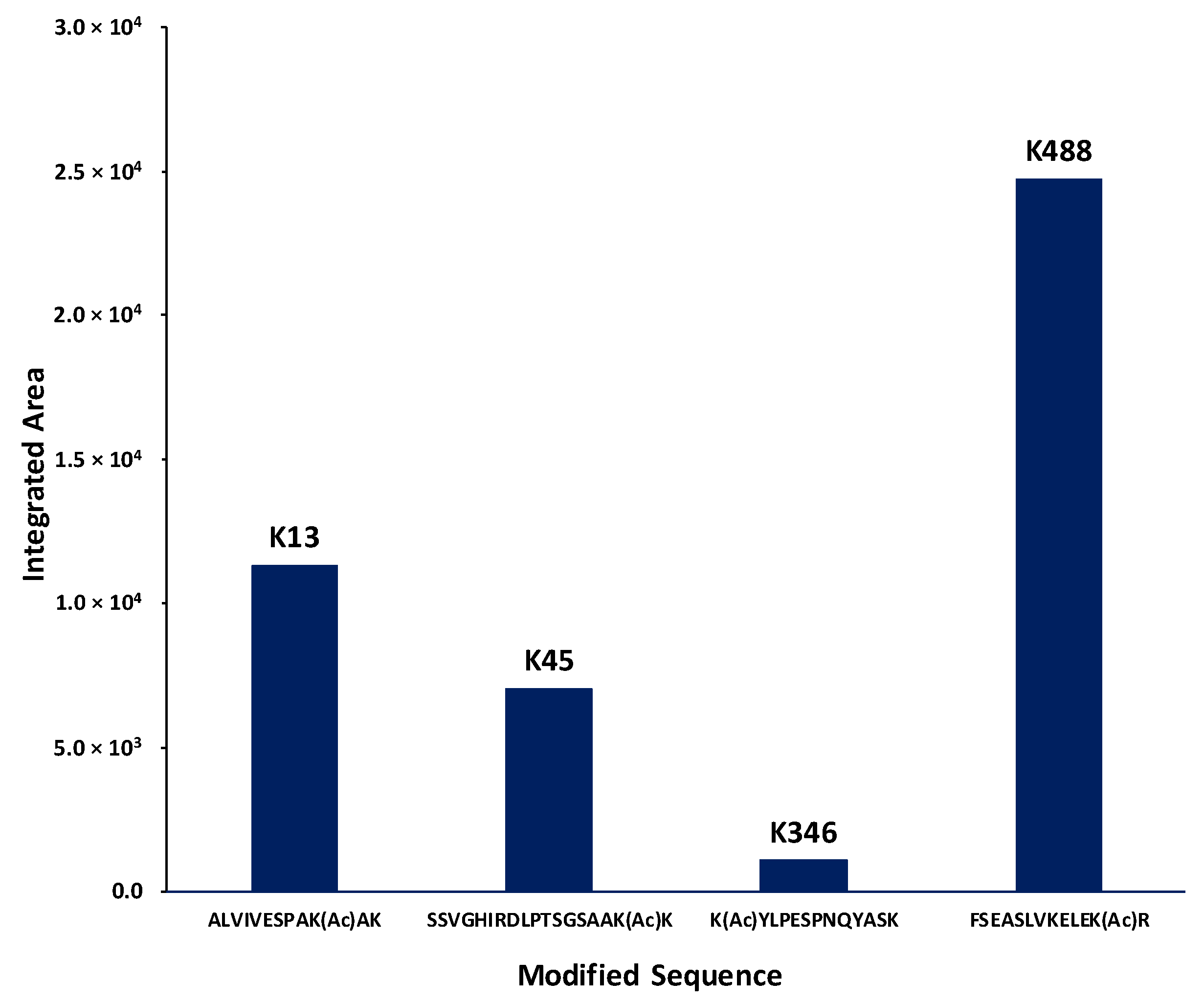

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Q.; Gomez Hernandez, M.E.; Fernandez-Lima, F.; Tse-Dinh, Y.-C. Biochemical Basis of E. coli Topoisomerase I Relaxation Activity Reduction by Nonenzymatic Lysine Acetylation. Int. J. Mol. Sci. 2018, 19, 1439. https://doi.org/10.3390/ijms19051439
Zhou Q, Gomez Hernandez ME, Fernandez-Lima F, Tse-Dinh Y-C. Biochemical Basis of E. coli Topoisomerase I Relaxation Activity Reduction by Nonenzymatic Lysine Acetylation. International Journal of Molecular Sciences. 2018; 19(5):1439. https://doi.org/10.3390/ijms19051439
Chicago/Turabian StyleZhou, Qingxuan, Mario E. Gomez Hernandez, Francisco Fernandez-Lima, and Yuk-Ching Tse-Dinh. 2018. "Biochemical Basis of E. coli Topoisomerase I Relaxation Activity Reduction by Nonenzymatic Lysine Acetylation" International Journal of Molecular Sciences 19, no. 5: 1439. https://doi.org/10.3390/ijms19051439



