Genome-Wide Identification and Characterization of Tyrosine Kinases in the Silkworm, Bombyx mori
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of Silkworm TK Proteins
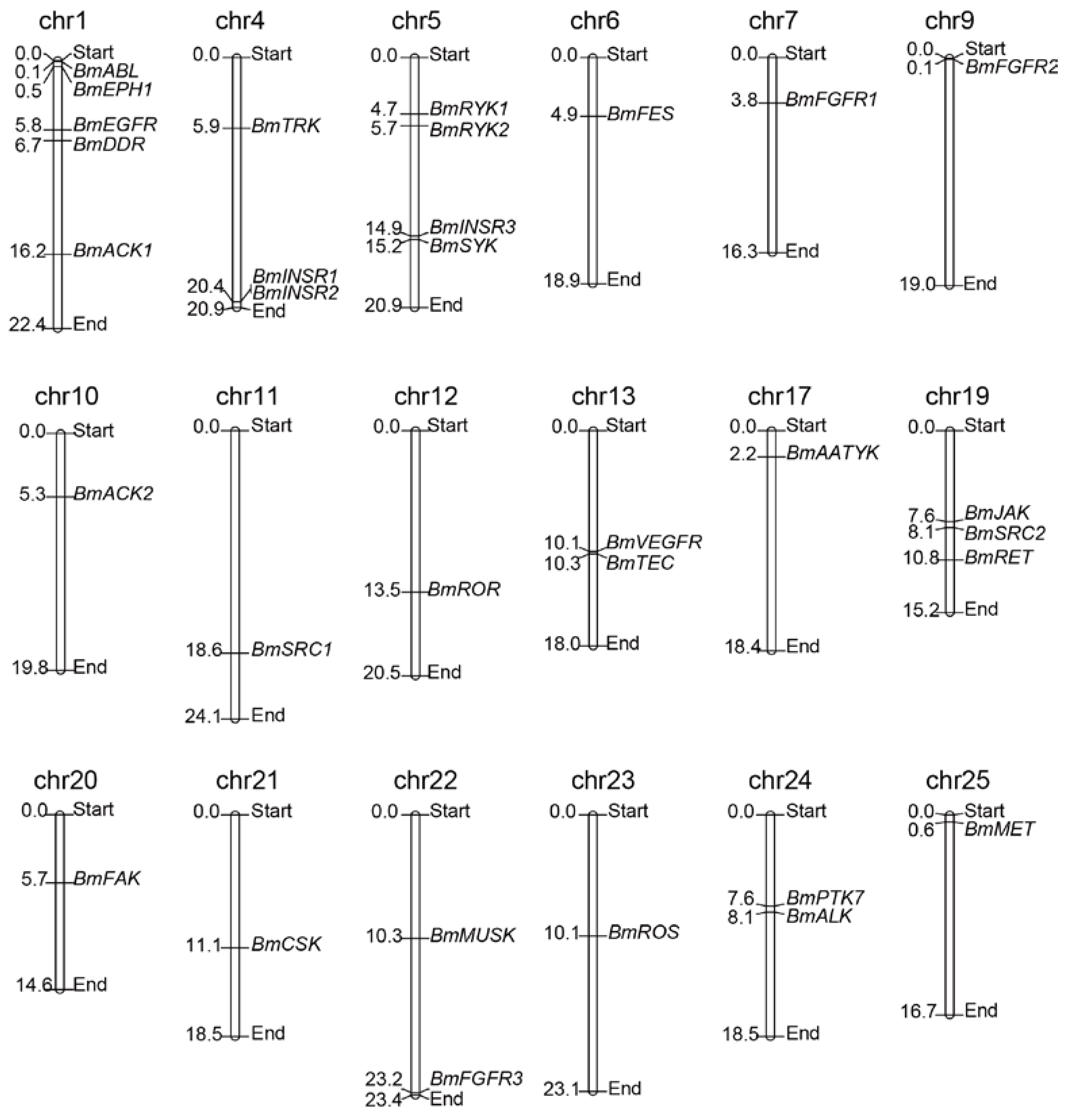
2.2. Chromosomal Distribution of the BmTK Gene Family
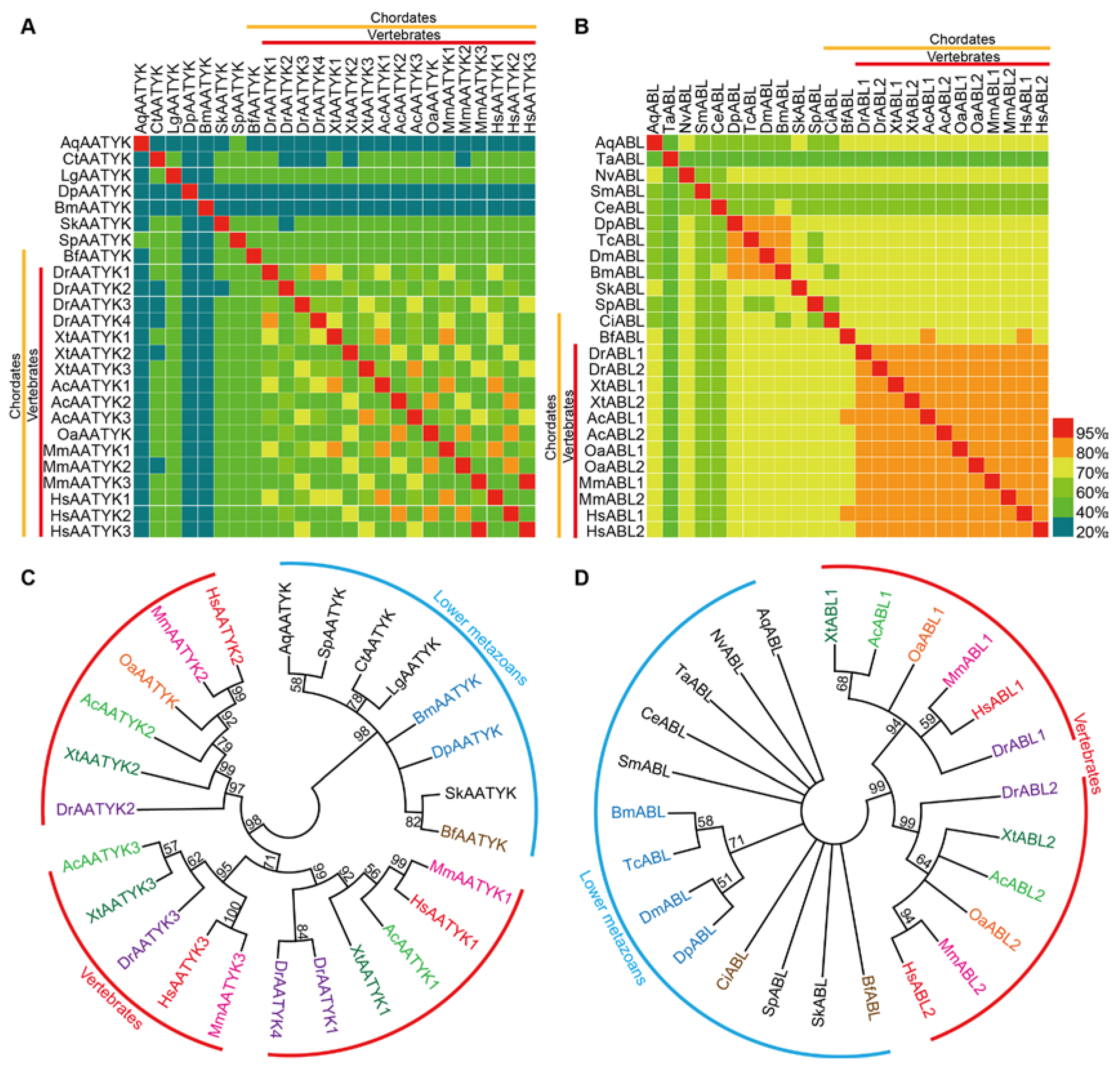
2.3. Comparative Analysis of TKs within Metazoans
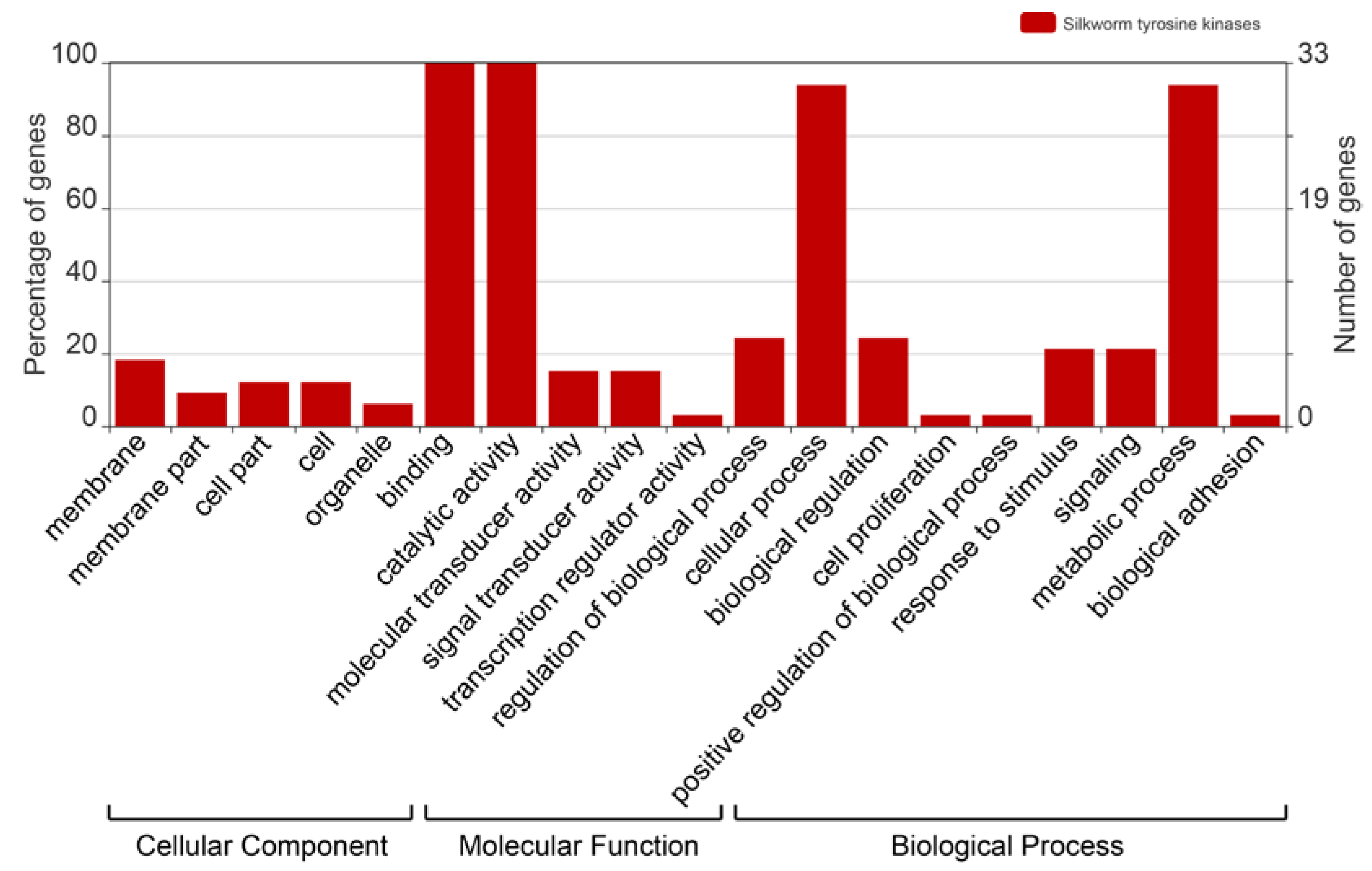
2.4. Gene Ontology Analysis of the BmTKs
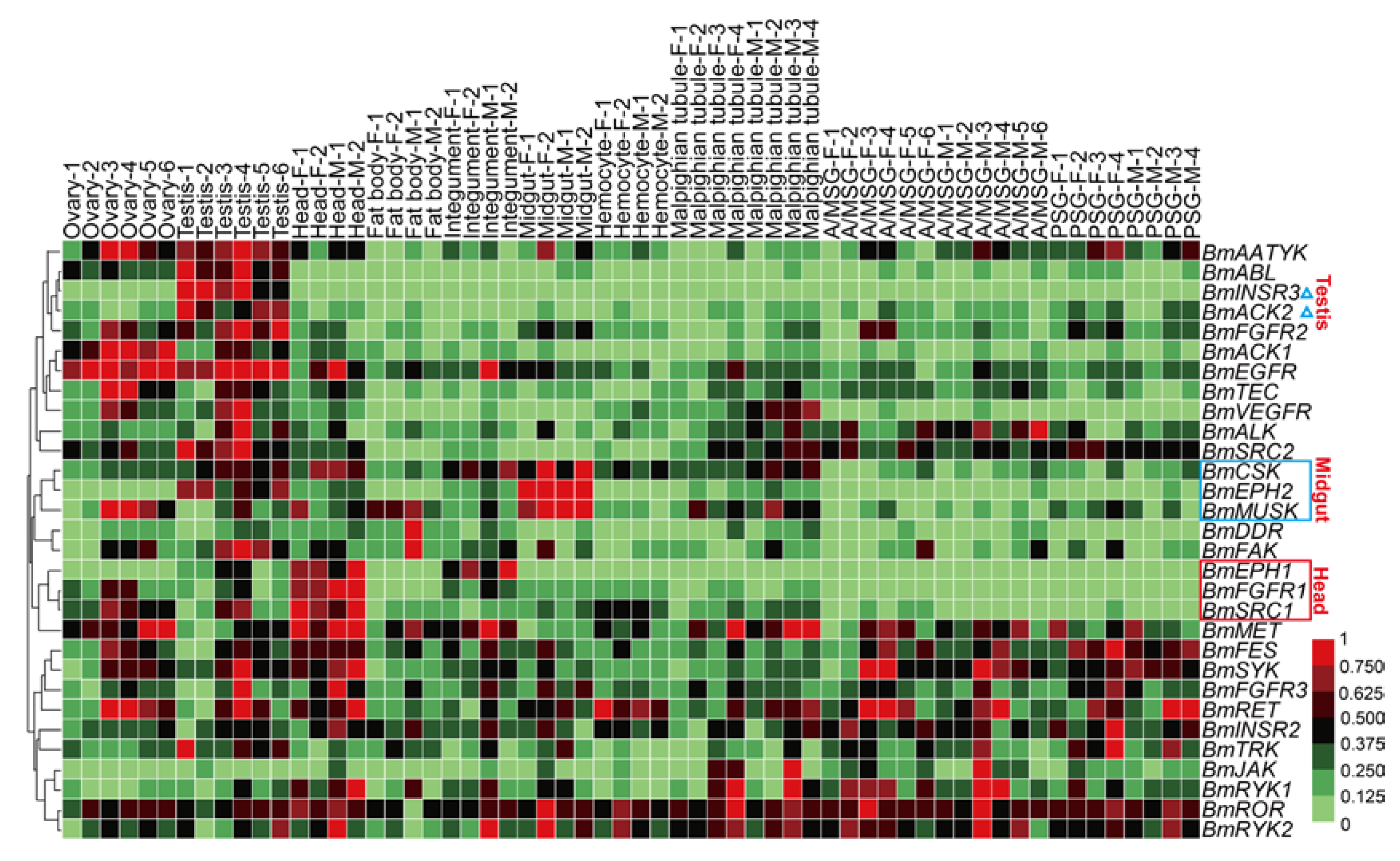
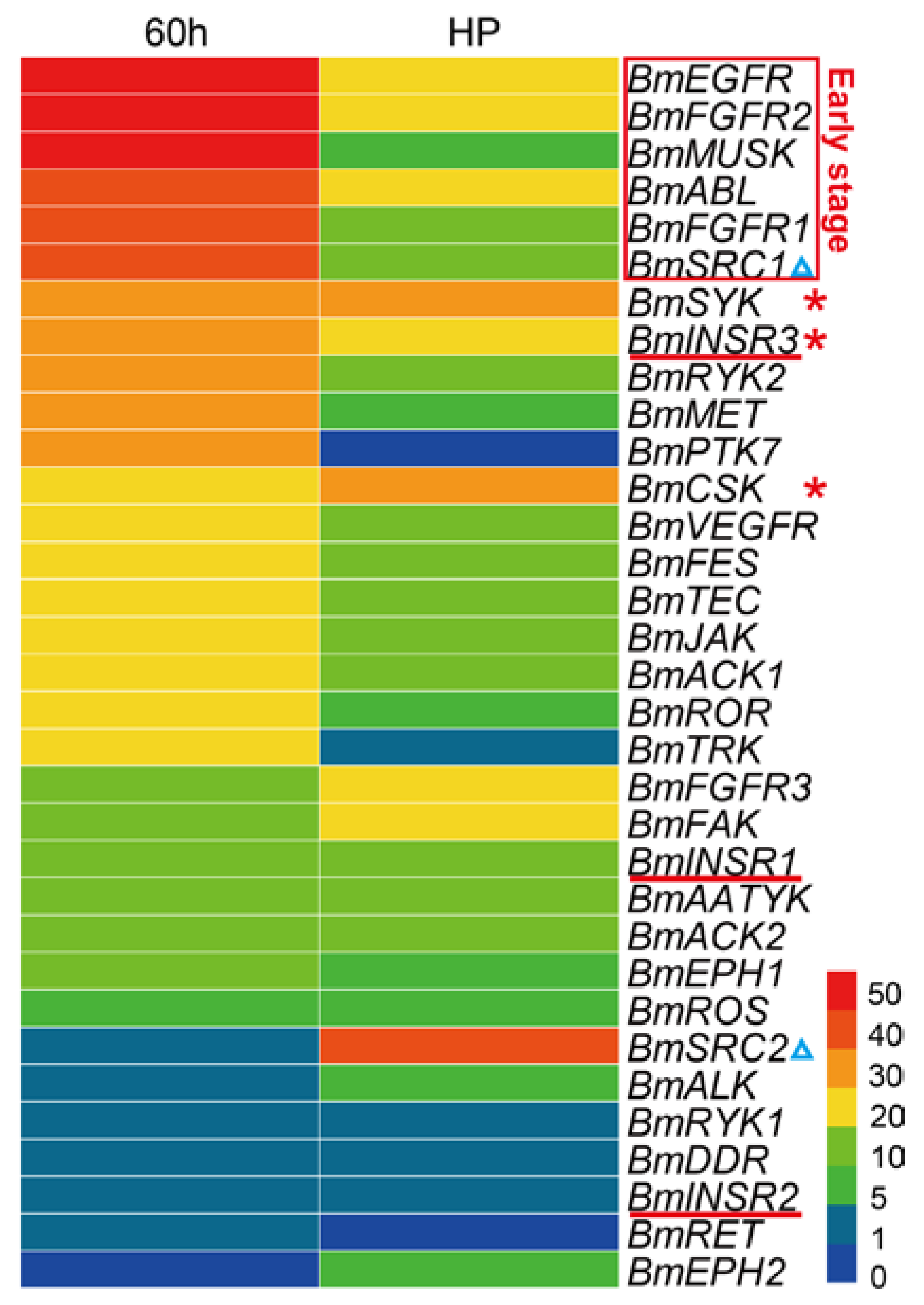
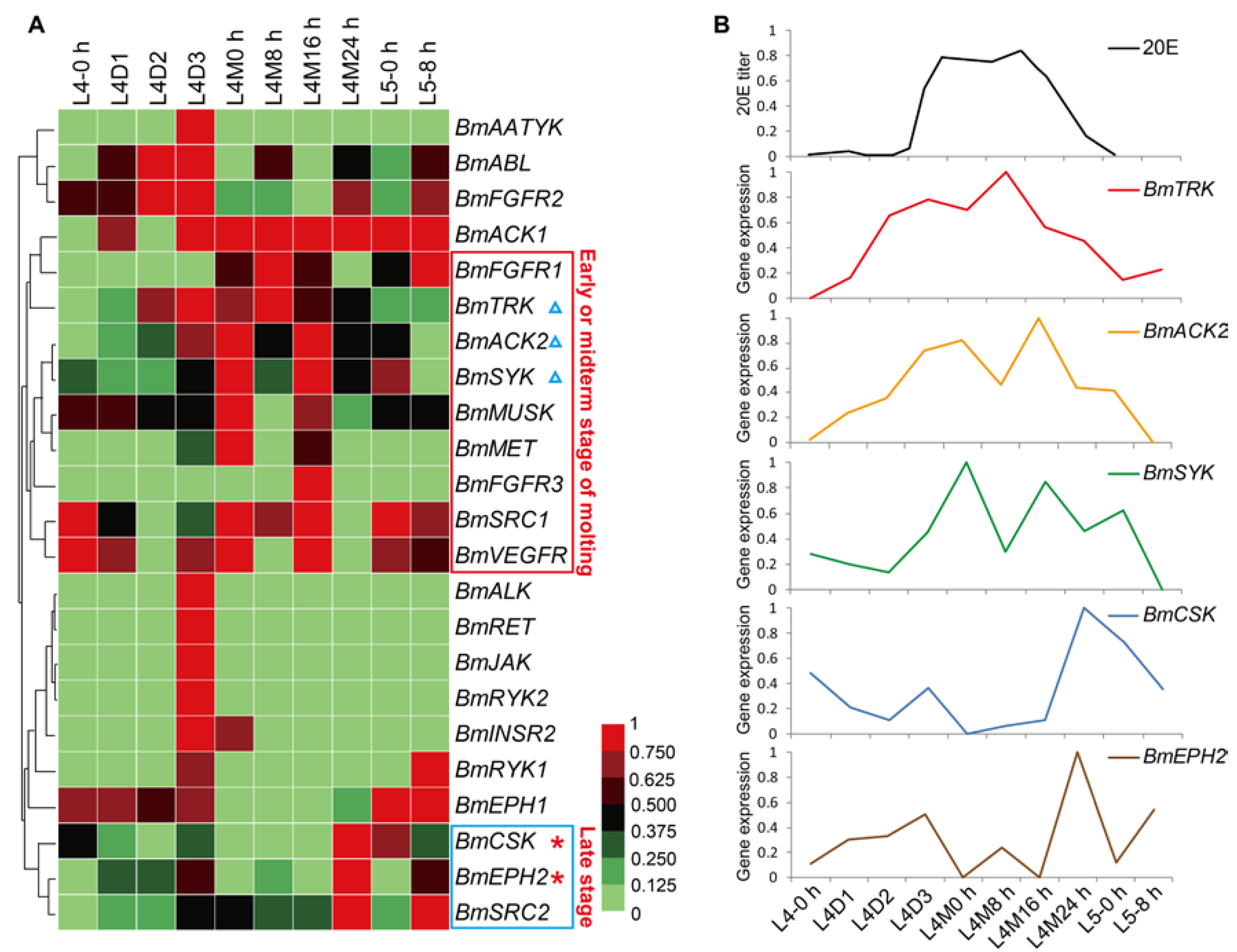
2.5. Spatial and Temporal Expression Profile of the BmTK Genes
3. Materials and Methods
3.1. Genome-Wide Identification of TK Proteins
3.2. Chromosomal Distribution and Gene Basic Information
3.3. Sequence Identity Matrix and Phylogenetic Analysis
3.4. Gene Ontology Annotation
3.5. Expression Profiling Based on DGE Sequencing and Microarray Data
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| TK | Tyrosine Kinase |
| Bm | Bombyx mori |
| kDa | kilo-Dalton |
| RTK | Receptor TK |
| CTK | Cytoplasmic TK (nonreceptor TK) |
References
- Mohanty, S.; Oruganty, K.; Kwon, A.; Byrne, D.P.; Ferries, S.; Ruan, Z.; Hanold, L.E.; Katiyar, S.; Kennedy, E.J.; Eyers, P.A.; et al. Hydrophobic Core Variations Provide a Structural Framework for Tyrosine Kinase Evolution and Functional Specialization. PLoS Genet. 2016, 12, e1005885. [Google Scholar]
- Eckhart, W.; Hutchinson, M.A.; Hunter, T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell 1979, 18, 925–933. [Google Scholar] [CrossRef]
- Sefton, B.M.; Hunter, T.; Beemon, K. Product of in vitro translation of the Rous sarcoma virus src gene has protein kinase activity. J. Virol. 1979, 30, 311–318. [Google Scholar] [PubMed]
- Collett, M.S.; Purchio, A.F.; Erikson, R.L. Avian sarcoma virus-transforming protein, pp60src shows protein kinase activity specific for tyrosine. Nature 1980, 285, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Hanks, S.K.; Quinn, A.M. Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Methods Enzymol. 1991, 200, 38–62. [Google Scholar] [PubMed]
- Hunter, T.; Eckhart, W. The discovery of tyrosine phosphorylation: It’s all in the buffer! Cell 2004, 116, S35–S39. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Annenkov, A. Receptor tyrosine kinase (RTK) signalling in the control of neural stem and progenitor cell (NSPC) development. Mol. Neurobiol. 2014, 49, 440–471. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.R.; Till, J.H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000, 69, 373–398. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.R.; Wu, Y.M.; Lin, S.F. The protein tyrosine kinase family of the human genome. Oncogene 2000, 19, 5548–5557. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; He, F.; Gu, X. Identification and characterization of tyrosine kinases in anole lizard indicate the conserved tyrosine kinase repertoire in vertebrates. Mol. Genet. Genom. 2017, 292, 1405–1418. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, K.; Mahajan, N.P. Cross talk of tyrosine kinases with the DNA damage signaling pathways. Nucleic Acids Res. 2015, 43, 10588–10601. [Google Scholar] [CrossRef] [PubMed]
- Pires-daSilva, A.; Sommer, R.J. The evolution of signalling pathways in animal development. Nat. Rev. Genet. 2003, 4, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Van der Geer, P.; Hunter, T.; Lindberg, R.A. Receptor protein-tyrosine kinases and their signal transduction pathways. Annu. Rev. Cell Biol. 1994, 10, 251–337. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. The Croonian Lecture 1997. The phosphorylation of proteins on tyrosine: Its role in cell growth and disease. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1998, 353, 583–605. [Google Scholar] [CrossRef] [PubMed]
- Robertson, S.C.; Tynan, J.; Donoghue, D.J. RTK mutations and human syndromes: When good receptors turn bad. Trends Genet. 2000, 16, 265–271. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.H.; Schlessinger, J. Asymmetric tyrosine kinase arrangements in activation or autophosphorylation of receptor tyrosine kinases. Mol. Cells 2010, 29, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Brunet, F.G.; Volff, J.N.; Schartl, M. Whole Genome Duplications Shaped the Receptor Tyrosine Kinase Repertoire of Jawed Vertebrates. Genome Biol. Evol. 2016, 8, 1600–1613. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Lu, D.; Yuan, Y.; Xu, S. Signature of positive selection of PTK6 gene in East Asian populations: A cross talk for Helicobacter pylori invasion and gastric cancer endemicity. Mol. Genet. Genom. 2015, 290, 1741–1752. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Ball-Kell, S.M.; Tyner, A.L. Protein tyrosine kinase 6 promotes ERBB2-induced mammary gland tumorigenesis in the mouse. Cell Death Dis. 2015, 6, e1848. [Google Scholar] [CrossRef] [PubMed]
- Valiaho, J.; Faisal, I.; Ortutay, C.; Smith, C.I.; Vihinen, M. Characterization of all possible single-nucleotide change caused amino acid substitutions in the kinase domain of Bruton tyrosine kinase. Hum. Mutat. 2015, 36, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Maxson, J.E.; Abel, M.L.; Wang, J.; Deng, X.; Reckel, S.; Luty, S.B.; Sun, H.; Gorenstein, J.; Hughes, S.B.; Bottomly, D.; et al. Identification and Characterization of Tyrosine Kinase Nonreceptor 2 Mutations in Leukemia through Integration of Kinase Inhibitor Screening and Genomic Analysis. Cancer Res. 2016, 76, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.M.; Lee, J.K.; Witte, O.N. Clinical targeting of mutated and wild-type protein tyrosine kinases in cancer. Mol. Cell. Biol. 2014, 34, 1722–1732. [Google Scholar] [CrossRef] [PubMed]
- Giordani, E.; Zoratto, F.; Strudel, M.; Papa, A.; Rossi, L.; Minozzi, M.; Caruso, D.; Zaccarelli, E.; Verrico, M.; Tomao, S. Old Tyrosine Kinase Inhibitors and Newcomers in Gastrointestinal Cancer Treatment. Curr. Cancer Drug Targets 2016, 16, 175–185. [Google Scholar] [PubMed]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.M.; Kung, H.J.; Evans, C.P. Nonreceptor tyrosine kinases in prostate cancer. Neoplasia 2007, 9, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Nelson, E.G.; Grandis, J.R. Aberrant kinase signaling: Lessons from head and neck cancer. Future Oncol. 2007, 3, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Young, S.L.; King, N.; Miller, W.T. Signaling properties of a non-metazoan Src kinase and the evolutionary history of Src negative regulation. J. Biol. Chem. 2008, 283, 15491–15501. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Young, S.L.; Miller, W.T.; Zhai, Y. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc. Natl. Acad. Sci. USA 2008, 105, 9674–9679. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.H.; Li, W.H. Origins, lineage-specific expansions, and multiple losses of tyrosine kinases in eukaryotes. Mol. Biol. Evol. 2004, 21, 828–840. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, S.; Irimia, M.; Maeso, I.; Pascual-Anaya, J.; Jimenez-Delgado, S.; Bertrand, S.; Garcia-Fernandez, J. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol. Biol. Evol. 2008, 25, 1841–1854. [Google Scholar] [CrossRef] [PubMed]
- Challa, A.K.; Chatti, K. Conservation and early expression of zebrafish tyrosine kinases support the utility of zebrafish as a model for tyrosine kinase biology. Zebrafish 2013, 10, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hua, X.; Song, L.; Xia, Q.; Wang, F. Identification of Tyrosine Kinase Src Responsible for Antimicrobial Peptides Production in Bombyx mori. Protein Pept. Lett. 2017, 24, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Shobahah, J.; Xue, S.; Hu, D.; Zhao, C.; Wei, M.; Quan, Y.; Yu, W. Quantitative phosphoproteome on the silkworm (Bombyx mori) cells infected with baculovirus. Virol. J. 2017, 14, 117. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, M.R.; Shimada, T.; Abe, H. The genetics and genomics of the silkworm, Bombyx mori. Annu. Rev. Entomol. 2005, 50, 71–100. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Suga, H.; Dacre, M.; de Mendoza, A.; Shalchian-Tabrizi, K.; Manning, G.; Ruiz-Trillo, I. Genomic survey of premetazoans shows deep conservation of cytoplasmic tyrosine kinases and multiple radiations of receptor tyrosine kinases. Sci. Signal. 2012, 5, ra35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lu, H.H.; Chung, W.Y.; Yang, J.; Li, W.H. Patterns of segmental duplication in the human genome. Mol. Biol. Evol. 2005, 22, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Shoja, V.; Zhang, L. A roadmap of tandemly arrayed genes in the genomes of human, mouse, and rat. Mol. Biol. Evol. 2006, 23, 2134–2141. [Google Scholar] [CrossRef] [PubMed]
- Iwabe, N.; Kuma, K.; Miyata, T. Evolution of gene families and relationship with organismal evolution: Rapid divergence of tissue-specific genes in the early evolution of chordates. Mol. Biol. Evol. 1996, 13, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Suga, H.; Kuma, K.; Iwabe, N.; Nikoh, N.; Ono, K.; Koyanagi, M.; Hoshiyama, D.; Miyata, T. Intermittent divergence of the protein tyrosine kinase family during animal evolution. FEBS Lett. 1997, 412, 540–546. [Google Scholar] [CrossRef]
- Suga, H.; Koyanagi, M.; Hoshiyama, D.; Ono, K.; Iwabe, N.; Kuma, K.; Miyata, T. Extensive gene duplication in the early evolution of animals before the parazoan-eumetazoan split demonstrated by G proteins and protein tyrosine kinases from sponge and hydra. J. Mol. Evol. 1999, 48, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Truman, J.W.; Riddiford, L.M. Endocrine insights into the evolution of metamorphosis in insects. Annu. Rev. Entomol. 2002, 47, 467–500. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, H.; Kitamoto, T. The steroid molting hormone Ecdysone regulates sleep in adult Drosophila melanogaster. Genetics 2010, 185, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Kiguchi, K.; Agui, N. Ecdysteroid levels and developmental events during larval moulting in the silkworm, Bombyx mori. J. Insect Physiol. 1981, 27, 805–807. [Google Scholar] [CrossRef]
- Sharma, D.; Kinsey, W.H. Fertilization triggers localized activation of Src-family protein kinases in the zebrafish egg. Dev. Biol. 2006, 295, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Gordon, L.R.; Shtibin, E.; Granato, M. Temporal and spatial requirements of unplugged/MuSK function during zebrafish neuromuscular development. PLoS ONE 2010, 5, e8843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budi, E.H.; Patterson, L.B.; Parichy, D.M. Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation. Development 2008, 135, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Barrios, A.; Poole, R.J.; Durbin, L.; Brennan, C.; Holder, N.; Wilson, S.W. Eph/Ephrin signaling regulates the mesenchymal-to-epithelial transition of the paraxial mesoderm during somite morphogenesis. Curr. Biol. 2003, 13, 1571–1582. [Google Scholar] [CrossRef] [PubMed]
- Julich, D.; Mould, A.P.; Koper, E.; Holley, S.A. Control of extracellular matrix assembly along tissue boundaries via Integrin and Eph/Ephrin signaling. Development 2009, 136, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Oates, A.C.; Lackmann, M.; Power, M.A.; Brennan, C.; Down, L.M.; Do, C.; Evans, B.; Holder, N.; Boyd, A.W. An early developmental role for eph-ephrin interaction during vertebrate gastrulation. Mech. Dev. 1999, 83, 77–94. [Google Scholar] [CrossRef]
- Wagle, M.; Grunewald, B.; Subburaju, S.; Barzaghi, C.; Le Guyader, S.; Chan, J.; Jesuthasan, S. EphrinB2a in the zebrafish retinotectal system. J. Neurobiol. 2004, 59, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Elsen, G.E.; Choi, L.Y.; Prince, V.E.; Ho, R.K. The autism susceptibility gene met regulates zebrafish cerebellar development and facial motor neuron migration. Dev. Biol. 2009, 335, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Latimer, A.J.; Jessen, J.R. Hgf/c-met expression and functional analysis during zebrafish embryogenesis. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2008, 237, 3904–3915. [Google Scholar]
- Tallafuss, A.; Eisen, J.S. The Met receptor tyrosine kinase prevents zebrafish primary motoneurons from expressing an incorrect neurotransmitter. Neural Dev. 2008, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Schultz, J.; Milpetz, F.; Bork, P. SMART: Identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 1999, 27, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, H. Protein phylogenetic inference using maximum likelihood with a genetic algorithm. Pac. Symp. Biocomput. 1996, 96, 512–523. [Google Scholar]
- Salter, L.A.; Pearl, D.K. Stochastic search strategy for estimation of maximum likelihood phylogenetic trees. Syst. Biol. 2001, 50, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Cheng, D.; Duan, J.; Wang, G.; Cheng, T.; Zha, X.; Liu, C.; Zhao, P.; Dai, F.; Zhang, Z.; et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007, 8, R162. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Tong, X.; Han, M.; Hu, H.; Dai, F. Genome-Wide Identification and Characterization of WD40 Protein Genes in the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 19, 527. [Google Scholar] [CrossRef] [PubMed]

| Type | Gene | Protein Length | Molecular Weight | Isoelectric Point | Chromosome |
|---|---|---|---|---|---|
| RTKs | BmDDR | 656 | 75,616.43 | 8.24 | chr1 |
| BmEGFR | 1448 | 161,311.64 | 5.82 | chr1 | |
| BmEPH1 | 1006 | 111,746 | 5.78 | chr1 | |
| BmTRK | 578 | 64,646.3 | 5.43 | chr4 | |
| BmRYK1 | 597 | 65,058.48 | 8.49 | chr5 | |
| BmRYK2 | 542 | 60,476.73 | 6.98 | chr5 | |
| BmFGFR1 | 856 | 97,292.72 | 5.86 | chr7 | |
| BmFGFR2 | 616 | 69,437.85 | 5.61 | chr9 | |
| BmROR | 971 | 109,667.49 | 8.05 | chr12 | |
| BmVEGFR | 1366 | 155,204.04 | 5.85 | chr13 | |
| BmAATYK | 3012 | 332,254.39 | 4.41 | chr17 | |
| BmRET | 807 | 91,551.54 | 6.24 | chr19 | |
| BmFGFR3 | 910 | 134,082.47 | 8.65 | chr22 | |
| BmMUSK | 638 | 72,945.69 | 5.88 | chr22 | |
| BmROS | 2464 | 276,014.61 | 6.56 | chr23 | |
| BmALK | 1698 | 187,163.1 | 6.67 | chr24 | |
| BmPTK7 | 184 | 21,040.15 | 6.31 | chr24 | |
| BmMET | 739 | 83,289.74 | 5.83 | chr25 | |
| BmEPH2 | 184 | 20,469.51 | 4.85 | unknown | |
| CTKs | BmABL | 1247 | 136,157.76 | 8.31 | chr1 |
| BmACK1 | 1136 | 127,807.11 | 6.84 | chr1 | |
| BmINSR1 | 985 | 107,739.52 | 8.53 | chr4 | |
| BmINSR2 | 1022 | 112,004.68 | 6.67 | chr4 | |
| BmINSR3 | 1472 | 164,583.02 | 6.4 | chr5 | |
| BmSYK | 803 | 90,867.3 | 6.34 | chr5 | |
| BmFES | 867 | 98,133.68 | 8.38 | chr6 | |
| BmACK2 | 911 | 102,923.58 | 6.89 | chr10 | |
| BmSRC1 | 521 | 59,837.59 | 7.22 | chr11 | |
| BmTEC | 657 | 74,694.1 | 8.49 | chr13 | |
| BmJAK | 1074 | 123,869.62 | 6.75 | chr19 | |
| BmSRC2 | 507 | 58,373.31 | 6.21 | chr19 | |
| BmFAK | 1524 | 169,553.79 | 7.39 | chr20 | |
| BmCSK | 589 | 64,243.5 | 8.31 | chr21 |
| Type | Silkworm Gene Name | Protein Accession | Human Gene Name | Protein Accession | Identity |
|---|---|---|---|---|---|
| RTKs | BmAATYK | XP_021203756.1 | AATYK | NP_004911 | 29% |
| BmALK | XP_012552890.1 | ALK | NP_004295 | 61% | |
| BmDDR | NP_001108469.1 | DDR2 | NP_006173 | 42% | |
| BmEGFR | XP_004929799.2 | ERBB4 | NP_005226 | 65% | |
| BmEPH1 | XP_021207962.1 | EPHA5 | P54756 | 72% | |
| BmEPH2 | Gene016321 | EPHA2 | NP_004422 | 47% | |
| BmFGFR1 | NP_001037558.1 | FGFR3 | NP_000133 | 64% | |
| BmFGFR2 | XP_004922880.2 | FGFR1 | AAA35835 | 48% | |
| BmFGFR3 | BGIBMGA010869 | FGFR1 | AAA35835 | 37% | |
| BmMET | XP_004925752.1 | RON | NP_002438 | 40% | |
| BmMUSK | NP_001186538.1 | MUSK | NP_005583 | 62% | |
| BmPTK7 | XP_021209072.1 | PTK7 | NP_002812 | 39% | |
| BmRET | NP_001164049.1 | RET | P07949 | 44% | |
| BmROR | XP_021203906.1 | ROR1 | NP_005003 | 55% | |
| BmROS | XP_012551361.1 | ROS1 | NP_002935 | 59% | |
| BmRYK1 | XP_021207735.1 | RYK | AAB26341 | 44% | |
| BmRYK2 | XP_004932386.1 | RYK | AAB26341 | 50% | |
| BmTRK | XP_004929369.2 | NTRK2 | NP_006171 | 46% | |
| BmVEGFR | XP_012547297.1 | VEGFR1 | NP_002010 | 43% | |
| CTKs | BmABL | XP_012551486.1 | ABL1 | NP_005148 | 77% |
| BmACK1 | XP_004933204.2 | ACK1 | NP_005772 | 47% | |
| BmACK2 | XP_004930430.1 | ACK1 | NP_005772 | 58% | |
| BmCSK | XP_012548803.1 | CSK | NP_004374 | 57% | |
| BmFAK | XP_021207025.1 | FAK | NP_005598 | 59% | |
| BmFES | XP_012549644.1 | FES | NP_001996 | 57% | |
| BmINSR1 | XP_021203435.1 | INSR | NP_000199 | 44% | |
| BmINSR2 | XP_021203450.1 | INSRR | AAC31759 | 35% | |
| BmINSR3 | NP_001037011.1 | IGF1R | NP_000866 | 71% | |
| BmJAK | XP_021206252.1 | JAK2 | NP_004963 | 34% | |
| BmSRC1 | XP_004932912.1 | FYN | NP_002028 | 57% | |
| BmSRC2 | XP_012549151.1 | FYN | NP_002028 | 70% | |
| BmSYK | XP_004930074.1 | ZAP70 | A44266 | 48% | |
| BmTEC | XP_021204775.1 | TEC | NP_003206 | 58% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, S.; Tong, X.; Han, M.; Bai, Y.; Dai, F. Genome-Wide Identification and Characterization of Tyrosine Kinases in the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2018, 19, 934. https://doi.org/10.3390/ijms19040934
He S, Tong X, Han M, Bai Y, Dai F. Genome-Wide Identification and Characterization of Tyrosine Kinases in the Silkworm, Bombyx mori. International Journal of Molecular Sciences. 2018; 19(4):934. https://doi.org/10.3390/ijms19040934
Chicago/Turabian StyleHe, Songzhen, Xiaoling Tong, Minjin Han, Yanmin Bai, and Fangyin Dai. 2018. "Genome-Wide Identification and Characterization of Tyrosine Kinases in the Silkworm, Bombyx mori" International Journal of Molecular Sciences 19, no. 4: 934. https://doi.org/10.3390/ijms19040934
APA StyleHe, S., Tong, X., Han, M., Bai, Y., & Dai, F. (2018). Genome-Wide Identification and Characterization of Tyrosine Kinases in the Silkworm, Bombyx mori. International Journal of Molecular Sciences, 19(4), 934. https://doi.org/10.3390/ijms19040934





