A Combined NMR-Computational Study of the Interaction between Influenza Virus Hemagglutinin and Sialic Derivatives from Human and Avian Receptors on the Surface of Transfected Cells
Abstract
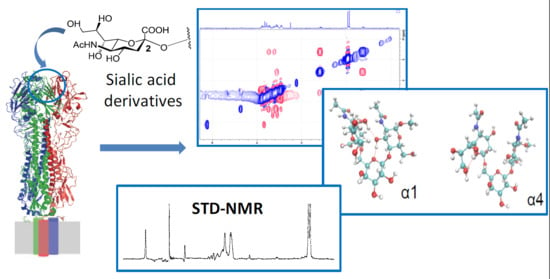
:1. Introduction
2. Results
2.1. Conformational Analysis of the Ligands in Solution
2.2. NMR Interaction Studies
2.2.1 Interaction Studies in the Presence of H5
2.2.2 Interaction Studies in the Presence of H1
3. Discussion
4. Materials and Methods
4.1. Construction of Stable Hemagglutinin-Expressing Transfectant Cell Lines
4.2. NMR Experiments
4.3. Conformational Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| NMR | Nuclear Magnetic Resonance |
| STD | Saturation Transfer Difference |
| NOE | Nuclear Overhauser Effect |
| IAV | Influenza A virus |
| HA | Haemagglutinin |
| NA | Neuroaminidase |
References
- Stevens, J.; Blixt, O.; Paulson, J.C.; Wilson, I.A. Glycan microarray technologies: Tools to survey host specificity of influenza viruses. Nat. Rev. Microbiol. 2006, 4, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.C.; Blixt, O.; Collins, B.E. Sweet spots in functional glycomics. Nat. Chem. Biol. 2006, 2, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Layne, S.P.; Monto, A.S.; Taubenberger, J.K. Pandemic influenza: An inconvenient mutation. Science 2009, 323, 1560–1561. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, K.; Chandrasekaran, A.; Srinivasan, A.; Raman, R.; Sasisekharan, V.; Sasisekharan, R. Glycans as receptors for influenza pathogenesis. Glycoconj. J. 2010, 27, 561–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, J.; Blixt, O.; Tumpey, T.M.; Taubenberger, J.K.; Paulson, J.C.; Wilson, I.A. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 2006, 312, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Haselhorst, T.; Garcia, J.M.; Islam, T.; Lai, J.C.; Rose, F.J.; Nicholls, J.M.; Peiris, J.S.; von Itzstein, M. Avian influenza H5-containing virus-like particles (VLPs): Host-cell receptor specificity by STD NMR spectroscopy. Angew. Chem. 2008, 47, 1910–1912. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Li, R.; Du, L.; Liu, S. Roles of the hemagglutinin of influenza A virus in viral entry and development of antiviral therapeutics and vaccines. Protein Cell 2010, 1, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Onishi, A.; Saito, T.; Shimada, A.; Inoue, H.; Taki, T.; Nagata, K.; Okahata, Y.; Sato, T. Sialic acid-mimic peptides as hemagglutinin inhibitors for anti-influenza therapy. J. Med. Chem. 2010, 53, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Puryear, W.B.; Seifried, B.M.; Dong, X.; Runstadler, J.A.; Ribbeck, K.; Olsen, B.D. Antiviral Agents from Multivalent Presentation of Sialyl Oligosaccharides on Brush Polymers. ACS Macro Lett. 2016, 5, 413–418. [Google Scholar] [CrossRef]
- Vasile, F.; Gubinelli, F.; Panigada, M.; Soprana, E.; Siccardi, A.; Potenza, D. NMR interaction studies of Neu5Ac-α-(2,6)-Gal-β-(1-4)-GlcNAc with influenza-virus hemagglutinin expressed in transfected human cells. Glycobiology 2018, 28, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Vasile, F.; Civera, M.; Belvisi, L.; Potenza, D.; Tiana, G. Thermodynamically-weighted conformational ensemble of cyclic RGD peptidomimetics from NOE data. J. Phys. Chem. B 2016, 120, 7098–7107. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, A.; Camilloni, C.; Vendruscolo, M. Molecular dynamics simulations with replica-averaged structural restraints generate structural ensembles according to the maximum entropy principle. J. Chem. Phys. 2013, 138, 094112. [Google Scholar] [CrossRef] [PubMed]
- Olsson, S.; Frellsen, I.; Boomsma, W.; Mardia, K.V.; Hamelrycka, T. Inference of Structure Ensembles of Flexible Biomolecules from Sparse, Averaged Data. PLoS ONE 2013, 8, e79439. [Google Scholar] [CrossRef] [PubMed]
- Olsson, S.; Wu, H.; Paul, F.; Clementi, C.; Noé, F. Combining experimental and simulation data of molecular processes via augmented Markov models. Proc. Natl. Acad. Sci. USA 2017, 114, 8265–8270. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.; Peters, T. NMR Spectroscopy Techniques for Screening and Identifying Ligand Binding to Protein Receptors. Angew. Chem. 2003, 42, 864–890. [Google Scholar] [CrossRef] [PubMed]
- Vasile, F.; Rossi, D.; Collina, S.; Potenza, D. Diffusion-Ordered Spectroscopy and Saturation Transfer Difference NMR Spectroscopy Studies of Selective Interactions between ELAV Protein Fragments and an mRNA target. Eur. J. Org. Chem. 2014, 6399–6404. [Google Scholar] [CrossRef]
- Heggelund, J.E.; Haugen, E.; Lygren, B.; Mackenzie, A.; Holmner, A.; Vasile, F.; Reina, J.J.; Bernardi, A.; Krengel, U. Both El Tor and classical cholera toxin bind blood group determinants. Biochem. Biophys. Res. Commun. 2012, 418, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Mari, S.; Invernizzi, C.; Spitaleri, A.; Alberici, L.; Ghitti, M.; Bordignon, C.; Traversari, C.; Rizzardi, G.; Musco, G. 2D TR-NOESY Experiments Interrogate and Rank Ligand–Receptor Interactions in Living Human Cancer Cells. Angew. Chem. 2010, 49, 1071–1074. [Google Scholar] [CrossRef] [PubMed]
- Vasile, F.; Reina, J.J.; Potenza, D.; Heggelund, J.E.; Mackenzie, A.; Krengel, U.; Bernardi, A. Comprehensive analysis of blood group antigen binding to classical and El Tor cholera toxin B-pentamers by NMR. Glycobiology 2014, 24, 766–778. [Google Scholar] [CrossRef] [PubMed]
- Potenza, D.; Vasile, F.; Belvisi, L.; Civera, M.; Araldi, E.M. STD and trNOESY NMR Study of Receptor–Ligand Interactions in Living Cancer Cells. ChemBioChem 2011, 12, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Guzzetti, I.; Civera, M.; Vasile, F.; Araldi, E.M.; Belvisi, L.; Gennari, C.; Potenza, D.; Fanelli, R.; Piarulli, U. Determination of the binding epitope of RGD-peptidomimetics to αvβ3 and α(IIb)β3 integrin-rich intact cells by NMR and computational studies. Org. Biomol. Chem. 2013, 11, 3886–3893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasile, F.; Menchi, G.; Lenci, E.; Guarna, A.; Potenza, D.; Trabocchi, A. Insight to the binding mode of triazole RGD-peptidomimetics to integrin-rich cancer cells by NMR and molecular modeling. Bioorg. Med. Chem. 2016, 24, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Guzzetti, I.; Civera, M.; Vasile, F.; Arosio, D.; Tringali, C.; Piarulli, U.; Gennari, C.; Pignataro, L.; Belvisi, L.; Potenza, D. Insights into the binding of cyclic RGD peptidomimetics to α5β1 integrin by live cell NMR and computational studies. ChemistryOpen 2016, 6, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Gambaryan, A.; Tuzikov, A.; Pazynina, G.; Bovin, N.; Balish, A.; Klimov, A. Evolution of the receptor binding phenotype of influenza A (H5) viruses. Virology 2006, 344, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Stevens, D.J.; Haire, L.F.; Walker, P.A.; Coombs, P.J.; Russell, R.J.; Gamblin, S.J.; Skehel, J.J. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc. Natl. Acad. Sci. USA 2009, 106, 17175–17180. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.; Srinivasan, A.; Raman, R.; Viswanathan, K.; Raguram, S.; Tumpey, T.M.; Sasisekharan, R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat. Biotechnol. 2008, 26, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Sassaki, G.L.; Elli, S.; Rudd, T.R.; Macchi, E.; Yates, E.A.; Naggi, A.; Shriver, Z.; Raman, R.; Sasisekharan, R.; Torri, G.; et al. Human (α2→6) and Avian (α2→3) Sialylated Receptors of Influenza A Virus Show Distinct Conformations and Dynamics in Solution. Biochemistry 2013, 52, 7217–7230. [Google Scholar] [CrossRef] [PubMed]
- Elli, S.; Macchi, E.; Rudd, T.R.; Raman, R.; Sassaki, G.; Viswanathan, K.; Yates, E.A.; Shriver, Z.; Naggi, A.; Torri, G.; et al. Insights into the Human Glycan Receptor Conformation of 1918 Pandemic Hemagglutinin−Glycan Complexes Derived from Nuclear Magnetic Resonance and Molecular Dynamics Studies. Biochemistry 2014, 53, 4122–4135. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Viswanathan, K.; Raman, R.; Yu, W.; Sasisekharan, R.; Wilson, I.A. Structural Basis for a Switch in Receptor Binding Specificity of Two H5N1 Hemagglutinin Mutants. Cell Rep. 2015, 13, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Di Lullo, G.; Soprana, E.; Panigada, M.; Palini, A.; Erfle, V.; Staib, C.; Sutter, G.; Siccardi, A.G. Marker gene swapping facilitates recombinant Modified Vaccinia Virus Ankara production by host-range selection. J. Virol. Methods 2009, 156, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Friesen, R.H.E.; Koudstaal, W.; Koldijk, M.H.; Weverling, G.J.; Brakenhoff, J.P.J.; Lenting, P.J.; Stittelaar, K.J.; Osterhaus, A.D.M.E.; Kompier, R.; Goudsmit, J. New Class of Monoclonal Antibodies against Severe Influenza: Prophylactic and Therapeutic Efficacy in Ferrets. PLoS ONE 2010, 5, 9106. [Google Scholar] [CrossRef] [PubMed]
- Wuthrich, K. NMR of Proteins and Nucleic Acids; Wiley-Interscience: New York, NY, USA, 1986; ISBN 0471119172. [Google Scholar]
- Vasile, F.; Pechkova, E.; Nicolini, C. Solution structure of the β-subunit of the translation initiation factor aIF2 from archaebacteria Sulfolobus solfataricus. Protein Struct. Funct. Bioinform. 2008, 70, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Kutzner, C.; van der Spoel, D.; Lindahl, E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory Comput. 2008, 4, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Peter, C.; Daura, X.; van Gunsteren, W.F. Calculation of NMR-relaxation parameters for flexible molecules from molecular dynamics simulations. J. Biomol. NMR 2001, 20, 297–310. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasile, F.; Panigada, M.; Siccardi, A.; Potenza, D.; Tiana, G. A Combined NMR-Computational Study of the Interaction between Influenza Virus Hemagglutinin and Sialic Derivatives from Human and Avian Receptors on the Surface of Transfected Cells. Int. J. Mol. Sci. 2018, 19, 1267. https://doi.org/10.3390/ijms19051267
Vasile F, Panigada M, Siccardi A, Potenza D, Tiana G. A Combined NMR-Computational Study of the Interaction between Influenza Virus Hemagglutinin and Sialic Derivatives from Human and Avian Receptors on the Surface of Transfected Cells. International Journal of Molecular Sciences. 2018; 19(5):1267. https://doi.org/10.3390/ijms19051267
Chicago/Turabian StyleVasile, Francesca, Maddalena Panigada, Antonio Siccardi, Donatella Potenza, and Guido Tiana. 2018. "A Combined NMR-Computational Study of the Interaction between Influenza Virus Hemagglutinin and Sialic Derivatives from Human and Avian Receptors on the Surface of Transfected Cells" International Journal of Molecular Sciences 19, no. 5: 1267. https://doi.org/10.3390/ijms19051267