gga-miR-451 Negatively Regulates Mycoplasma gallisepticum (HS Strain)-Induced Inflammatory Cytokine Production via Targeting YWHAZ
Abstract
:1. Introduction
2. Results
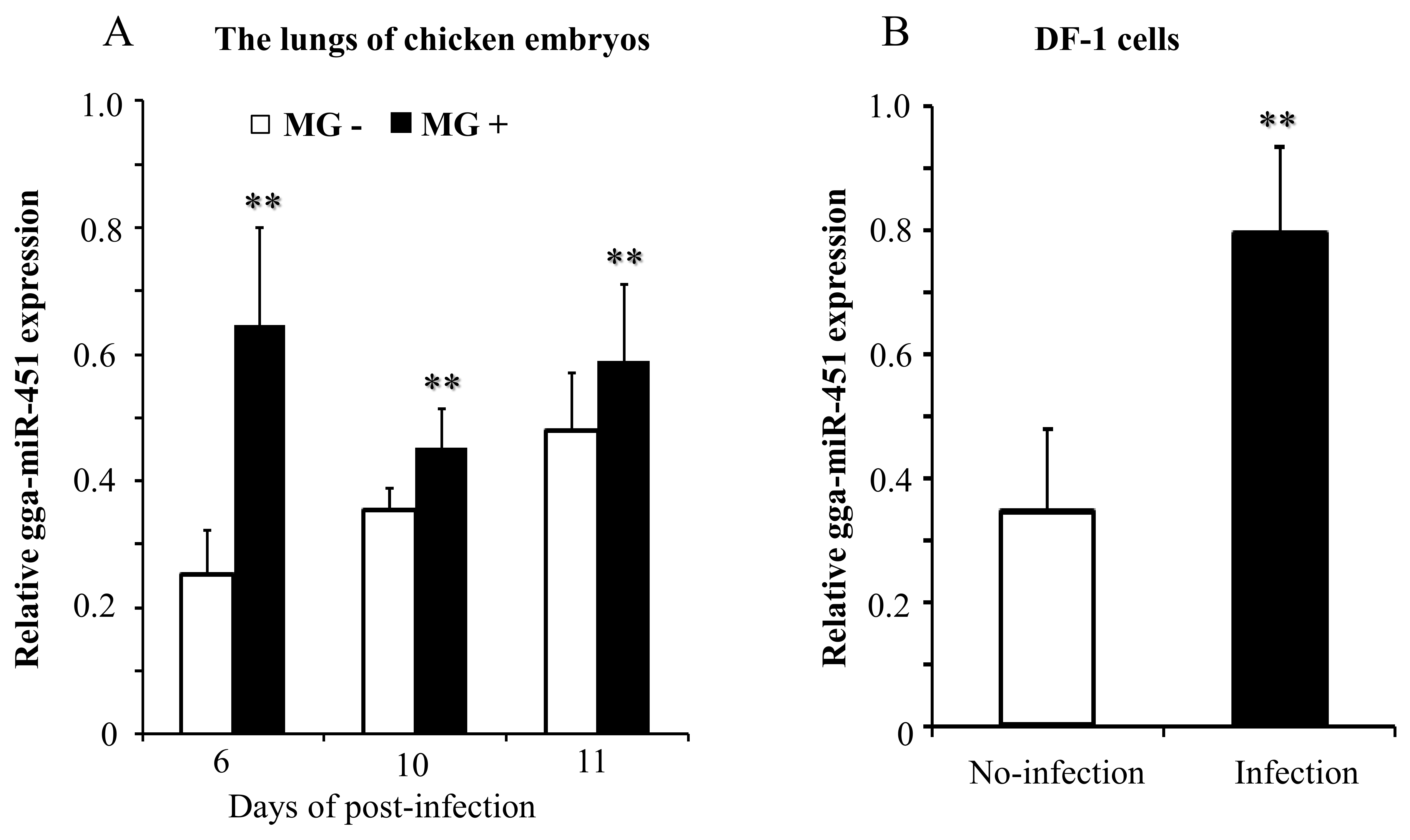
2.1. MG Infection Significantly Upregulates gga-miR-451 Expression
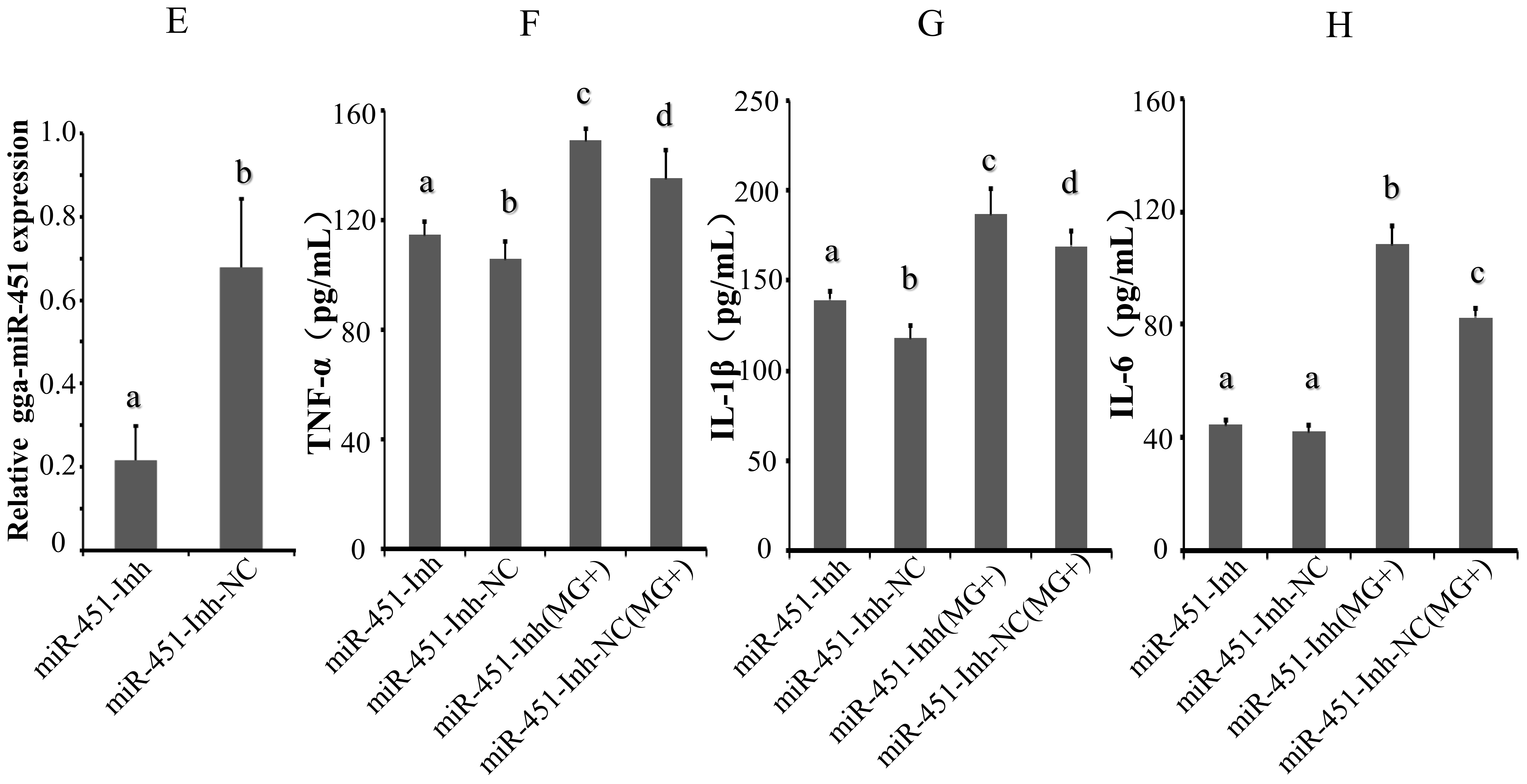
2.2. gga-miR-451 Reduces MG–Mediated Inflammatory Cytokine Production
2.3. YWHAZ Is a Direct Target of gga-miR-451
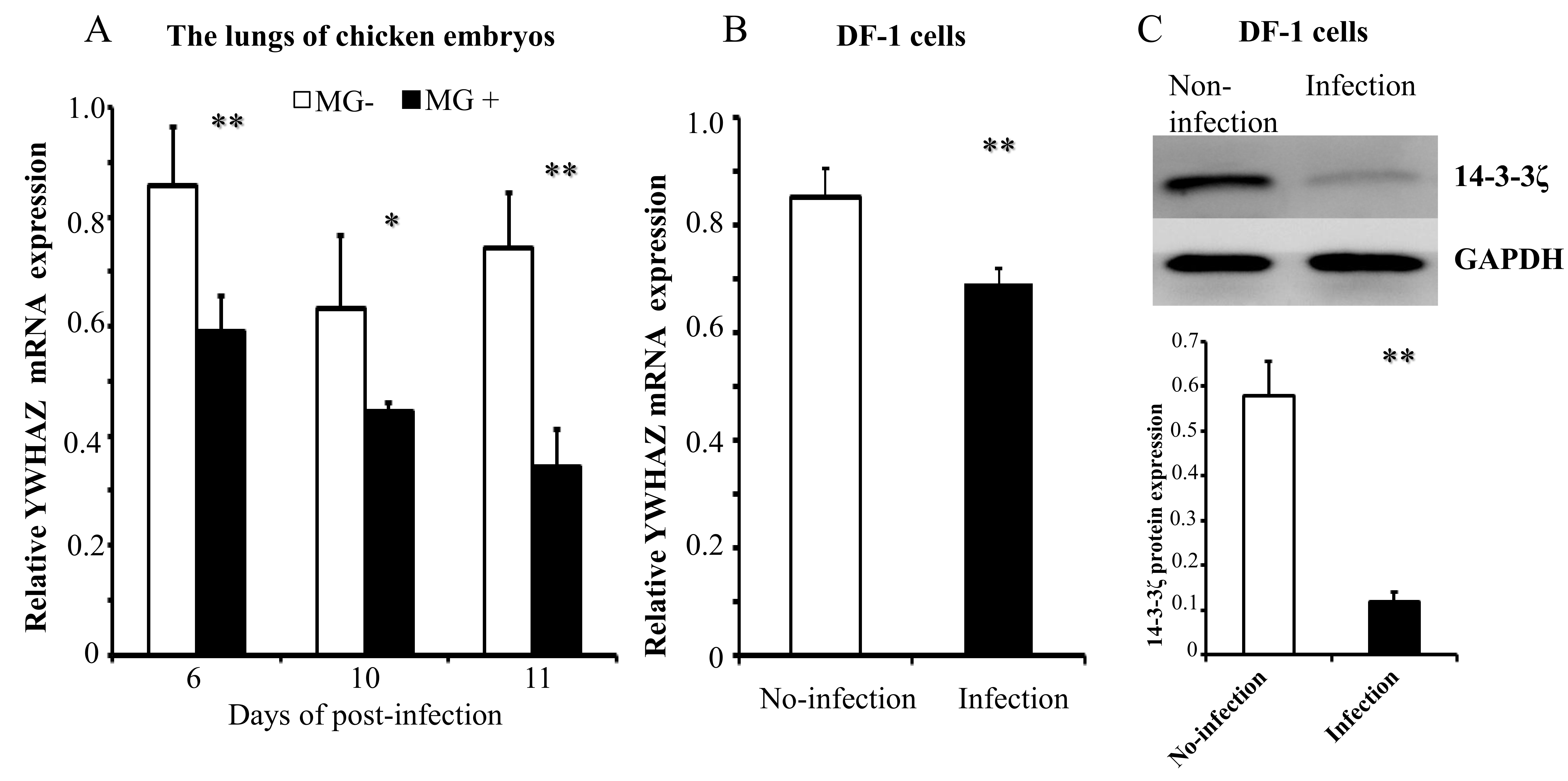
2.4. gga-miR-451 Negatively Regulates YWHAZ Expression
2.5. MG Infection Downregulates YWHAZ Expression
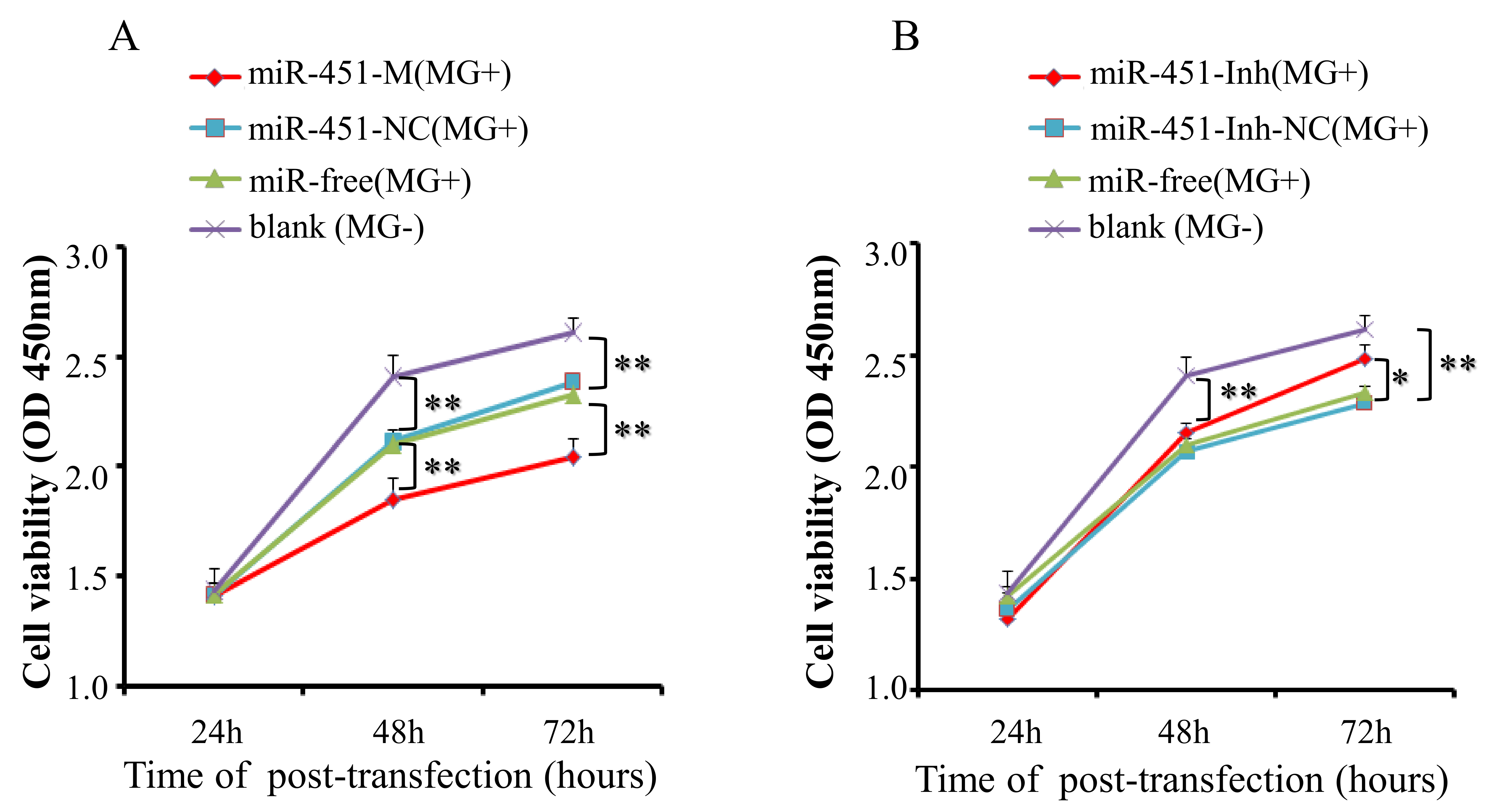
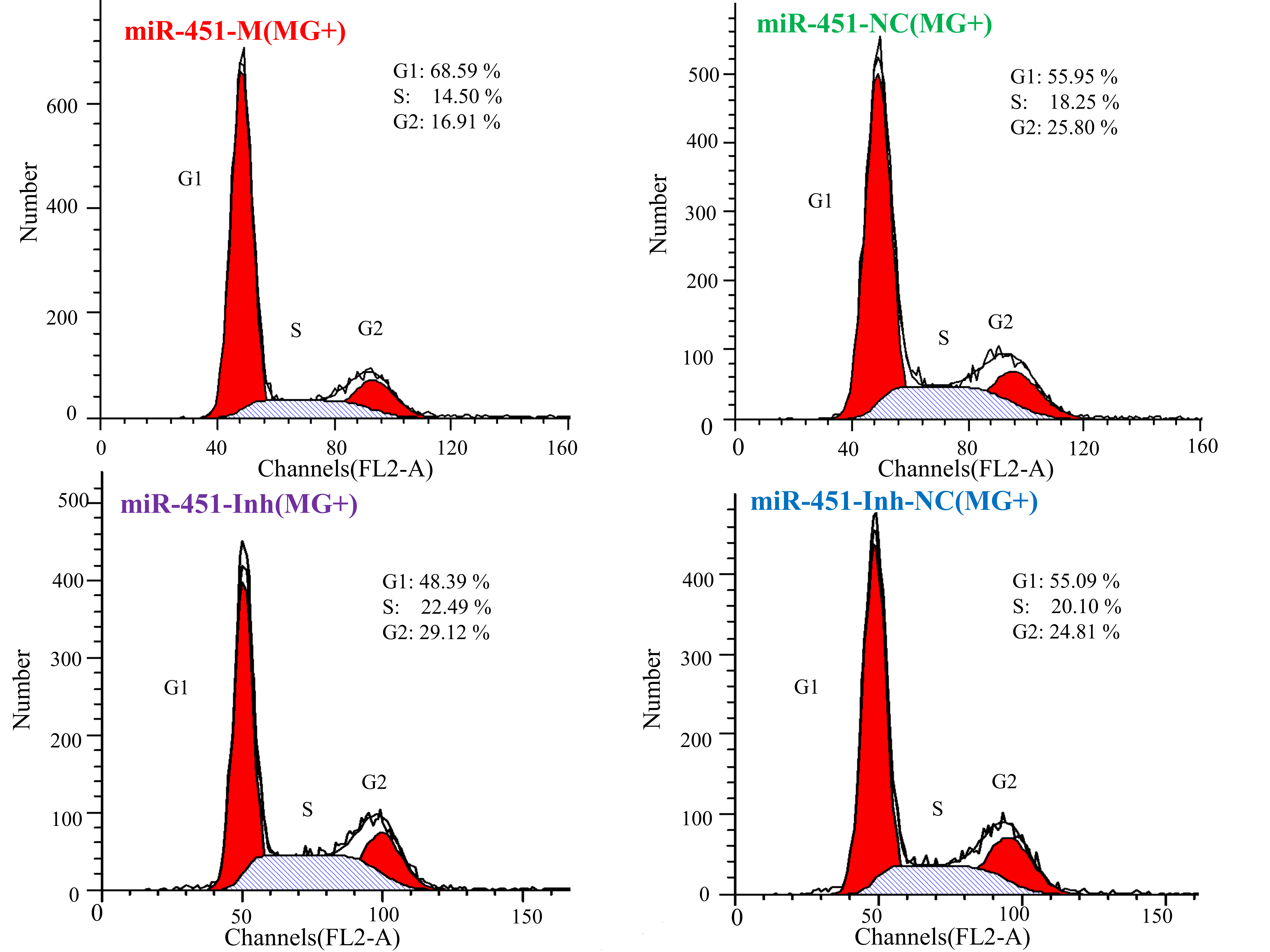
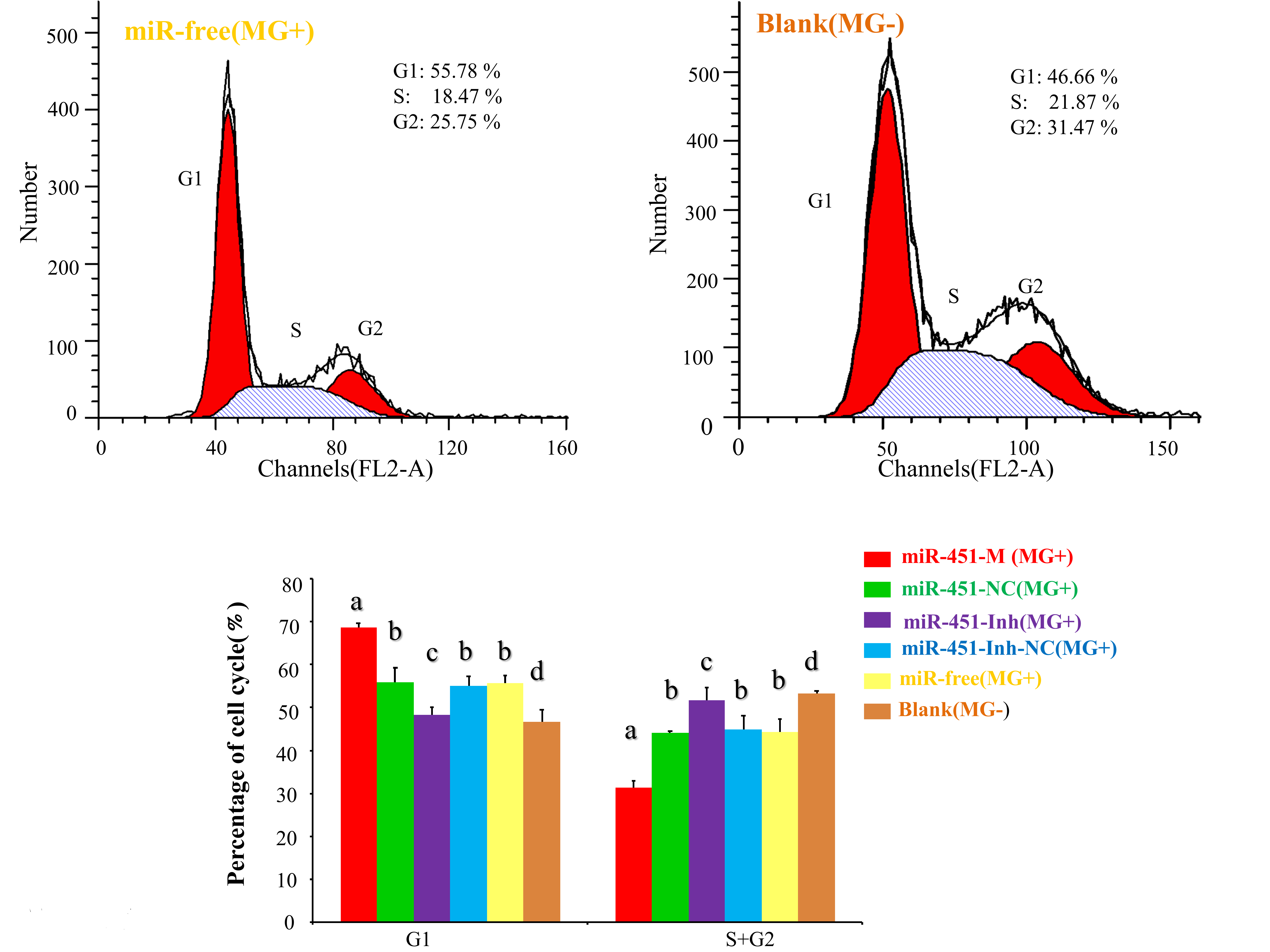
2.6. gga-miR-451 Inhibits Cell Proliferation by Affecting the Cell Cycle and Cell Apoptosis
3. Discussion
4. Materials and Methods
4.1. MG-HS Culture
4.2. Infection Experiments
4.3. Cell Culture and Treatment
4.4. microRNA Target Prediction and Sequences
4.5. Constructs and Plasmids
4.6. Dual-Luciferase Reporter Assay
4.7. RNA Extraction and Quantitative Real-Time (RT-qPCR)
4.8. Western Blot
4.9. Electrophoretic Immunosorbent Assay (ELISA)
4.10. Cell Proliferation, Cell Cycle and Cell Apoptosis
4.11. Statistical Analysis
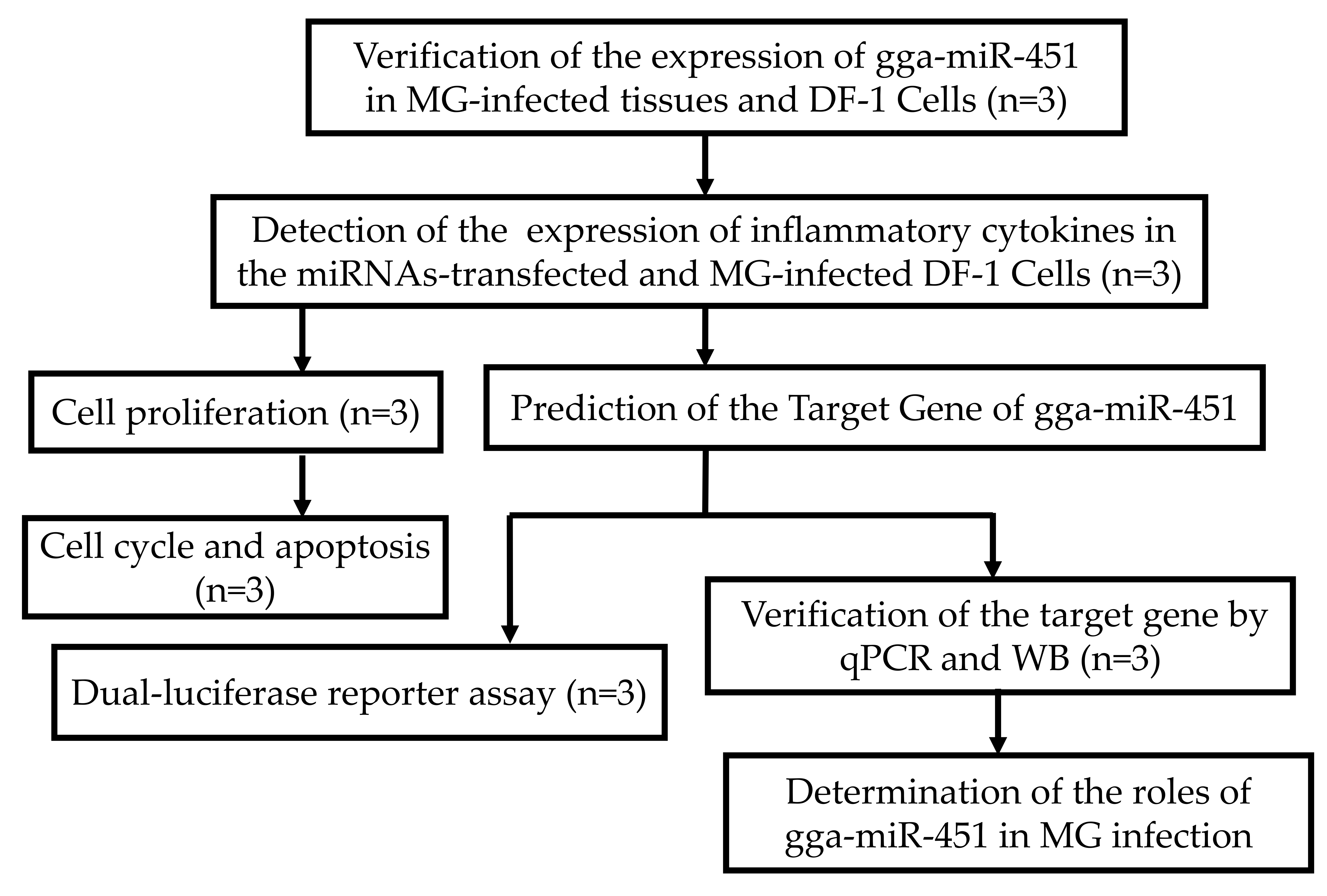
4.12. The Experimental Design
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Himmel, M.E.; Hardenberg, G.; Piccirillo, C.A.; Steiner, T.S.; Levings, M.K. The role of T-regulatory cells and toll-like receptors in the pathogenesis of human inflammatory bowel disease. Immunology 2008, 125, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Ye, X.; Wu, Y.; Huang, Z.; Gu, X.; Cai, Q.; Shen, X.; Jiang, H.; Ding, H. Determination of the mutant selection window and evaluation of the killing of Mycoplasma gallisepticum by danofloxacin, doxycycline, tilmicosin, tylvalosin and valnemulin. PLoS ONE 2017, 12, e0169134. [Google Scholar] [CrossRef] [PubMed]
- Stipkovits, L.; Egyed, L.; Palfi, V.; Beres, A.; Pitlik, E.; Somogyi, M.; Szathmary, S.; Denes, B. Effect of low-pathogenicity influenza virus H3N8 infection on Mycoplasma gallisepticum infection of chickens. Avian Pathol. 2012, 41, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Zohaib, A.; Li, Y.; Zhu, B.; Ye, J.; Wan, S.; Xu, Q.; Song, Y.; Chen, H.; Cao, S. MiR-206 modulates lipopolysaccharide-mediated inflammatory cytokine production in human astrocytes. Cell. Signal. 2015, 27, 61. [Google Scholar] [CrossRef] [PubMed]
- Sarfaraz, S.; Muhammad, K.; Yaqub, T.; Aslam, A.; Rabbani, M.; Khalil, M.; Riaz, R. Antibody response of broilers to oil based combined avian influenza (H-9 N-2) and Mycoplasma gallisepticum vaccine. J. Anim. Plant Sci. 2017, 27, 1150–1154. [Google Scholar]
- Ricketts, C.; Pickler, L.; Maurer, J.; Ayyampalayam, S.; García, M.; Fergusonnoel, N.M. Identification of strain-specific sequences that distinguish a Mycoplasma gallisepticum vaccine strain from field isolates. J. Clin. Microbiol. 2017, 55, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Vogl, G.; Plaickner, A.; Szathmary, S.; Stipkovits, L.; Rosengarten, R.; Szostak, M.P. Mycoplasma gallisepticum invades chicken erythrocytes during infection. Infect. Immun. 2008, 76, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Furnkranz, U.; Siebert-Gulle, K.; Rosengarten, R.; Szostak, M.P. Factors influencing the cell adhesion and invasion capacity of Mycoplasma gallisepticum. Acta Vet. Scand. 2013, 55, 63. [Google Scholar] [CrossRef] [PubMed]
- Winner, F.; Rosengarten, R.; Citti, C. In vitro cell invasion of Mycoplasma gallisepticum. Infect. Immun. 2000, 68, 4238–4244. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Zhao, C.; Hu, Q.; Sun, J.; Peng, X. Roles of toll-like receptors 2 and 6 in the inflammatory response to Mycoplasma gallisepticum infection in df-1 cells and in chicken embryos. Dev. Comp. Immunol. 2016, 59, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Ji, X. The isolation and identification of the Mycoplasma gallisepticum. Acta Vet. Zootechnol. Sin. 1988, 1, 146–148. [Google Scholar]
- Bi, D.; Xu, Q. Study on pathogenicity of HS strain Mycoplasma gallisepticum. Chin. J. Anim. Poult. Infec. Dis. 1997, 5, 24–26. [Google Scholar]
- Aw, S.S.; Tang, M.X.; Teo, Y.N.; Cohen, S.M. A conformation-induced fluorescence method for microrna detection. Nucleic Acids Res. 2016, 44, e92. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, S.; Ebert, M.S.; Zheng, G.X.; Tsang, J.S.; Sharp, P.A.; Van, O.A. MicroRNAs can generate thresholds in target gene expression. Nat. Genet. 2011, 43, 854–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, M.; Slack, F.J. MicroRNAs: Small molecules with big roles—C. elegans to human cancer. Biol. Cell 2008, 100, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Indiková, I.; Much, P.; Stipkovits, L.; Siebert-Gulle, K.; Szostak, M.P.; Rosengarten, R.; Citti, C. Role of the GapA and CrmA cytadhesins of Mycoplasma gallisepticum in promoting virulence and host colonization. Infec. Immun. 2013, 81, 1618–1624. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.; Geary, S.J.; Gladd, M.; Djordjevic, S.P. The Mycoplasma gallisepticum OsmC-like protein MG1142 resides on the cell surface and binds heparin. Microbiology 2007, 153, 1455–1463. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Huang, X.; Zhang, X.; Roensch, K.; Cao, Q.; Nakayama, K.I.; Blazar, B.R.; Zeng, Y.; Zhou, X. Mir-221 and mir-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood 2011, 117, 4293–4303. [Google Scholar] [CrossRef] [PubMed]
- West, C.; Mcdermott, M. Effects of microrna-146a on the proliferation and apoptosis of human osteochondrocytes by targeting traf6 through the NF-κB signalling pathway. Biosci. Rep. 2017, 37, BSR20170180. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, J.; Wang, H.; Shi, J.; Wu, K.; Liu, S.; Liu, Y.; Wu, J. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 2013, 9, e1003248. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Zhao, Y.; Wang, Z.; Hou, Y.; Bi, D.; Sun, J.; Peng, X. Chicken gga-miR-19a targets ZMYND11 and plays an important role in host defense against Mycoplasma gallisepticum (HS strain) infection. Front. Cell. Infect. Microbiol. 2016, 6, 102. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Z.; Bi, D.; Hou, Y.; Zhao, Y.; Sun, J.; Peng, X. Gga-miR-101-3p plays a key role in Mycoplasma gallisepticum (HS strain) infection of chicken. Int. J. Mol. Sci. 2015, 16, 28669–28682. [Google Scholar] [CrossRef] [PubMed]
- Rosenberger, C.M.; Podyminogin, R.L.; Navarro, G.; Zhao, G.W.; Askovich, P.S.; Weiss, M.J.; Aderem, A. MiR-451 regulates dendritic cell cytokine responses to influenza infection. J. Immunol. 2012, 189, 5965–5975. [Google Scholar] [CrossRef] [PubMed]
- Bergamaschi, A.; Katzenellenbogen, B.S. Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes breast cancer cell survival and endocrine resistance. Oncogene 2012, 31, 39. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; dos Santos, C.O.; Zhao, G.; Jiang, J.; Amigo, J.D.; Khandros, E.; Dore, L.C.; Yao, Y.; D’Souza, J.; Zhang, Z. MiR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010, 24, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Luo, X.; Ding, S.; Chen, J.; Chen, T.; Chen, X.; Zha, H.; Yao, L.; He, X.; Peng, H. MicroRNA-451 regulates p38 MAPK signaling by targeting of YWHAZ and suppresses the mesangial hypertrophy in early diabetic nephropathy. FEBS Lett. 2012, 586, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, L.; Wang, Y.; Ding, Y.; Chen, T.; Wang, Y.; Wang, H.; Li, Y.; Duan, K.; Chen, S. Involvement of miR-451 in resistance to paclitaxel by regulating YWHAZ in breast cancer. Cell Death Dis. 2017, 8, e3071. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hou, Y.; Zhang, K.; Yuan, B.; Peng, X. Identification of differentially expressed miRNAs through high-throughput sequencing in the chicken lung in response to Mycoplasma gallisepticum HS. Comp. Biochem. Physiol. Part D 2017, 22, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Dejean, A.S.; Beisner, D.R.; Ch’En, I.L.; Kerdiles, Y.M.; Babour, A.; Arden, K.C.; Castrillon, D.H.; Depinho, R.A.; Hedrick, S.M. Transcription factor Foxo3 controls the magnitude of T cell immune responses by modulating the function of dendritic cells. Nat. Immunol. 2009, 10, 504. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Kim, T.S.; Basu, J.; Jo, E.K. MicroRNA in innate immunity and autophagy during mycobacterial infection. Cell. Microbiol. 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Jiang, W.; Yu, Q.; Liu, W.; Zhou, P.; Li, J.; Xu, J.; Xu, B.; Wang, F.; Shao, F. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 2017, 551, 378. [Google Scholar] [CrossRef] [PubMed]
- Cullen, B.R. MicroRNAs as mediators of viral evasion of the immune system. Nat. Immunol. 2013, 14, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Xu, Z.; Yuan, M.; Zhang, Y.; Zhao, B.; Wang, J.; Zhang, A.; Li, G. MicroRNA-16 suppresses the activation of inflammatory macrophages in atherosclerosis by targeting PDCD4. Int. J. Mol. Med. 2016, 37, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Morgan, M.J.; Choksi, S.; Zhang, Y.; Kim, Y.S.; Liu, Z.G. MicroRNAs modulate the noncanonical transcription factor NF-κB pathway by regulating expression of the kinase IKKα during macrophage differentiation. Nat. Immunol. 2010, 11, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Z.; Hou, Y.; Zhang, K.; Peng, X. Gga-miR-99a targets SMARCA5 to regulate Mycoplasma gallisepticum (HS strain) infection by depressing cell proliferation in chicken. Gene 2017, 627, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, X.; Du, Z.; Li, N. Identification of microRNAs from different tissues of chicken embryo and adult chicken. FEBS Lett. 2006, 580, 3610–3616. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Lu, J.; Kulbokas, E.J.; Golub, T.R.; Mootha, V.; Lindblad-Toh, K.; Lander, E.S.; Kellis, M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 2005, 434, 338–345. [Google Scholar] [CrossRef] [PubMed]
- Joshi, N.R.; Su, R.W.; Chandramouli, G.V.; Khoo, S.K.; Jeong, J.W.; Young, S.L.; Lessey, B.A.; Fazleabas, A.T. Altered expression of microRNA-451 in eutopic endometrium of baboons (Papio anubis) with endometriosis. Hum. Reprod. 2015, 30, 2881–2891. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.B.; He, X.; Huang, Y.F.; Wu, Q.N.; Zhou, Y.C.; Hao, D.J. Long non-coding RNA crnde promotes multiple myeloma cell growth by suppressing miR-451. Oncol. Res. 2017, 25, 1207–1214. [Google Scholar] [CrossRef] [PubMed]
- Chapman, L.M.; Ture, S.K.; Field, D.J.; Morrell, C.N. MiR-451 limits CD4 + T cell proliferative responses to infection in mice. Immunol. Res. 2017, 65, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Burnside, J.; Ouyang, M.; Anderson, A.; Bernberg, E.; Lu, C.; Meyers, B.C.; Green, P.J.; Markis, M.; Isaacs, G.; Huang, E.; et al. Deep sequencing of chicken microRNAs. BMC Genom. 2008, 9, 185. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: Microrna biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Barton, G.M.; Medzhitov, R. Toll-like receptor signaling pathways. Science 2003, 300, 1524–1525. [Google Scholar] [CrossRef] [PubMed]
- Beutler, B. Inferences, questions and possibilities in toll-like receptor signalling. Nature 2004, 430, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Muzio, M.; Polentarutti, N.; Bosisio, D.; Manoj, K.P.; Mantovani, A. Toll-like receptor family and signalling pathway. Biochem. Soc. Trans. 2000, 28, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Zhao, C.; Bi, D.; Tian, W.; Chen, J.; Sun, J.; Peng, X. Mycoplasma gallisepticum (HS strain) surface lipoprotein pMGA interacts with host apolipoprotein A-I during infection in chicken. Appl. Microbiol. Biotechnol. 2016, 100, 1343–1354. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Zhou, Q.; Zhang, T.; Wang, X.; Song, L. Changes and role evaluation of TNF-α and IL-1β in lung tissues of ARDS mice. Chin. J. Cell. Mol. Immunol. 2017, 33, 159–163. [Google Scholar]
- Berzat, A.; Hall, A. Cellular responses to extracellular guidance cues. EMBO J. 2010, 29, 2734–2745. [Google Scholar] [CrossRef] [PubMed]
- Hur, W.; Lee, J.H.; Kim, S.W.; Kim, J.H.; Si, H.B.; Kim, M.; Hwang, D.; Kim, Y.S.; Park, T.; Um, S.J. Downregulation of microRNA-451 in non-alcoholic steatohepatitis inhibits fatty acid-induced proinflammatory cytokine production through the AMPK/AKT pathway. Int. J. Biochem. Cell Biol. 2015, 64, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.C.; Lu, H.; Zhou, Q.; Yu, S.M.; Mao, Y.L.; Zhang, H.J.; Zhang, P.C.; Yan, W.J. MiR-451 inhibits synovial fibroblasts proliferation and inflammatory cytokines secretion in rheumatoid arthritis through mediating p38MAPK signaling pathway. Int. J. Clin. Exp. Pathol. 2015, 8, 14562. [Google Scholar] [PubMed]
- Tzivion, G.; Gupta, V.S.; Kaplun, L.; Balan, V. 14-3-3 proteins as potential oncogenes. Semin. Cancer Biol. 2006, 16, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Takayama, K.; Urano, T.; Fujimura, T.; Ashikari, D.; Obinata, D.; Horie-Inoue, K.; Takahashi, S.; Ouchi, Y.; Homma, Y. 14-3-3ζ, a novel androgen-responsive gene, is upregulated in prostate cancer and promotes prostate cancer cell proliferation and survival. Clin. Cancer Res. 2012, 18, 5617–5627. [Google Scholar] [CrossRef] [PubMed]
- Kratochvill, F.; Machacek, C.; Vogl, C.; Ebner, F.; Sedlyarov, V.; Gruber, A.R.; Hartweger, H.; Vielnascher, R.; Karaghiosoff, M.; Rülicke, T. Tristetraprolin-driven regulatory circuit controls quality and timing of mrna decay in inflammation. Mol. Syst. Biol. 2011, 7, 560. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.G.; Amar, M.J.; Remaley, A.T.; Kwon, J.; Blackshear, P.J.; Wang, P.Y.; Hwang, P.M. Zinc finger protein tristetraprolin interacts with CCL3 mRNA and regulates tissue inflammation. J. Immunol. 2011, 187, 2696–2701. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Han, L.; Zhang, A.; Wang, G.; Jia, Z.; Yang, Y.; Xiao, Y.; Pu, P.; Zhong, Y.; Kang, C. MiRNA-451 plays a role as tumor suppressor in human glioma cells. Brain Res. 2010, 1359, 14–21. [Google Scholar]
- Pozuelo-Rubio, M. Proteomic and biochemical analysis of 14-3-3-binding proteins during C2-ceramide-induced apoptosis. FEBS J. 2010, 277, 3321–3342. [Google Scholar] [CrossRef] [PubMed]
- Calus, D.; Maes, D.; Vranckx, K.; Villareal, I.; Pasmans, F.; Haesebrouck, F. Validation of ATP luminometry for rapid and accurate titration of Mycoplasma hyopneumoniae in friis medium and a comparison with the color changing units assay. J. Microbiol. Methods 2010, 83, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]





| Name | Primer Sequence (5′-3′) | Accession No. |
|---|---|---|
| Primers for 3′-UTR Cloning | ||
| YWHAZ 3′-UTR-F | GCGCTCGAGCAAGCGAGGAAGACT | NM_001031343.1 |
| YWHAZ 3′-UTR-R | ATTGCGGCCGCAAGCAACAGCAGGTA | |
| Mut-P-Y 3′-UTR-F | GAAGGACAGAGAAAGTCATCACATTCCATTATTTGTAAAGTTACCTGCTGTTGC | NM_001031343.1 |
| Mut-P-R 3′-UTR-R | AATAATGGAATGTGATGACTTTCTCTGTCCTTCTCTAGAGAAAGTTGACTGG | |
| Primers for RT-qPCR | ||
| GAPDH-F | GAGGGTAGTGAAGGCTGCTG | NM 204305 |
| GAPDH-R | CACAACACGGTTGCTGTATC | |
| YWHAZ-F | AAAATGTTGTAGGAGCCCGTAGG | NM_001031343.1 |
| YWHAZ-R | TTGCTTTCTGCTTGCGAAGC | |
| RT-gga-miR-451 | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGAAACTCAG | MIMAT0003775 |
| gga-miR-451-F | GTAGGAAACCGTTACCATTACTGAG | MIMAT0003775 |
| gga-miR-451-R | ACTGGTGTCGTGGAGTCGGC | |
| gga-5s-rRNA-F | CCATACCACCCTGGAAACGC | |
| gga-5s-rRNA-R | TACTAACCGAGCCCGACCCT | |
| Name | Sequences (5′-3′) |
|---|---|
| gga-miR-451 mimics | AAACCGUUACCAUUACUGAGUUUACUCAGUAAUGGUAACGGUUUUU |
| gga-miR-451 NC | UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT |
| gga-miR-451 inhibitor | AAACUCAGUAAUGGUAACGGUUU |
| gga-miR-451 inhibitor NC | CAGUACUUUUGUGUAGUACAA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Zhang, K.; Zou, M.; Sun, Y.; Peng, X. gga-miR-451 Negatively Regulates Mycoplasma gallisepticum (HS Strain)-Induced Inflammatory Cytokine Production via Targeting YWHAZ. Int. J. Mol. Sci. 2018, 19, 1191. https://doi.org/10.3390/ijms19041191
Zhao Y, Zhang K, Zou M, Sun Y, Peng X. gga-miR-451 Negatively Regulates Mycoplasma gallisepticum (HS Strain)-Induced Inflammatory Cytokine Production via Targeting YWHAZ. International Journal of Molecular Sciences. 2018; 19(4):1191. https://doi.org/10.3390/ijms19041191
Chicago/Turabian StyleZhao, Yabo, Kang Zhang, Mengyun Zou, Yingfei Sun, and Xiuli Peng. 2018. "gga-miR-451 Negatively Regulates Mycoplasma gallisepticum (HS Strain)-Induced Inflammatory Cytokine Production via Targeting YWHAZ" International Journal of Molecular Sciences 19, no. 4: 1191. https://doi.org/10.3390/ijms19041191




