Polyamine Metabolism and Oxidative Protein Folding in the ER as ROS-Producing Systems Neglected in Virology
Abstract
:1. Introduction
2. Biogenic Polyamines at the Crossroads of Redox Status
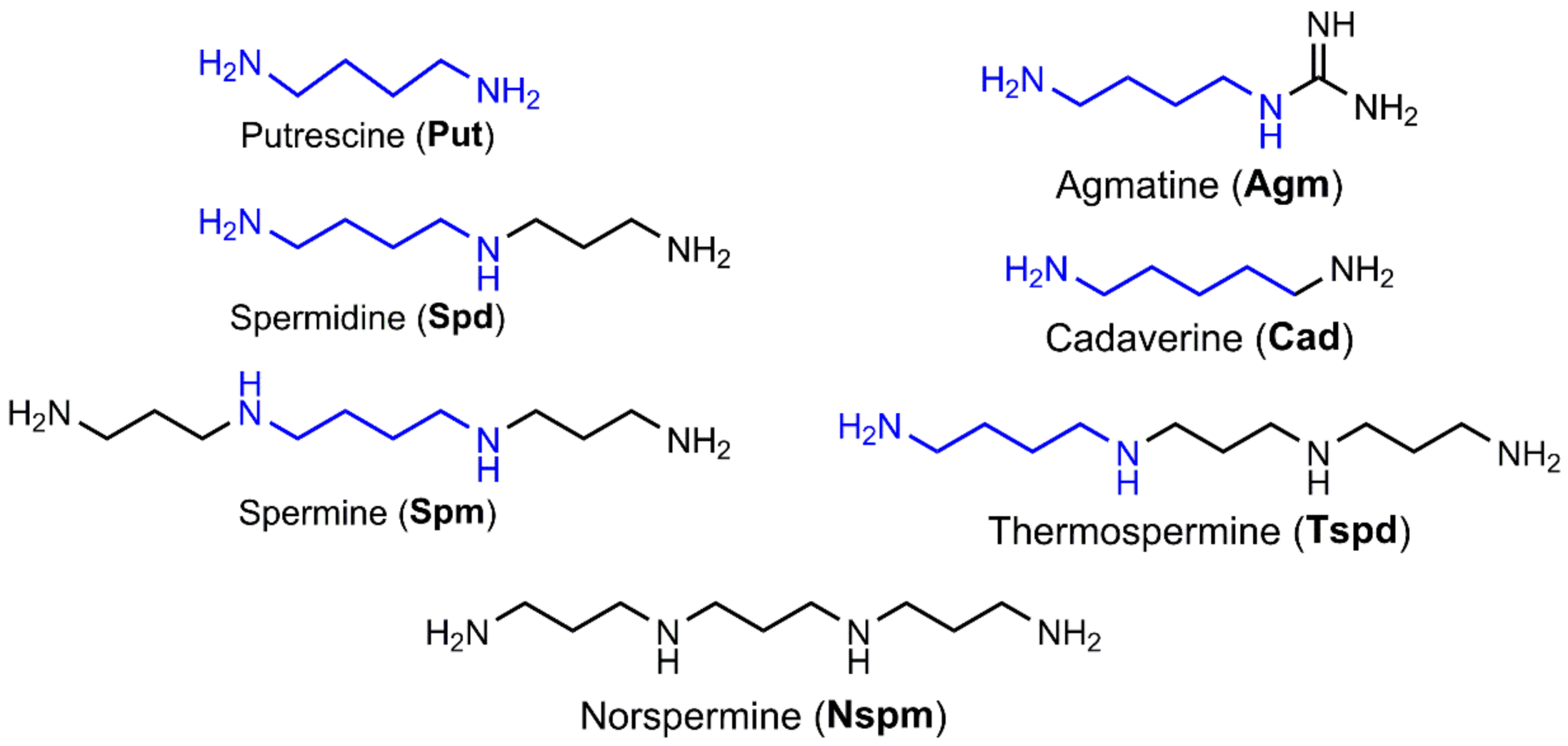
2.1. Polyamine Biosynthesis and Catabolism
2.2. Regulation of Polyamine Metabolism
2.3. Polyamines Can Act as Antioxidants
2.4. Polyamines in Health and Disease
2.5. Polyamines and Viral Infections
3. ER Oxidoreductins as Key Players in Oxidative Protein Folding, Calcium Efflux from the ER, and Evasion of Cancer Cells from Immune Responses
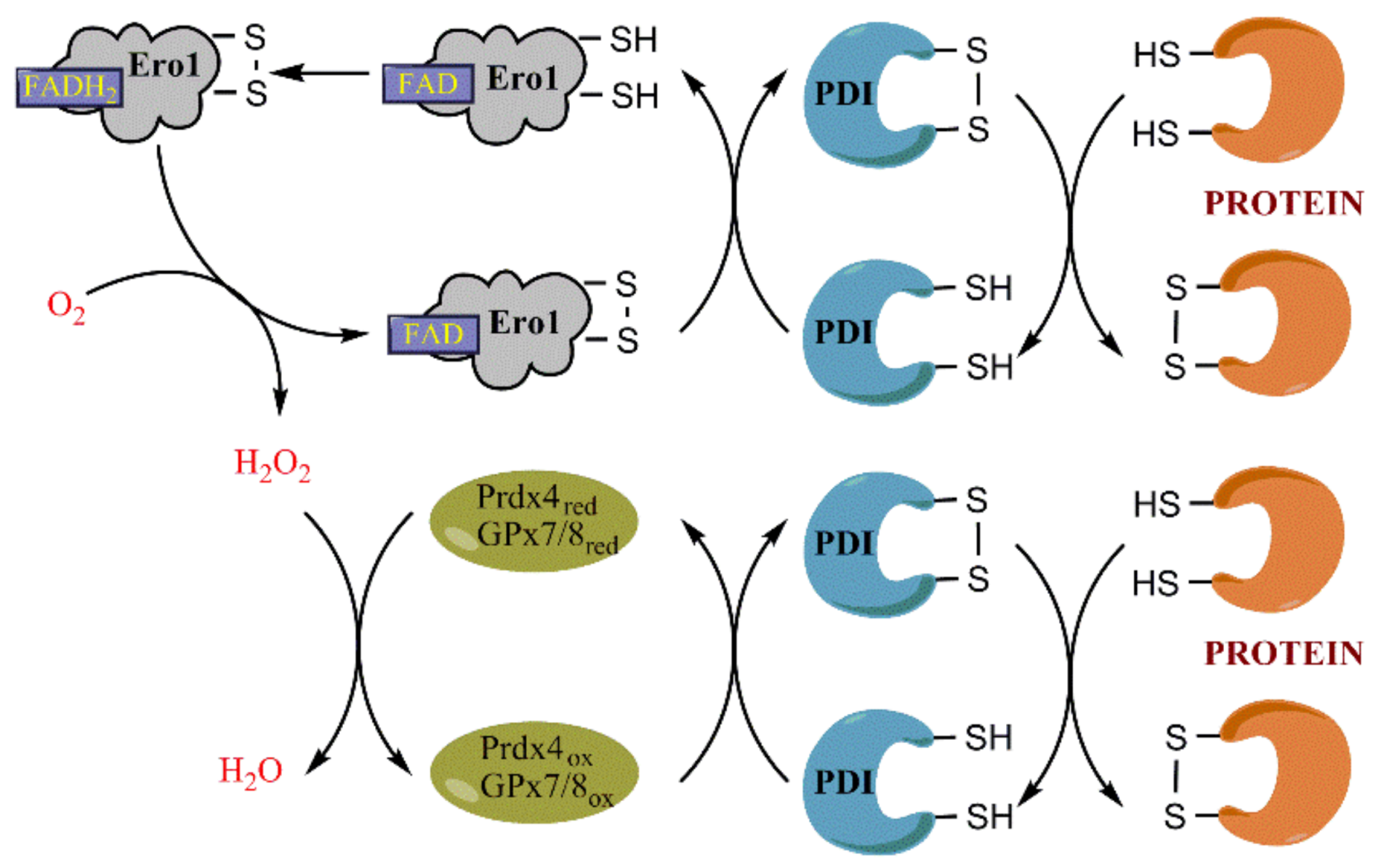
3.1. Ero1 Is the Key Player in Oxidative Protein Folding
3.2. Regulation of Ero1 Expression
3.3. Role of Prdx4 and GPx7/8 in Scavenging H2O2 in the ER and in Protein Folding
3.4. Ero1 Controls Efflux of Calcium Ions from the ER
3.5. Role of Ero1 in Pathology
3.6. Ero1α in Viral Infections
4. Interplay between Polyamine Metabolism and ER Stress/Unfolded Protein Response
5. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| ROS | Reactive oxygen species |
| ER | Endoplasmic reticulum |
| Ero1 | ER oxidoreductin 1 |
| Ero1L | Ero1-like |
| HIV | Human immunodeficiency virus |
| HBV | Hepatitis B virus |
| HCV | Hepatitis C virus |
| HDV | Hepatitis D virus |
| EBV | Epstein-Barr virus |
| HPV | Human papilloma virus |
| Nox | NADPH-oxidase |
| MAO | Monoaminooxidase |
| ODC | Ornithine decarboxylase |
| ADC | Arginine decarboxylase |
| AZInI | Antizyme inhibitor |
| ARG | Arginase |
| OTC | Ornithine transcarbamylase |
| AGMAT | Agmatinase |
| SRM | Spermidine synthase |
| SMS | Spermine synthase |
| 3-AAP | N-acetyl-3-aminopropanal |
| 3-AP | 3-Aminopropanal |
| dcAdoMet | Decarboxylated S-adenosylmethionine |
| AdoMetDC | S-Adenosylmethionine decarboxylase |
| SSAT | Spermidine/spermine-N1-acetyltransferase |
| FAD | Flavin adenine dinucleotide |
| PAOX | Acetylpolyamine oxidase |
| SMOX | Spermine oxidase |
| DHPS | Deoxyhypusine synthase |
| DOHH | Deoxyhypusine hydroxylase |
| NF-κB | Nuclear factor-kappa B |
| Nrf2 | nuclear factor erythroid 2 (NFE2)-related factor 2 |
| PI3K | Phosphatidylinositol-4,5-bisphosphate 3-kinase |
| MAPK | Mitogen-activated protein kinase |
| SLC | Solute carrier |
| PRE | Polyamine response element |
| ARE | Antioxidant response element |
| AZ | Antizyme |
| Nqo1 | NADPH:quinone oxidoreductase 1 |
| PERK | Protein kinase R (PKR)-like ER kinase |
| CHOP | CCAAT/Enhancer-Binding Protein Homologous Protein |
| ATF4 | Activating transcription factor 4 |
| HIF-1α | Hypoxia inducible factor 1α |
| G-CSF | Granulocyte colony-stimulating factor |
| CXCL | Chemokine (C-X-C motif) ligand |
| FDMO | d,l-difluoromethylornithine |
| DPPH | 1,1-Diphenyl-2-picrylhydrazyl |
| PBMC | Peripheral blood mononuclear cells |
| eIF5A | Eukaryotic translation initiation factor 5A |
| HAT | Histone acetyl transferase |
| HSV | Human herpes virus |
| HCMV | Human cytomegalovirus |
| PDI | Protein disulfide isomerase |
| GSSG | Oxidized glutathione |
| GSH | Reduced glutathione |
| UPR | Unfolded protein response |
| Prdx4 | Peroxiredoxin 4 |
| GPx | Glutathione peroxidase |
| SERCA | Sarco/endoplasmic reticulum Ca2+-ATPase |
| IP3R | Inositol 1,4,5-trisphosphate receptor |
| RyR | Ryanodine receptor |
| SEPN1 | Selenoprotein N |
| VDAC | Voltage-dependent anion channel |
| MCU | Mitochondrial calcium uniporter channel |
| MICU1 | Mitochondrial calcium uptake 1 |
| MAMs | Mitochondria-associated membranes |
| PD-L1 | Programmed cell-death 1 ligand 1 |
| MDSCs | Myeloid-derived suppressor cells |
| MHC-1 | Major histocompatibility complex 1 |
References
- Fields, B.N.; Knipe, D.M.; Howley, P.M. Fields Virology, 5th ed.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- Lemon, S.M.; Walker, C.M.; Alter, M.J.; Yi, M.-K. Hepatitis C virus. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott, Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 1, pp. 1253–1304. [Google Scholar]
- Kuritzkes, D.R.; Walker, B.D. HIV-1 pathigenesis, clinical manifestations and treatment. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott, Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 1, pp. 2187–2214. [Google Scholar]
- Oh, J.K.; Weiderpass, E. Infection and cancer: Global distribution and burden of diseases. Ann. Glob. Health 2014, 80, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Biagioli, M.C.; Kaul, P.; Singh, I.; Turner, R.B. The role of oxidative stress in rhinovirus induced elaboration of IL-8 by respiratory epithelial cells. Free Radic. Biol. Med. 1999, 26, 454–462. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Valuev-Ellison, V.T.; Ivanova, O.N.; Kochetkov, S.N.; Starodubova, E.S.; Bartosch, B.; Isaguliants, M.G. Oxidative stress during HIV infection: Mechanisms and consequences. Oxid. Med. Cell. Longev. 2016, 2016, 8910396. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Tyurina, D.A.; Ivanova, O.N.; Kochetkov, S.N.; Bartosch, B.; Isaguliants, M.G. Oxidative stress, a trigger of hepatitis C and B virus-induced liver carcinogenesis. Oncotarget 2017, 8, 3895–3932. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenko, Y.V.; Smirnova, O.A.; Ivanov, A.V.; Starodubova, E.S.; Karpov, V.L. Nonstructural Protein 1 of Tick-Borne Encephalitis Virus Induces Oxidative Stress and Activates Antioxidant Defense by the Nrf2/ARE Pathway. Intervirology 2016, 59, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.P.; Lee, J.C.; Loke, W.M.; Yeo, L.L.; Quek, A.M.; Lim, E.C.; Halliwell, B.; Seet, R.C. Does influenza A infection increase oxidative damage? Antioxid. Redox Signal. 2014, 21, 1025–1031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olagnier, D.; Peri, S.; Steel, C.; van Montfoort, N.; Chiang, C.; Beljanski, V.; Slifker, M.; He, Z.; Nichols, C.N.; Lin, R.; et al. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog. 2014, 10, e1004566. [Google Scholar] [CrossRef] [PubMed]
- Jamaluddin, M.; Tian, B.; Boldogh, I.; Garofalo, R.P.; Brasier, A.R. Respiratory syncytial virus infection induces a reactive oxygen species-MSK1-phospho-Ser-276 RelA pathway required for cytokine expression. J. Virol. 2009, 83, 10605–10615. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Oxidative stress: A concept in redox biology and medicine. Redox Biol. 2015, 4, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.J. Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 2016, 473, 2315–2329. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwenhoek, A. Observationes, D. Anthonii Leeuwenhoek, de natis e semine genitali animalculis. Philos. Trans. R. Soc. Lond. 1678, 12, 1040–1046. [Google Scholar] [CrossRef]
- Dudley, H.W.; Rosenheim, O.; Starling, W.W. The Chemical Constitution of Spermine: Structure and Synthesis. Biochem. J. 1926, 20, 1082–1094. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Mei, Y.H.; Wilson, T. A proposed function for spermine and spermidine: Protection of replicating DNA against damage by singlet oxygen. Proc. Natl. Acad. Sci. USA 1992, 89, 11426–11427. [Google Scholar] [CrossRef] [PubMed]
- Tabor, H.; Tabor, C.W.; Irreverre, F. Quantitative determination of aliphatic diamines and polyamines by an automated liquid chromatography procedure. Anal. Biochem. 1973, 55, 457–467. [Google Scholar] [CrossRef]
- Pegg, A.E.; Wechter, R.S.; Clark, R.S.; Wiest, L.; Erwin, B.G. Acetylation of decarboxylated S-adenosylmethionine by mammalian cells. Biochemistry 1986, 25, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Burtin, D.; Martin-Tanguy, J.; Paynot, M.; Rossin, N. Effects of the suicide inhibitors of arginine and ornithine decarboxylase activities on organogenesis, growth, free polyamine and hydroxycinnamoyl putrescine levels in leaf explants of Nicotiana Xanthi n.c. Cultivated in vitro in a medium producing callus formation. Plant Physiol. 1989, 89, 104–110. [Google Scholar] [PubMed]
- Satishchandran, C.; Boyle, S.M. Purification and properties of agmatine ureohydrolyase, a putrescine biosynthetic enzyme in Escherichia coli. J. Bacteriol. 1986, 165, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Nakada, Y.; Itoh, Y. Identification of the putrescine biosynthetic genes in Pseudomonas aeruginosa and characterization of agmatine deiminase and N-carbamoylputrescine amidohydrolase of the arginine decarboxylase pathway. Microbiology 2003, 149, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Mistry, S.K.; Burwell, T.J.; Chambers, R.M.; Rudolph-Owen, L.; Spaltmann, F.; Cook, W.J.; Morris, S.M., Jr. Cloning of human agmatinase. An alternate path for polyamine synthesis induced in liver by hepatitis B virus. Am. J. Physiol. Gastrointest. Liver Physiol. 2002, 282, G375–G381. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Regunathan, S.; Barrow, C.J.; Eshraghi, J.; Cooper, R.; Reis, D.J. Agmatine: An endogenous clonidine-displacing substance in the brain. Science 1994, 263, 966–969. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Regunathan, S.; Youngson, C.; Bramwell, S.; Reis, D.J. An antibody to agmatine localizes the amine in bovine adrenal chromaffin cells. Neurosci. Lett. 1995, 183, 17–21. [Google Scholar] [CrossRef]
- Regunathan, S.; Youngson, C.; Raasch, W.; Wang, H.; Reis, D.J. Imidazoline receptors and agmatine in blood vessels: A novel system inhibiting vascular smooth muscle proliferation. J. Pharmacol. Exp. Ther. 1996, 276, 1272–1282. [Google Scholar] [PubMed]
- Raasch, W.; Regunathan, S.; Li, G.; Reis, D.J. Agmatine, the bacterial amine, is widely distributed in mammalian tissues. Life Sci. 1995, 56, 2319–2330. [Google Scholar] [CrossRef]
- Bence, A.K.; Worthen, D.R.; Stables, J.P.; Crooks, P.A. An in vivo evaluation of the antiseizure activity and acute neurotoxicity of agmatine. Pharmacol. Biochem. Behav. 2003, 74, 771–775. [Google Scholar] [CrossRef]
- Regunathan, S.; Reis, D.J. Characterization of arginine decarboxylase in rat brain and liver: Distinction from ornithine decarboxylase. J. Neurochem. 2000, 74, 2201–2208. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Molina, B.; Lopez-Contreras, A.J.; Lambertos, A.; Dardonville, C.; Cremades, A.; Penafiel, R. Influence of ornithine decarboxylase antizymes and antizyme inhibitors on agmatine uptake by mammalian cells. Amino Acids 2015, 47, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.S.; Hu, G.; Pegg, A.E. Putrescine biosynthesis in mammalian tissues. Biochem. J. 2004, 379, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Balasundaram, D.; Xie, Q.W.; Tabor, C.W.; Tabor, H. The presence of an active S-adenosylmethionine decarboxylase gene increases the growth defect observed in Saccharomyces cerevisiae mutants unable to synthesize putrescine, spermidine, and spermine. J. Bacteriol. 1994, 176, 6407–6409. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E.; Xiong, H.; Feith, D.J.; Shantz, L.M. S-adenosylmethionine decarboxylase: Structure, function and regulation by polyamines. Biochem. Soc. Trans. 1998, 26, 580–586. [Google Scholar] [CrossRef] [PubMed]
- Hillary, R.A.; Pegg, A.E. Decarboxylases involved in polyamine biosynthesis and their inactivation by nitric oxide. Biochim. Biophys. Acta 2003, 1647, 161–166. [Google Scholar] [CrossRef]
- Finney, J.; Moon, H.J.; Ronnebaum, T.; Lantz, M.; Mure, M. Human copper-dependent amine oxidases. Arch. Biochem. Biophys. 2014, 546, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E.; Casero, R.A., Jr. Current status of the polyamine research field. Methods Mol. Biol. 2011, 720, 3–35. [Google Scholar] [PubMed]
- Wang, Y.; Devereux, W.; Woster, P.M.; Stewart, T.M.; Hacker, A.; Casero, R.A., Jr. Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res. 2001, 61, 5370–5373. [Google Scholar] [PubMed]
- Yoshida, M.; Tomitori, H.; Machi, Y.; Hagihara, M.; Higashi, K.; Goda, H.; Ohya, T.; Niitsu, M.; Kashiwagi, K.; Igarashi, K. Acrolein toxicity: Comparison with reactive oxygen species. Biochem. Biophys. Res. Commun. 2009, 378, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Van den Munckhof, R.J.; Denyn, M.; Tigchelaar-Gutter, W.; Schipper, R.G.; Verhofstad, A.A.; Van Noorden, C.J.; Frederiks, W.M. In situ substrate specificity and ultrastructural localization of polyamine oxidase activity in unfixed rat tissues. J. Histochem. Cytochem. 1995, 43, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Murray-Stewart, T.; Wang, Y.; Goodwin, A.; Hacker, A.; Meeker, A.; Casero, R.A., Jr. Nuclear localization of human spermine oxidase isoforms—Possible implications in drug response and disease etiology. FEBS J. 2008, 275, 2795–2806. [Google Scholar] [CrossRef] [PubMed]
- Bonaiuto, E.; Grancara, S.; Martinis, P.; Stringaro, A.; Colone, M.; Agostinelli, E.; Macone, A.; Stevanato, R.; Vianello, F.; Toninello, A.; et al. A novel enzyme with spermine oxidase properties in bovine liver mitochondria: Identification and kinetic characterization. Free Radic. Biol. Med. 2015, 81, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Pledgie, A.; Huang, Y.; Hacker, A.; Zhang, Z.; Woster, P.M.; Davidson, N.E.; Casero, R.A., Jr. Spermine oxidase SMO(PAOh1), Not N1-acetylpolyamine oxidase PAO, is the primary source of cytotoxic H2O2 in polyamine analogue-treated human breast cancer cell lines. J. Biol. Chem. 2005, 280, 39843–39851. [Google Scholar] [CrossRef] [PubMed]
- Ceci, R.; Duranti, G.; Leonetti, A.; Pietropaoli, S.; Spinozzi, F.; Marcocci, L.; Amendola, R.; Cecconi, F.; Sabatini, S.; Mariottini, P.; et al. Adaptive responses of heart and skeletal muscle to spermine oxidase overexpression: Evaluation of a new transgenic mouse model. Free Radic. Biol. Med. 2017, 103, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Ou, Y.; Wang, S.J.; Li, D.; Chu, B.; Gu, W. Activation of SAT1 engages polyamine metabolism with p53-mediated ferroptotic responses. Proc. Natl. Acad. Sci. USA 2016, 113, E6806–E6812. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Mammalian polyamine metabolism and function. IUBMB Life 2009, 61, 880–894. [Google Scholar] [CrossRef] [PubMed]
- Abdulhussein, A.A.; Wallace, H.M. Polyamines and membrane transporters. Amino Acids 2014, 46, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Perez-Leal, O.; Merali, S. Regulation of polyamine metabolism by translational control. Amino Acids 2012, 42, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.J.; Meyers, C.; Laimins, L.A.; Hay, N. c-Myc induces the expression and activity of ornithine decarboxylase. Cell Growth Differ. 1993, 4, 879–883. [Google Scholar] [PubMed]
- Packham, G.; Cleveland, J.L. Ornithine decarboxylase is a mediator of c-Myc-induced apoptosis. Mol. Cell. Biol. 1994, 14, 5741–5747. [Google Scholar] [CrossRef] [PubMed]
- Tacchini, L.; De Ponti, C.; Matteucci, E.; Follis, R.; Desiderio, M.A. Hepatocyte growth factor-activated NF-κB regulates HIF-1 activity and ODC expression, implicated in survival, differently in different carcinoma cell lines. Carcinogenesis 2004, 25, 2089–2100. [Google Scholar] [CrossRef] [PubMed]
- Babbar, N.; Casero, R.A., Jr. Tumor necrosis factor-α increases reactive oxygen species by inducing spermine oxidase in human lung epithelial cells: A potential mechanism for inflammation-induced carcinogenesis. Cancer Res. 2006, 66, 11125–11130. [Google Scholar] [CrossRef] [PubMed]
- Babbar, N.; Gerner, E.W.; Casero, R.A., Jr. Induction of spermidine/spermine N1-acetyltransferase (SSAT) by aspirin in Caco-2 colon cancer cells. Biochem. J. 2006, 394, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Babbar, N.; Hacker, A.; Huang, Y.; Casero, R.A., Jr. Tumor necrosis factor α induces spermidine/spermine N1-acetyltransferase through nuclear factor κB in non-small cell lung cancer cells. J. Biol. Chem. 2006, 281, 24182–24192. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, L.; Thiagalingam, A.; Nelkin, B.D.; Casero, R.A., Jr. The identification of a cis-element and a trans-acting factor involved in the response to polyamines and polyamine analogues in the regulation of the human spermidine/spermine N1-acetyltransferase gene transcription. J. Biol. Chem. 1998, 273, 34623–34630. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Devereux, W.; Stewart, T.M.; Casero, R.A., Jr. Cloning and characterization of human polyamine-modulated factor-1, a transcriptional cofactor that regulates the transcription of the spermidine/spermine N1-acetyltransferase gene. J. Biol. Chem. 1999, 274, 22095–22101. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, O.A.; Isaguliants, M.G.; Hyvonen, M.T.; Keinanen, T.A.; Tunitskaya, V.L.; Vepsalainen, J.; Alhonen, L.; Kochetkov, S.N.; Ivanov, A.V. Chemically induced oxidative stress increases polyamine levels by activating the transcription of ornithine decarboxylase and spermidine/spermine-N1-acetyltransferase in human hepatoma HUH7 cells. Biochimie 2012, 94, 1876–1883. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.H.; Yang, Y.; Wu, G.; Ignarro, L.J. IL-4 and IL-13 upregulate ornithine decarboxylase expression by PI3K and MAP kinase pathways in vascular smooth muscle cells. Am. J. Physiol. Cell Physiol. 2008, 294, C1198–C1205. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Alm, K.; Vujcic, S.; Kramer, D.L.; Kee, K.; Diegelman, P.; Porter, C.W. The role of mitogen-activated protein kinase activation in determining cellular outcomes in polyamine analogue-treated human melanoma cells. Cancer Res. 2003, 63, 3619–3625. [Google Scholar] [PubMed]
- Wang, C.; Ruan, P.; Zhao, Y.; Li, X.; Wang, J.; Wu, X.; Liu, T.; Wang, S.; Hou, J.; Li, W.; et al. Spermidine/spermine N1-acetyltransferase regulates cell growth and metastasis via AKT/β-catenin signaling pathways in hepatocellular and colorectal carcinoma cells. Oncotarget 2017, 8, 1092–1109. [Google Scholar] [PubMed]
- Pegg, A.E.; McCann, P.P. Polyamine metabolism and function. Am. J. Physiol. 1982, 243, C212–C221. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Pan, H.; Verma, A.K. Non-AP-1 tumor promoter 12-O-tetradecanoylphorbol-13-acetate-responsive sequences in the human ornithine decarboxylase gene. Mol. Carcinog. 1994, 10, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Aleksunes, L.M.; Manautou, J.E. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol. Pathol. 2007, 35, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Forshell, T.P.; Rimpi, S.; Nilsson, J.A. Chemoprevention of B-cell lymphomas by inhibition of the Myc target spermidine synthase. Cancer Prev. Res. 2010, 3, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.A.; Keller, U.B.; Baudino, T.A.; Yang, C.; Norton, S.; Old, J.A.; Nilsson, L.M.; Neale, G.; Kramer, D.L.; Porter, C.W.; et al. Targeting ornithine decarboxylase in Myc-induced lymphomagenesis prevents tumor formation. Cancer Cell 2005, 7, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Regulation of ornithine decarboxylase. J. Biol. Chem. 2006, 281, 14529–14532. [Google Scholar] [CrossRef] [PubMed]
- Asher, G.; Bercovich, Z.; Tsvetkov, P.; Shaul, Y.; Kahana, C. 20S proteasomal degradation of ornithine decarboxylase is regulated by NQO1. Mol. Cell 2005, 17, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Contreras, A.J.; Ramos-Molina, B.; Martinez-de-la-Torre, M.; Penafiel-Verdu, C.; Puelles, L.; Cremades, A.; Penafiel, R. Expression of antizyme inhibitor 2 in male haploid germinal cells suggests a role in spermiogenesis. Int. J. Biochem. Cell Biol. 2009, 41, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Pitkanen, L.T.; Heiskala, M.; Andersson, L.C. Expression of a novel human ornithine decarboxylase-like protein in the central nervous system and testes. Biochem. Biophys. Res. Commun. 2001, 287, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hacker, A.; Murray-Stewart, T.; Fleischer, J.G.; Woster, P.M.; Casero, R.A., Jr. Induction of human spermine oxidase SMO(PAOh1) is regulated at the levels of new mRNA synthesis, mRNA stabilization and newly synthesized protein. Biochem. J. 2005, 386, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Casero, R.A., Jr. Mammalian polyamine catabolism: A therapeutic target, a pathological problem, or both? J. Biochem. 2006, 139, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, K.; Wang, Z.; Barone, S.; Prada, A.E.; Kelly, C.N.; Casero, R.A.; Yokota, N.; Porter, C.W.; Rabb, H.; Soleimani, M. Expression of SSAT, a novel biomarker of tubular cell damage, increases in kidney ischemia-reperfusion injury. Am. J. Physiol. Renal Physiol. 2003, 284, F1046–F1055. [Google Scholar] [CrossRef] [PubMed]
- Cervelli, M.; Fratini, E.; Amendola, R.; Bianchi, M.; Signori, E.; Ferraro, E.; Lisi, A.; Federico, R.; Marcocci, L.; Mariottini, P. Increased spermine oxidase (SMO) activity as a novel differentiation marker of myogenic C2C12 cells. Int. J. Biochem. Cell Biol. 2009, 41, 934–944. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.A.; Pegg, A.E. Polyamine catabolism and disease. Biochem. J. 2009, 421, 323–338. [Google Scholar] [CrossRef] [PubMed]
- Soulet, D.; Gagnon, B.; Rivest, S.; Audette, M.; Poulin, R. A fluorescent probe of polyamine transport accumulates into intracellular acidic vesicles via a two-step mechanism. J. Biol. Chem. 2004, 279, 49355–49366. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.C.; Sirisoma, N.S.; Kuppusamy, P.; Zweier, J.L.; Woster, P.M.; Casero, R.A., Jr. The natural polyamine spermine functions directly as a free radical scavenger. Proc. Natl. Acad. Sci. USA 1998, 95, 11140–11145. [Google Scholar] [CrossRef] [PubMed]
- Park, I.-H.; Lim, M.-M. Inhibitory Effect of Spermidine with Antioxidant Activity on Oxidative Stress in Human Dermal Fibroblasts. J. Life Sci. 2011, 21, 693–699. [Google Scholar] [CrossRef]
- Battaglia, V.; Rossi, C.A.; Colombatto, S.; Grillo, M.A.; Toninello, A. Different behavior of agmatine in liver mitochondria: Inducer of oxidative stress or scavenger of reactive oxygen species? Biochim. Biophys. Acta 2007, 1768, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Gaboriau, F.; Vaultier, M.; Moulinoux, J.P.; Delcros, J.G. Antioxidative properties of natural polyamines and dimethylsilane analogues. Redox Rep. 2005, 10, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Das, K.C.; Misra, H.P. Hydroxyl radical scavenging and singlet oxygen quenching properties of polyamines. Mol. Cell. Biochem. 2004, 262, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Sava, I.G.; Battaglia, V.; Rossi, C.A.; Salvi, M.; Toninello, A. Free radical scavenging action of the natural polyamine spermine in rat liver mitochondria. Free Radic. Biol. Med. 2006, 41, 1272–1281. [Google Scholar] [CrossRef] [PubMed]
- Rigobello, M.P.; Toninello, A.; Siliprandi, D.; Bindoli, A. Effect of spermine on mitochondrial glutathione release. Biochem. Biophys. Res. Commun. 1993, 194, 1276–1281. [Google Scholar] [CrossRef] [PubMed]
- Muscari, C.; Guarnieri, C.; Stefanelli, C.; Giaccari, A.; Caldarera, C.M. Protective effect of spermine on DNA exposed to oxidative stress. Mol. Cell. Biochem. 1995, 144, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.C.; Yager, J.D.; Woster, P.A.; Casero, R.A., Jr. Structural specificity of polyamines and polyamine analogues in the protection of DNA from strand breaks induced by reactive oxygen species. Biochem. Biophys. Res. Commun. 1998, 244, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Rider, J.E.; Hacker, A.; Mackintosh, C.A.; Pegg, A.E.; Woster, P.M.; Casero, R.A. Spermine and spermidine mediate protection against oxidative damage caused by hydrogen peroxide. Amino Acids 2007, 33, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Jung, I.L.; Oh, T.J.; Kim, I.G. Abnormal growth of polyamine-deficient Escherichia coli mutant is partially caused by oxidative stress-induced damage. Arch. Biochem. Biophys. 2003, 418, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.K.; Tabor, C.W.; Tabor, H. Polyamine deficiency leads to accumulation of reactive oxygen species in a spe2Delta mutant of Saccharomyces cerevisiae. Yeast 2006, 23, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Buttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Von Deutsch, A.W.; Mitchell, C.D.; Williams, C.E.; Dutt, K.; Silvestrov, N.A.; Klement, B.J.; Abukhalaf, I.K.; von Deutsch, D.A. Polyamines protect against radiation-induced oxidative stress. Gravit. Space Biol. Bull. 2005, 18, 109–110. [Google Scholar] [PubMed]
- Marzabadi, M.R.; Llvaas, E. Spermine prevent iron accumulation and depress lipofuscin accumulation in cultured myocardial cells. Free Radic. Biol. Med. 1996, 21, 375–381. [Google Scholar] [CrossRef]
- Minois, N.; Carmona-Gutierrez, D.; Bauer, M.A.; Rockenfeller, P.; Eisenberg, T.; Brandhorst, S.; Sigrist, S.J.; Kroemer, G.; Madeo, F. Spermidine promotes stress resistance in Drosophila melanogaster through autophagy-dependent and -independent pathways. Cell Death Dis. 2012, 3, e401. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.W.; Cha, H.J.; Han, M.H.; Hwang, S.J.; Lee, D.S.; Yoo, J.S.; Choi, I.W.; Kim, S.; Kim, H.S.; Kim, G.Y.; et al. Spermidine Protects against Oxidative Stress in Inflammation Models Using Macrophages and Zebrafish. Biomol. Ther. 2018, 26, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Childs, A.C.; Mehta, D.J.; Gerner, E.W. Polyamine-dependent gene expression. Cell. Mol. Life Sci. 2003, 60, 1394–1406. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Tomitori, H.; Mizuno, S.; Higashi, K.; Full, C.; Fukiwake, T.; Terui, Y.; Leewanich, P.; Nishimura, K.; Toida, T.; et al. Binding of spermine and ifenprodil to a purified, soluble regulatory domain of the N-methyl-d-aspartate receptor. J. Neurochem. 2008, 107, 1566–1577. [Google Scholar] [CrossRef] [PubMed]
- Mott, D.D.; Washburn, M.S.; Zhang, S.; Dingledine, R.J. Subunit-dependent modulation of kainate receptors by extracellular protons and polyamines. J. Neurosci. 2003, 23, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Yan, D.H. Low-affinity spermine block mediating outward currents through Kir2.1 and Kir2.2 inward rectifier potassium channels. J. Physiol. 2007, 583, 891–908. [Google Scholar] [CrossRef] [PubMed]
- Desiderio, M.A.; Tacchini, L.; Anzon, E.; Pogliaghi, G.; Radice, L.; Bernelli-Zazzera, A. Effects of polyamine imbalance on the induction of stress genes in hepatocarcinoma cells exposed to heat shock. Hepatology 1996, 24, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Guo, X.; Rao, J.N.; Zou, T.; Marasa, B.S.; Chen, J.; Greenspon, J.; Casero, R.A., Jr.; Wang, J.Y. Polyamine-modulated c-Myc expression in normal intestinal epithelial cells regulates p21Cip1 transcription through a proximal promoter region. Biochem. J. 2006, 398, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Rao, J.N.; Zou, T.; Liu, L.; Marasa, B.S.; Chen, J.; Turner, D.J.; Passaniti, A.; Wang, J.Y. Induced JunD in intestinal epithelial cells represses CDK4 transcription through its proximal promoter region following polyamine depletion. Biochem. J. 2007, 403, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Monti, M.G.; Ghiaroni, S.; Pernecco, L.; Barbieri, D.; Marverti, G.; Franceschi, C. Polyamine depletion protects HL-60 cells from 2-deoxy-d-ribose-induced apoptosis. Life Sci. 1998, 62, 799–806. [Google Scholar] [CrossRef]
- Monti, M.G.; Ghiaroni, S.; Marverti, G.; Montanari, M.; Moruzzi, M.S. Polyamine depletion switches the form of 2-deoxy-d-ribose-induced cell death from apoptosis to necrosis in HL-60 cells. Int. J. Biochem. Cell Biol. 2004, 36, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Chen, Y.; Huang, Y.P.; Hsu, Y.C.; Chiang, L.H.; Chen, T.Y.; Wang, G.J. Exogenous polyamines promote osteogenic differentiation by reciprocally regulating osteogenic and adipogenic gene expression. J. Cell. Biochem. 2013, 114, 2718–2728. [Google Scholar] [CrossRef] [PubMed]
- Quemener, V.; Blanchard, Y.; Lescoat, D.; Havouis, R.; Moulinoux, J.P. Depletion in nuclear spermine during human spermatogenesis, a natural process of cell differentiation. Am. J. Physiol. 1992, 263, C343–C347. [Google Scholar] [CrossRef] [PubMed]
- Pietila, M.; Pirinen, E.; Keskitalo, S.; Juutinen, S.; Pasonen-Seppanen, S.; Keinanen, T.; Alhonen, L.; Janne, J. Disturbed keratinocyte differentiation in transgenic mice and organotypic keratinocyte cultures as a result of spermidine/spermine N-acetyltransferase overexpression. J. Investig. Dermatol. 2005, 124, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Cooper, H.L.; Folk, J.E. Identification of hypusine, an unusual amino acid, in a protein from human lymphocytes and of spermidine as its biosynthetic precursor. Proc. Natl. Acad. Sci. USA 1981, 78, 2869–2873. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H. The post-translational synthesis of a polyamine-derived amino acid, hypusine, in the eukaryotic translation initiation factor 5A (eIF5A). J. Biochem. 2006, 139, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Eyler, D.E.; Green, R.; Dever, T.E. Hypusine-containing protein eIF5A promotes translation elongation. Nature 2009, 459, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, E.; Shin, B.S.; Woolstenhulme, C.J.; Kim, J.R.; Saini, P.; Buskirk, A.R.; Dever, T.E. eIF5A promotes translation of polyproline motifs. Mol. Cell 2013, 51, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Nakatsu, F.; Kashiwagi, K.; Ohno, H.; Saito, T.; Igarashi, K. Essential role of S-adenosylmethionine decarboxylase in mouse embryonic development. Genes Cells 2002, 7, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Abdellatif, M.; Schroeder, S.; Primessnig, U.; Stekovic, S.; Pendl, T.; Harger, A.; Schipke, J.; Zimmermann, A.; Schmidt, A.; et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 2016, 22, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.; Li, W.; Zou, J.; Jiang, X.; Xu, G.; Huang, H.; Liu, L. Spermidine Prolongs Lifespan and Prevents Liver Fibrosis and Hepatocellular Carcinoma by Activating MAP1S-Mediated Autophagy. Cancer Res. 2017, 77, 2938–2951. [Google Scholar] [CrossRef] [PubMed]
- Rahman, I. Oxidative stress, transcription factors and chromatin remodelling in lung inflammation. Biochem. Pharmacol. 2002, 64, 935–942. [Google Scholar] [CrossRef]
- Rahman, I.; Gilmour, P.S.; Jimenez, L.A.; MacNee, W. Oxidative stress and TNF-α induce histone acetylation and NF-κB/AP-1 activation in alveolar epithelial cells: Potential mechanism in gene transcription in lung inflammation. Mol. Cell. Biochem. 2002, 234–235, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Rahman, I.; Marwick, J.; Kirkham, P. Redox modulation of chromatin remodeling: Impact on histone acetylation and deacetylation, NF-κB and pro-inflammatory gene expression. Biochem. Pharmacol. 2004, 68, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, M.; Park, P.H.; Jackson, D.; Shukla, S.D. Evidence for the role of oxidative stress in the acetylation of histone H3 by ethanol in rat hepatocytes. Alcohol 2010, 44, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gibson, S.B. Is mitochondrial generation of reactive oxygen species a trigger for autophagy? Autophagy 2008, 4, 246–248. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.F.; Ma, Z.; Liu, Z.; Terada, L.S. Nox4-derived H2O2 mediates endoplasmic reticulum signaling through local Ras activation. Mol. Cell. Biol. 2010, 30, 3553–3568. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Luo, H.; Kan, G.; Zhao, Y.; Lin, L.; Tang, Q.; Yu, C.; Sun, W.; Cai, L.; Cui, S. The protective role of autophagy in Heterocephalus glaber hepatic stellate cells exposed to H2O2 or nutritional stress. Cell. Physiol. Biochem. 2014, 34, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Cho, K.S.; Koh, J.Y. Oxidative injury triggers autophagy in astrocytes: The role of endogenous zinc. Glia 2009, 57, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Megosh, L.; Gilmour, S.K.; Rosson, D.; Soler, A.P.; Blessing, M.; Sawicki, J.A.; O’Brien, T.G. Increased frequency of spontaneous skin tumors in transgenic mice which overexpress ornithine decarboxylase. Cancer Res. 1995, 55, 4205–4209. [Google Scholar] [PubMed]
- Nowotarski, S.L.; Woster, P.M.; Casero, R.A., Jr. Polyamines and cancer: Implications for chemotherapy and chemoprevention. Expert Rev. Mol. Med. 2013, 15, e3. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T.G.; Megosh, L.C.; Gilliard, G.; Soler, A.P. Ornithine decarboxylase overexpression is a sufficient condition for tumor promotion in mouse skin. Cancer Res. 1997, 57, 2630–2637. [Google Scholar] [PubMed]
- Hayes, C.S.; DeFeo, K.; Lan, L.; Paul, B.; Sell, C.; Gilmour, S.K. Elevated levels of ornithine decarboxylase cooperate with Raf/ERK activation to convert normal keratinocytes into invasive malignant cells. Oncogene 2006, 25, 1543–1553. [Google Scholar] [CrossRef] [PubMed]
- Shukla-Dave, A.; Castillo-Martin, M.; Chen, M.; Lobo, J.; Gladoun, N.; Collazo-Lorduy, A.; Khan, F.M.; Ponomarev, V.; Yi, Z.; Zhang, W.; et al. Ornithine Decarboxylase Is Sufficient for Prostate Tumorigenesis via Androgen Receptor Signaling. Am. J. Pathol. 2016, 186, 3131–3145. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Hayes, C.S.; Laury-Kleintop, L.; Gilmour, S.K. Suprabasal induction of ornithine decarboxylase in adult mouse skin is sufficient to activate keratinocytes. J. Investig. Dermatol. 2005, 124, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, R.; Asim, M.; Romero-Gallo, J.; Barry, D.P.; Hoge, S.; de Sablet, T.; Delgado, A.G.; Wroblewski, L.E.; Piazuelo, M.B.; Yan, F.; et al. Spermine oxidase mediates the gastric cancer risk associated with Helicobacter pylori CagA. Gastroenterology 2011, 141. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, R.; de Sablet, T.; Asim, M.; Piazuelo, M.B.; Barry, D.P.; Verriere, T.G.; Sierra, J.C.; Hardbower, D.M.; Delgado, A.G.; Schneider, B.G.; et al. Increased Helicobacter pylori-associated gastric cancer risk in the Andean region of Colombia is mediated by spermine oxidase. Oncogene 2015, 34, 3429–3440. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef] [PubMed]
- Snezhkina, A.V.; Krasnov, G.S.; Lipatova, A.V.; Sadritdinova, A.F.; Kardymon, O.L.; Fedorova, M.S.; Melnikova, N.V.; Stepanov, O.A.; Zaretsky, A.R.; Kaprin, A.D.; et al. The Dysregulation of Polyamine Metabolism in Colorectal Cancer Is Associated with Overexpression of c-Myc and C/EBPβ rather than Enterotoxigenic Bacteroides fragilis Infection. Oxid. Med. Cell. Longev. 2016, 2016, 2353560. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.C.; Barwise, A.H.; Walker, I.O. DNA contained by two densonucleosis viruses. J. Virol. 1977, 21, 396–407. [Google Scholar] [PubMed]
- Lanzer, W.; Holowczak, J.A. Polyamines in vaccinia virions and polypeptides released from viral cores by acid extraction. J. Virol. 1975, 16, 1254–1264. [Google Scholar] [PubMed]
- Fukuma, I.; Cohen, S.S. Polyamines in bacteriophage R17 and its RNA. J. Virol. 1975, 16, 222–227. [Google Scholar] [PubMed]
- Sheppard, S.L.; Burness, A.T.; Boyle, S.M. Polyamines in encephalomyocarditis virus. J. Virol. 1980, 34, 266–267. [Google Scholar] [PubMed]
- Gibson, W.; Roizman, B. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. USA 1971, 68, 2818–2821. [Google Scholar] [CrossRef] [PubMed]
- Bachrach, U.; Don, S.; Wiener, H. Occurrence of polyamines in myxoviruses. J. Gen. Virol. 1974, 22, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Fout, G.S.; Medappa, K.C.; Mapoles, J.E.; Rueckert, R.R. Radiochemical determination of polyamines in poliovirus and human rhinovirus 14. J. Biol. Chem. 1984, 259, 3639–3643. [Google Scholar] [PubMed]
- Raina, A.; Tuomi, K.; Mantyjarvi, R. Roles of polyamines in the replication of animal viruses. Med. Biol. 1981, 59, 428–432. [Google Scholar] [PubMed]
- Mounce, B.C.; Cesaro, T.; Moratorio, G.; Hooikaas, P.J.; Yakovleva, A.; Werneke, S.W.; Smith, E.C.; Poirier, E.Z.; Simon-Loriere, E.; Prot, M.; et al. Inhibition of Polyamine Biosynthesis Is a Broad-Spectrum Strategy against RNA Viruses. J. Virol. 2016, 90, 9683–9692. [Google Scholar] [CrossRef] [PubMed]
- Mounce, B.C.; Poirier, E.Z.; Passoni, G.; Simon-Loriere, E.; Cesaro, T.; Prot, M.; Stapleford, K.A.; Moratorio, G.; Sakuntabhai, A.; Levraud, J.P.; et al. Interferon-Induced Spermidine-Spermine Acetyltransferase and Polyamine Depletion Restrict Zika and Chikungunya Viruses. Cell Host Microbe 2016, 20, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Olsen, M.E.; Filone, C.M.; Rozelle, D.; Mire, C.E.; Agans, K.N.; Hensley, L.; Connor, J.H. Polyamines and Hypusination Are Required for Ebolavirus Gene Expression and Replication. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Greco, A.; Calle, A.; Morfin, F.; Thouvenot, D.; Cayre, M.; Kindbeiter, K.; Martin, L.; Levillain, O.; Diaz, J.J. S-adenosyl methionine decarboxylase activity is required for the outcome of herpes simplex virus type 1 infection and represents a new potential therapeutic target. FASEB J. 2005, 19, 1128–1130. [Google Scholar] [CrossRef] [PubMed]
- Gibson, W.; van Breemen, R.; Fields, A.; LaFemina, R.; Irmiere, A. d,l-α-Difluoromethylornithine inhibits human cytomegalovirus replication. J. Virol. 1984, 50, 145–154. [Google Scholar] [PubMed]
- Tyms, A.S.; Scamans, E.; Williamson, J.D. Polyamine metabolism in MRC5 cells infected with different herpesviruses. Biochem. Biophys. Res. Commun. 1979, 86, 312–318. [Google Scholar] [CrossRef]
- Tyms, A.S.; Williamson, J.D. Inhibitors of polyamine biosynthesis block human cytomegalovirus replication. Nature 1982, 297, 690–691. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.; Hanauske-Abel, H.M.; Palumbo, P.; Saxena, D.; D’Alliessi Gandolfi, D.; Park, M.H.; Pe’ery, T.; Mathews, M.B. Inhibition of HIV-1 gene expression by Ciclopirox and Deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology 2009, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Mounce, B.C.; Cesaro, T.; Vlajnic, L.; Vidina, A.; Vallet, T.; Weger-Lucarelli, J.; Passoni, G.; Stapleford, K.A.; Levraud, J.P.; Vignuzzi, M. Chikungunya Virus Overcomes Polyamine Depletion by Mutation of nsP1 and the Opal Stop Codon to Confer Enhanced Replication and Fitness. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- McCormick, F. Polyamine turnover and leakage during infection of HeLa and L-cells with herpes simplex virus type 1. Virology 1978, 91, 496–503. [Google Scholar] [CrossRef]
- Gibson, W.; Roizman, B. The structural role and metabolic involvement of polyamines with herpes simplex virus. In Polyamines in Normal and Neoplastic Growth; Russell, D.H., Ed.; Raven Press, Inc.: New York, NY, USA, 1973. [Google Scholar]
- McCormick, F.P.; Newton, A.A. Polyamine metabolism in cells infected with herpes simplex virus. J. Gen. Virol. 1975, 27, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Isom, H.C. Stimulation of ornithine decarboxylase by human cytomegalovirus. J. Gen. Virol. 1979, 42, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.R.; Tyms, A.S. Polyamine biosynthesis in cells infected with different clinical isolates of human cytomegalovirus. J. Med. Virol. 1991, 34, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Garnett, H.M. Altered polyamine concentrations in cytomegalovirus-infected human cells. S. Afr. Med. J. 1988, 73, 209–211. [Google Scholar] [PubMed]
- Hodgson, J.; Williamson, J.D. Ornithine decarboxylase activity in uninfected and vaccinia virus-infected HeLa cells. Biochem. Biophys. Res. Commun. 1975, 63, 308–312. [Google Scholar] [CrossRef]
- Colombatto, S.; De Agostini, M.; Corsi, D.; Sinicco, A. Polyamines in lymphocytes from patients infected by human immunodeficiency virus. Biol. Chem. Hoppe Seyler 1989, 370, 745–748. [Google Scholar] [CrossRef] [PubMed]
- White, E.L.; Rose, L.M.; Allan, P.W.; Buckheit, R.W., Jr.; Shannon, W.M.; Secrist, J.A., III. Polyamine pools in HIV-infected cells. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1998, 17, 101–103. [Google Scholar] [CrossRef] [PubMed]
- Capone, C.; Cervelli, M.; Angelucci, E.; Colasanti, M.; Macone, A.; Mariottini, P.; Persichini, T. A role for spermine oxidase as a mediator of reactive oxygen species production in HIV-Tat-induced neuronal toxicity. Free Radic. Biol. Med. 2013, 63, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, O.A.; Keinanen, T.A.; Ivanova, O.N.; Hyvonen, M.T.; Khomutov, A.R.; Kochetkov, S.N.; Bartosch, B.; Ivanov, A.V. Hepatitis C virus alters metabolism of biogenic polyamines by affecting expression of key enzymes of their metabolism. Biochem. Biophys. Res. Commun. 2017, 483, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Korovina, A.N.; Tunitskaya, V.L.; Khomutov, M.A.; Simonian, A.R.; Khomutov, A.R.; Ivanov, A.I.; Kochetkov, S.N. Biogenic Polyamines Spermine and Spermidine Activate RNA Polymerase and Inhibit RNA Helicase of Hepatitis C Virus. Biochemistry 2012, 77, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, A.; Vollmert, M.; Tholl, D.; Graves, M.V.; Gurnon, J.R.; Xing, W.; Lisec, A.D.; Nickerson, K.W.; Van Etten, J.L. Chlorella virus PBCV-1 encodes a functional homospermidine synthase. Virology 1999, 263, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Morehead, T.A.; Gurnon, J.R.; Adams, B.; Nickerson, K.W.; Fitzgerald, L.A.; Van Etten, J.L. Ornithine decarboxylase encoded by chlorella virus PBCV-1. Virology 2002, 301, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Ellgaard, L.; Ruddock, L.W. The human protein disulphide isomerase family: Substrate interactions and functional properties. EMBO Rep. 2005, 6, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Kojima, R.; Okumura, M.; Hagiwara, M.; Masui, S.; Maegawa, K.; Saiki, M.; Horibe, T.; Suzuki, M.; Inaba, K. Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci. Rep. 2013, 3, 2456. [Google Scholar] [CrossRef] [PubMed]
- Oka, O.B.; Bulleid, N.J. Forming disulfides in the endoplasmic reticulum. Biochim. Biophys. Acta 2013, 1833, 2425–2429. [Google Scholar] [CrossRef] [PubMed]
- Jessop, C.E.; Watkins, R.H.; Simmons, J.J.; Tasab, M.; Bulleid, N.J. Protein disulphide isomerase family members show distinct substrate specificity: P5 is targeted to BiP client proteins. J. Cell Sci. 2009, 122, 4287–4295. [Google Scholar] [CrossRef] [PubMed]
- Frand, A.R.; Kaiser, C.A. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell 1998, 1, 161–170. [Google Scholar] [CrossRef]
- Pollard, M.G.; Travers, K.J.; Weissman, J.S. Ero1p: A novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1998, 1, 171–182. [Google Scholar] [CrossRef]
- Cabibbo, A.; Pagani, M.; Fabbri, M.; Rocchi, M.; Farmery, M.R.; Bulleid, N.J.; Sitia, R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J. Biol. Chem. 2000, 275, 4827–4833. [Google Scholar] [CrossRef] [PubMed]
- Pagani, M.; Fabbri, M.; Benedetti, C.; Fassio, A.; Pilati, S.; Bulleid, N.J.; Cabibbo, A.; Sitia, R. Endoplasmic reticulum oxidoreductin 1-Lβ (ERO1-Lβ), a human gene induced in the course of the unfolded protein response. J. Biol. Chem. 2000, 275, 23685–23692. [Google Scholar] [CrossRef] [PubMed]
- Dias-Gunasekara, S.; Gubbens, J.; van Lith, M.; Dunne, C.; Williams, J.A.; Kataky, R.; Scoones, D.; Lapthorn, A.; Bulleid, N.J.; Benham, A.M. Tissue-specific expression and dimerization of the endoplasmic reticulum oxidoreductase Ero1β. J. Biol. Chem. 2005, 280, 33066–33075. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, L.; Wang, C.C. The endoplasmic reticulum sulfhydryl oxidase Ero1β drives efficient oxidative protein folding with loose regulation. Biochem. J. 2011, 434, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.; Sevier, C.S.; Heldman, N.; Vitu, E.; Bentzur, M.; Kaiser, C.A.; Thorpe, C.; Fass, D. Generating disulfides enzymatically: Reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. USA 2006, 103, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Tu, B.P.; Weissman, J.S. The FAD- and O2-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol. Cell 2002, 10, 983–994. [Google Scholar] [CrossRef]
- Tu, B.P.; Ho-Schleyer, S.C.; Travers, K.J.; Weissman, J.S. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science 2000, 290, 1571–1574. [Google Scholar] [CrossRef] [PubMed]
- Sevier, C.S.; Kaiser, C.A. Disulfide transfer between two conserved cysteine pairs imparts selectivity to protein oxidation by Ero1. Mol. Biol. Cell 2006, 17, 2256–2266. [Google Scholar] [CrossRef] [PubMed]
- Frand, A.R.; Kaiser, C.A. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol. Cell 1999, 4, 469–477. [Google Scholar] [CrossRef]
- Sevier, C.S.; Qu, H.; Heldman, N.; Gross, E.; Fass, D.; Kaiser, C.A. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell 2007, 129, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Ramming, T.; Okumura, M.; Kanemura, S.; Baday, S.; Birk, J.; Moes, S.; Spiess, M.; Jeno, P.; Berneche, S.; Inaba, K.; et al. A PDI-catalyzed thiol-disulfide switch regulates the production of hydrogen peroxide by human Ero1. Free Radic. Biol. Med. 2015, 83, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, G.; Simmen, T.; Anelli, T.; Molteni, S.N.; Fesce, R.; Sitia, R. Two conserved cysteine triads in human Ero1α cooperate for efficient disulfide bond formation in the endoplasmic reticulum. J. Biol. Chem. 2004, 279, 30047–30052. [Google Scholar] [CrossRef] [PubMed]
- Hansen, H.G.; Soltoft, C.L.; Schmidt, J.D.; Birk, J.; Appenzeller-Herzog, C.; Ellgaard, L. Biochemical evidence that regulation of Ero1β activity in human cells does not involve the isoform-specific cysteine 262. Biosci. Rep. 2014, 34, e00103. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, C.; Oka, O.B.; Bulleid, N.J. Inactivation of mammalian Ero1α is catalysed by specific protein disulfide-isomerases. Biochem. J. 2014, 461, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Nagata, K. Functional in vitro analysis of the ERO1 protein and protein-disulfide isomerase pathway. J. Biol. Chem. 2011, 286, 32705–32712. [Google Scholar] [CrossRef] [PubMed]
- Mezghrani, A.; Fassio, A.; Benham, A.; Simmen, T.; Braakman, I.; Sitia, R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001, 20, 6288–6296. [Google Scholar] [CrossRef] [PubMed]
- Appenzeller-Herzog, C.; Riemer, J.; Zito, E.; Chin, K.T.; Ron, D.; Spiess, M.; Ellgaard, L. Disulphide production by Ero1α-PDI relay is rapid and effectively regulated. EMBO J. 2010, 29, 3318–3329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araki, K.; Iemura, S.; Kamiya, Y.; Ron, D.; Kato, K.; Natsume, T.; Nagata, K. Ero1-α and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J. Cell Biol. 2013, 202, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Anelli, T.; Alessio, M.; Bachi, A.; Bergamelli, L.; Bertoli, G.; Camerini, S.; Mezghrani, A.; Ruffato, E.; Simmen, T.; Sitia, R. Thiol-mediated protein retention in the endoplasmic reticulum: The role of ERp44. EMBO J. 2003, 22, 5015–5022. [Google Scholar] [CrossRef] [PubMed]
- Kakihana, T.; Araki, K.; Vavassori, S.; Iemura, S.; Cortini, M.; Fagioli, C.; Natsume, T.; Sitia, R.; Nagata, K. Dynamic regulation of Ero1α and peroxiredoxin 4 localization in the secretory pathway. J. Biol. Chem. 2013, 288, 29586–29594. [Google Scholar] [CrossRef] [PubMed]
- Cuozzo, J.W.; Kaiser, C.A. Competition between glutathione and protein thiols for disulphide-bond formation. Nat. Cell Biol. 1999, 1, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Molteni, S.N.; Fassio, A.; Ciriolo, M.R.; Filomeni, G.; Pasqualetto, E.; Fagioli, C.; Sitia, R. Glutathione limits Ero1-dependent oxidation in the endoplasmic reticulum. J. Biol. Chem. 2004, 279, 32667–32673. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.M.; Chakravarthi, S.; Langton, K.P.; Sheppard, A.M.; Lu, H.; Bulleid, N.J. Low reduction potential of Ero1α regulatory disulphides ensures tight control of substrate oxidation. EMBO J. 2008, 27, 2988–2997. [Google Scholar] [CrossRef] [PubMed]
- Ponsero, A.J.; Igbaria, A.; Darch, M.A.; Miled, S.; Outten, C.E.; Winther, J.R.; Palais, G.; D’Autreaux, B.; Delaunay-Moisan, A.; Toledano, M.B. Endoplasmic Reticulum Transport of Glutathione by Sec61 Is Regulated by Ero1 and Bip. Mol. Cell 2017, 67, 67. [Google Scholar] [CrossRef] [PubMed]
- Birk, J.; Meyer, M.; Aller, I.; Hansen, H.G.; Odermatt, A.; Dick, T.P.; Meyer, A.J.; Appenzeller-Herzog, C. Endoplasmic reticulum: Reduced and oxidized glutathione revisited. J. Cell Sci. 2013, 126, 1604–1617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakravarthi, S.; Bulleid, N.J. Glutathione is required to regulate the formation of native disulfide bonds within proteins entering the secretory pathway. J. Biol. Chem. 2004, 279, 39872–39879. [Google Scholar] [CrossRef] [PubMed]
- Gess, B.; Hofbauer, K.H.; Wenger, R.H.; Lohaus, C.; Meyer, H.E.; Kurtz, A. The cellular oxygen tension regulates expression of the endoplasmic oxidoreductase ERO1-Lα. Eur. J. Biochem. 2003, 270, 2228–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marciniak, S.J.; Yun, C.Y.; Oyadomari, S.; Novoa, I.; Zhang, Y.; Jungreis, R.; Nagata, K.; Harding, H.P.; Ron, D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004, 18, 3066–3077. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Mongillo, M.; Chin, K.T.; Harding, H.; Ron, D.; Marks, A.R.; Tabas, I. Role of ERO1-α-mediated stimulation of inositol 1,4,5-triphosphate receptor activity in endoplasmic reticulum stress-induced apoptosis. J. Cell Biol. 2009, 186, 783–792. [Google Scholar] [CrossRef] [PubMed]
- May, D.; Itin, A.; Gal, O.; Kalinski, H.; Feinstein, E.; Keshet, E. Ero1-Lα plays a key role in a HIF-1-mediated pathway to improve disulfide bond formation and VEGF secretion under hypoxia: Implication for cancer. Oncogene 2005, 24, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Takei, N.; Yoneda, A.; Sakai-Sawada, K.; Kosaka, M.; Minomi, K.; Tamura, Y. Hypoxia-inducible ERO1α promotes cancer progression through modulation of integrin-β1 modification and signalling in HCT116 colorectal cancer cells. Sci. Rep. 2017, 7, 9389. [Google Scholar] [CrossRef] [PubMed]
- Townsend, D.M.; Manevich, Y.; He, L.; Xiong, Y.; Bowers, R.R., Jr.; Hutchens, S.; Tew, K.D. Nitrosative stress-induced S-glutathionylation of protein disulfide isomerase leads to activation of the unfolded protein response. Cancer Res. 2009, 69, 7626–7634. [Google Scholar] [CrossRef] [PubMed]
- Tavender, T.J.; Bulleid, N.J. Peroxiredoxin IV protects cells from oxidative stress by removing H2O2 produced during disulphide formation. J. Cell Sci. 2010, 123, 2672–2679. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, L.; Niu, Y.; Sitia, R.; Wang, C.C. Glutathione peroxidase 7 utilizes hydrogen peroxide generated by Ero1α to promote oxidative protein folding. Antioxid. Redox Signal. 2014, 20, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Ramming, T.; Hansen, H.G.; Nagata, K.; Ellgaard, L.; Appenzeller-Herzog, C. GPx8 peroxidase prevents leakage of H2O2 from the endoplasmic reticulum. Free Radic. Biol. Med. 2014, 70, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Enyedi, B.; Varnai, P.; Geiszt, M. Redox state of the endoplasmic reticulum is controlled by Ero1L-alpha and intraluminal calcium. Antioxid. Redox Signal. 2010, 13, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.D.; Saaranen, M.J.; Karala, A.R.; Lappi, A.K.; Wang, L.; Raykhel, I.B.; Alanen, H.I.; Salo, K.E.; Wang, C.C.; Ruddock, L.W. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J. Mol. Biol. 2011, 406, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Tavender, T.J.; Springate, J.J.; Bulleid, N.J. Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 2010, 29, 4185–4197. [Google Scholar] [CrossRef] [PubMed]
- Konno, T.; Pinho Melo, E.; Lopes, C.; Mehmeti, I.; Lenzen, S.; Ron, D.; Avezov, E. ERO1-independent production of H2O2 within the endoplasmic reticulum fuels Prdx4-mediated oxidative protein folding. J. Cell Biol. 2015, 211, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Zito, E.; Melo, E.P.; Yang, Y.; Wahlander, A.; Neubert, T.A.; Ron, D. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Mol. Cell 2010, 40, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Bagur, R.; Hajnoczky, G. Intracellular Ca2+ Sensing: Its Role in Calcium Homeostasis and Signaling. Mol. Cell 2017, 66, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Baba-Aissa, F.; Raeymaekers, L.; Wuytack, F.; Dode, L.; Casteels, R. Distribution and isoform diversity of the organellar Ca2+ pumps in the brain. Mol. Chem. Neuropathol. 1998, 33, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Adachi, T.; Weisbrod, R.M.; Pimentel, D.R.; Ying, J.; Sharov, V.S.; Schoneich, C.; Cohen, R.A. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat. Med. 2004, 10, 1200–1207. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Thompson, M.D.; Shiraishi, Y.; Cohen, R.A.; Tong, X. Sarcoplasmic/endoplasmic reticulum Ca2+ ATPase C674 promotes ischemia- and hypoxia-induced angiogenesis via coordinated endothelial cell and macrophage function. J. Mol. Cell. Cardiol. 2014, 76, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.P.; Han, S.; Li, G.; Tabas, I.; Tall, A.R. Impaired MEK signaling and SERCA expression promote ER stress and apoptosis in insulin-resistant macrophages and are reversed by exenatide treatment. Diabetes 2012, 61, 2609–2620. [Google Scholar] [CrossRef] [PubMed]
- Marino, M.; Stoilova, T.; Giorgi, C.; Bachi, A.; Cattaneo, A.; Auricchio, A.; Pinton, P.; Zito, E. SEPN1, an endoplasmic reticulum-localized selenoprotein linked to skeletal muscle pathology, counteracts hyperoxidation by means of redox-regulating SERCA2 pump activity. Hum. Mol. Genet. 2015, 24, 1843–1855. [Google Scholar] [CrossRef] [PubMed]
- Szabadkai, G.; Bianchi, K.; Varnai, P.; De Stefani, D.; Wieckowski, M.R.; Cavagna, D.; Nagy, A.I.; Balla, T.; Rizzuto, R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 2006, 175, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Shoshan-Barmatz, V.; Ben-Hail, D.; Admoni, L.; Krelin, Y.; Tripathi, S.S. The mitochondrial voltage-dependent anion channel 1 in tumor cells. Biochim. Biophys. Acta 2015, 1848, 2547–2575. [Google Scholar] [CrossRef] [PubMed]
- Csordas, G.; Golenar, T.; Seifert, E.L.; Kamer, K.J.; Sancak, Y.; Perocchi, F.; Moffat, C.; Weaver, D.; de la Fuente Perez, S.; Bogorad, R.; et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca2+ uniporter. Cell Metab. 2013, 17, 976–987. [Google Scholar] [CrossRef] [PubMed]
- Csordas, G.; Varnai, P.; Golenar, T.; Roy, S.; Purkins, G.; Schneider, T.G.; Balla, T.; Hajnoczky, G. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell 2010, 39, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Gilady, S.Y.; Bui, M.; Lynes, E.M.; Benson, M.D.; Watts, R.; Vance, J.E.; Simmen, T. Ero1α requires oxidizing and normoxic conditions to localize to the mitochondria-associated membrane (MAM). Cell Stress Chaperones 2010, 15, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Anelli, T.; Bergamelli, L.; Margittai, E.; Rimessi, A.; Fagioli, C.; Malgaroli, A.; Pinton, P.; Ripamonti, M.; Rizzuto, R.; Sitia, R. Ero1α regulates Ca2+ fluxes at the endoplasmic reticulum-mitochondria interface (MAM). Antioxid. Redox Signal. 2012, 16, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Higo, T.; Hattori, M.; Nakamura, T.; Natsume, T.; Michikawa, T.; Mikoshiba, K. Subtype-specific and ER lumenal environment-dependent regulation of inositol 1,4,5-trisphosphate receptor type 1 by ERp44. Cell 2005, 120, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Camacho, P. Ca2+-dependent redox modulation of SERCA 2b by ERp57. J. Cell Biol. 2004, 164, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Kutomi, G.; Tamura, Y.; Tanaka, T.; Kajiwara, T.; Kukita, K.; Ohmura, T.; Shima, H.; Takamaru, T.; Satomi, F.; Suzuki, Y.; et al. Human endoplasmic reticulum oxidoreductin 1-α is a novel predictor for poor prognosis of breast cancer. Cancer Sci. 2013, 104, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, G.; Gao, S.; Chen, Y.; Jin, C.; Wang, Z.; Yang, Y.; Ma, Z.; Zhang, W.; Feng, X. Expression of ERO1L in gastric cancer and its association with patient prognosis. Exp. Ther. Med. 2017, 14, 2298–2302. [Google Scholar] [CrossRef] [PubMed]
- Battle, D.M.; Gunasekara, S.D.; Watson, G.R.; Ahmed, E.M.; Saysell, C.G.; Altaf, N.; Sanusi, A.L.; Munipalle, P.C.; Scoones, D.; Walker, J.; et al. Expression of the endoplasmic reticulum oxidoreductase Ero1α in gastro-intestinal cancer reveals a link between homocysteine and oxidative protein folding. Antioxid. Redox Signal. 2013, 19, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Kukita, K.; Tamura, Y.; Tanaka, T.; Kajiwara, T.; Kutomi, G.; Saito, K.; Okuya, K.; Takaya, A.; Kanaseki, T.; Tsukahara, T.; et al. Cancer-Associated Oxidase ERO1-α Regulates the Expression of MHC Class I Molecule via Oxidative Folding. J. Immunol. 2015, 194, 4988–4996. [Google Scholar] [CrossRef] [PubMed]
- Seol, S.Y.; Kim, C.; Lim, J.Y.; Yoon, S.O.; Hong, S.W.; Kim, J.W.; Choi, S.H.; Cho, J.Y. Overexpression of Endoplasmic Reticulum Oxidoreductin 1-α (ERO1L) Is Associated with Poor Prognosis of Gastric Cancer. Cancer Res. Treat. 2016, 48, 1196–1209. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kutomi, G.; Kajiwara, T.; Kukita, K.; Kochin, V.; Kanaseki, T.; Tsukahara, T.; Hirohashi, Y.; Torigoe, T.; Okamoto, Y.; et al. Cancer-associated oxidoreductase ERO1-α drives the production of VEGF via oxidative protein folding and regulating the mRNA level. Br. J. Cancer 2016, 114, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kutomi, G.; Kajiwara, T.; Kukita, K.; Kochin, V.; Kanaseki, T.; Tsukahara, T.; Hirohashi, Y.; Torigoe, T.; Okamoto, Y.; et al. Cancer-associated oxidoreductase ERO1-α promotes immune escape through up-regulation of PD-L1 in human breast cancer. Oncotarget 2017, 8, 24706–24718. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Ngiow, S.F.; Ribas, A.; Teng, M.W. Combination cancer immunotherapies tailored to the tumour microenvironment. Nat. Rev. Clin. Oncol. 2016, 13, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Black, M.; Barsoum, I.B.; Truesdell, P.; Cotechini, T.; Macdonald-Goodfellow, S.K.; Petroff, M.; Siemens, D.R.; Koti, M.; Craig, A.W.; Graham, C.H. Activation of the PD-1/PD-L1 immune checkpoint confers tumor cell chemoresistance associated with increased metastasis. Oncotarget 2016, 7, 10557–10567. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kajiwara, T.; Torigoe, T.; Okamoto, Y.; Sato, N.; Tamura, Y. Cancer-associated oxidoreductase ERO1-α drives the production of tumor-promoting myeloid-derived suppressor cells via oxidative protein folding. J. Immunol. 2015, 194, 2004–2010. [Google Scholar] [CrossRef] [PubMed]
- Talmadge, J.E.; Gabrilovich, D.I. History of myeloid-derived suppressor cells. Nat. Rev. Cancer 2013, 13, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Leone, P.; Shin, E.C.; Perosa, F.; Vacca, A.; Dammacco, F.; Racanelli, V. MHC class I antigen processing and presenting machinery: Organization, function, and defects in tumor cells. J. Natl. Cancer Inst. 2013, 105, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- Kajiwara, T.; Tanaka, T.; Kukita, K.; Kutomi, G.; Saito, K.; Okuya, K.; Takaya, A.; Kochin, V.; Kanaseki, T.; Tsukahara, T.; et al. Hypoxia augments MHC class I antigen presentation via facilitation of ERO1-α-mediated oxidative folding in murine tumor cells. Eur. J. Immunol. 2016, 46, 2842–2851. [Google Scholar] [CrossRef] [PubMed]
- Ching, R.H.H.; Sze, K.M.F.; Lau, E.Y.T.; Chiu, Y.T.; Lee, J.M.F.; Ng, I.O.L.; Lee, T.K.W. C-terminal truncated hepatitis B virus X protein regulates tumorigenicity, self-renewal and drug resistance via STAT3/Nanog signaling pathway. Oncotarget 2017, 8, 23507–23516. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Waris, G.; Tanveer, R.; Siddiqui, A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-κB. Proc. Natl. Acad. Sci. USA 2001, 98, 9599–9604. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Smirnova, O.A.; Ivanova, O.N.; Masalova, O.V.; Kochetkov, S.N.; Isaguliants, M.G. Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells. PLoS ONE 2011, 6, e24957. [Google Scholar] [CrossRef] [PubMed]
- Okuda, M.; Li, K.; Beard, M.R.; Showalter, L.A.; Scholle, F.; Lemon, S.M.; Weinman, S.A. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology 2002, 122, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Boehning, D.F.; Qian, T.; Popov, V.L.; Weinman, S.A. Hepatitis C virus core protein increases mitochondrial ROS production by stimulation of Ca2+ uniporter activity. FASEB J. 2007, 21, 2474–2485. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.M.; Kim, S.J.; Kim, J.H.; Lee, W.; Kim, G.W.; Lee, K.H.; Choi, K.Y.; Oh, J.W. Interaction of hepatitis C virus core protein with Hsp60 triggers the production of reactive oxygen species and enhances TNF-α-mediated apoptosis. Cancer Lett. 2009, 279, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Benali-Furet, N.L.; Chami, M.; Houel, L.; De Giorgi, F.; Vernejoul, F.; Lagorce, D.; Buscail, L.; Bartenschlager, R.; Ichas, F.; Rizzuto, R.; et al. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 2005, 24, 4921–4933. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Smirnova, O.A.; Petrushanko, I.Y.; Ivanova, O.N.; Karpenko, I.L.; Alekseeva, E.; Sominskaya, I.; Makarov, A.A.; Bartosch, B.; Kochetkov, S.N.; et al. HCV core protein uses multiple mechanisms to induce oxidative stress in human hepatoma Huh7 cells. Viruses 2015, 7, 2745–2770. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, O.A.; Ivanova, O.N.; Bartosch, B.; Valuev-Elliston, V.T.; Mukhtarov, F.; Kochetkov, S.N.; Ivanov, A.V. Hepatitis C Virus NS5A Protein Triggers Oxidative Stress by Inducing NADPH Oxidases 1 and 4 and Cytochrome P450 2E1. Oxid. Med. Cell. Longev. 2016, 2016, 8341937. [Google Scholar] [CrossRef] [PubMed]
- Joubert, P.E.; Werneke, S.W.; de la Calle, C.; Guivel-Benhassine, F.; Giodini, A.; Peduto, L.; Levine, B.; Schwartz, O.; Lenschow, D.J.; Albert, M.L. Chikungunya virus-induced autophagy delays caspase-dependent cell death. J. Exp. Med. 2012, 209, 1029–1047. [Google Scholar] [CrossRef] [PubMed]
- Langsjoen, R.M.; Auguste, A.J.; Rossi, S.L.; Roundy, C.M.; Penate, H.N.; Kastis, M.; Schnizlein, M.K.; Le, K.C.; Haller, S.L.; Chen, R.; et al. Host oxidative folding pathways offer novel anti-chikungunya virus drug targets with broad spectrum potential. Antivir. Res. 2017, 143, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Nishikiori, M.; Ahlquist, P. Organelle luminal dependence of (+)strand RNA virus replication reveals a hidden druggable target. Sci. Adv. 2018, 4, eaap8258. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, R.C.; Jordan Steel, J.; Moon, S.L.; Soltani, E.; Geiss, B.J. Oxidative stress influences positive strand RNA virus genome synthesis and capping. Virology 2015, 475, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Hyodo, K.; Hashimoto, K.; Kuchitsu, K.; Suzuki, N.; Okuno, T. Harnessing host ROS-generating machinery for the robust genome replication of a plant RNA virus. Proc. Natl. Acad. Sci. USA 2017, 114, E1282–E1290. [Google Scholar] [CrossRef] [PubMed]
- Parkkinen, J.J.; Lammi, M.J.; Agren, U.; Tammi, M.; Keinanen, T.A.; Hyvonen, T.; Eloranta, T.O. Polyamine-dependent alterations in the structure of microfilaments, Golgi apparatus, endoplasmic reticulum, and proteoglycan synthesis in BHK cells. J. Cell. Biochem. 1997, 66, 165–174. [Google Scholar] [CrossRef]
- Prunotto, M.; Compagnone, A.; Bruschi, M.; Candiano, G.; Colombatto, S.; Bandino, A.; Petretto, A.; Moll, S.; Bochaton-Piallat, M.L.; Gabbiani, G.; et al. Endocellular polyamine availability modulates epithelial-to-mesenchymal transition and unfolded protein response in MDCK cells. Lab. Investig. 2010, 90, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Landau, G.; Ran, A.; Bercovich, Z.; Feldmesser, E.; Horn-Saban, S.; Korkotian, E.; Jacob-Hirsh, J.; Rechavi, G.; Ron, D.; Kahana, C. Expression profiling and biochemical analysis suggest stress response as a potential mechanism inhibiting proliferation of polyamine-depleted cells. J. Biol. Chem. 2012, 287, 35825–35837. [Google Scholar] [CrossRef] [PubMed]
- Hyvonen, M.T.; Merentie, M.; Uimari, A.; Keinanen, T.A.; Janne, J.; Alhonen, L. Mechanisms of polyamine catabolism-induced acute pancreatitis. Biochem. Soc. Trans. 2007, 35, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Mandal, A.; Park, M.H. Depletion of the polyamines spermidine and spermine by overexpression of spermidine/spermine N1-acetyltransferase 1 (SAT1) leads to mitochondria-mediated apoptosis in mammalian cells. Biochem. J. 2015, 468, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Park, J.H.; Woster, P.M.; Casero, R.A., Jr.; Park, M.H. Suppression of exogenous gene expression by spermidine/spermine N1-acetyltransferase 1 (SSAT1) cotransfection. J. Biol. Chem. 2010, 285, 15548–15556. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Mandal, A.; Johansson, H.E.; Orjalo, A.V.; Park, M.H. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc. Natl. Acad. Sci. USA 2013, 110, 2169–2174. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, K.; Barone, S.; Destefano-Shields, C.; Brooks, M.; Murray-Stewart, T.; Dunworth, M.; Li, W.; Doherty, J.R.; Hall, M.A.; Smith, R.D.; et al. Activation of endoplasmic reticulum stress response by enhanced polyamine catabolism is important in the mediation of cisplatin-induced acute kidney injury. PLoS ONE 2017, 12, e0184570. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Galvao, F.C.; Bellato, H.M.; Boldrin, P.E.; Andrews, B.J.; Valentini, S.R.; Zanelli, C.F. eIF5A has a function in the cotranslational translocation of proteins into the ER. Amino Acids 2014, 46, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Mandal, S.; Park, M.H. Global quantitative proteomics reveal up-regulation of endoplasmic reticulum stress response proteins upon depletion of eIF5A in HeLa cells. Sci. Rep. 2016, 6, 25795. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, W.C.; Hsu, P.C.; Liao, Y.F.; Young, S.T.; Wang, Z.W.; Lin, C.L.; Tsay, G.J.; Lee, H.; Hung, H.C.; Liu, G.Y. Overexpression of ornithine decarboxylase suppresses thapsigargin-induced apoptosis. Mol. Cells 2010, 30, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Robbins, R.D.; Tersey, S.A.; Ogihara, T.; Gupta, D.; Farb, T.B.; Ficorilli, J.; Bokvist, K.; Maier, B.; Mirmira, R.G. Inhibition of deoxyhypusine synthase enhances islet β cell function and survival in the setting of endoplasmic reticulum stress and type 2 diabetes. J. Biol. Chem. 2010, 285, 39943–39952. [Google Scholar] [CrossRef] [PubMed]
- Korenaga, M.; Wang, T.; Li, Y.; Showalter, L.A.; Chan, T.; Sun, J.; Weinman, S.A. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J. Biol. Chem. 2005, 280, 37481–37488. [Google Scholar] [CrossRef] [PubMed]
- Mitsuya, Y.; Takahashi, Y.; Berberich, T.; Miyazaki, A.; Matsumura, H.; Takahashi, H.; Terauchi, R.; Kusano, T. Spermine signaling plays a significant role in the defense response of Arabidopsis thaliana to cucumber mosaic virus. J. Plant Physiol. 2009, 166, 626–643. [Google Scholar] [CrossRef] [PubMed]
- Sagor, G.H.; Chawla, P.; Kim, D.W.; Berberich, T.; Kojima, S.; Niitsu, M.; Kusano, T. The polyamine spermine induces the unfolded protein response via the MAPK cascade in Arabidopsis. Front. Plant Sci. 2015, 6, 687. [Google Scholar] [CrossRef] [PubMed]
- Iwata, Y.; Koizumi, N. An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 2005, 102, 5280–5285. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.P.; Christopher, D.A. Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol. Genet. Genom. 2008, 280, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Morada, M.; Pendyala, L.; Wu, G.; Merali, S.; Yarlett, N. Cryptosporidium parvum induces an endoplasmic stress response in the intestinal adenocarcinoma HCT-8 cell line. J. Biol. Chem. 2013, 288, 30356–30364. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smirnova, O.A.; Bartosch, B.; Zakirova, N.F.; Kochetkov, S.N.; Ivanov, A.V. Polyamine Metabolism and Oxidative Protein Folding in the ER as ROS-Producing Systems Neglected in Virology. Int. J. Mol. Sci. 2018, 19, 1219. https://doi.org/10.3390/ijms19041219
Smirnova OA, Bartosch B, Zakirova NF, Kochetkov SN, Ivanov AV. Polyamine Metabolism and Oxidative Protein Folding in the ER as ROS-Producing Systems Neglected in Virology. International Journal of Molecular Sciences. 2018; 19(4):1219. https://doi.org/10.3390/ijms19041219
Chicago/Turabian StyleSmirnova, Olga A., Birke Bartosch, Natalia F. Zakirova, Sergey N. Kochetkov, and Alexander V. Ivanov. 2018. "Polyamine Metabolism and Oxidative Protein Folding in the ER as ROS-Producing Systems Neglected in Virology" International Journal of Molecular Sciences 19, no. 4: 1219. https://doi.org/10.3390/ijms19041219





