Abstract
Deoxyribonucleic acid (DNA) is the self-replicating hereditary material that provides a blueprint which, in collaboration with environmental influences, produces a structural and functional phenotype. As DNA coordinates and directs differentiation, growth, survival, and reproduction, it is responsible for life and the continuation of our species. Genome integrity requires the maintenance of DNA stability for the correct preservation of genetic information. This is facilitated by accurate DNA replication and precise DNA repair. DNA damage may arise from a wide range of both endogenous and exogenous sources but may be repaired through highly specific mechanisms. The most common mechanisms include mismatch, base excision, nucleotide excision, and double-strand DNA (dsDNA) break repair. Concurrent with regulation of the cell cycle, these mechanisms are precisely executed to ensure full restoration of damaged DNA. Failure or inaccuracy in DNA repair contributes to genome instability and loss of genetic information which may lead to mutations resulting in disease or loss of life. A detailed understanding of the mechanisms of DNA damage and its repair provides insight into disease pathogeneses and may facilitate diagnosis and the development of targeted therapies.
1. Deoxyribonucleic Acid as Hereditary Material
Deoxyribonucleic acid (DNA) is the hereditary material found in humans, other eukaryotes, and prokaryotes that carries instructions for structure and function [1]. Acting as a blueprint in collaboration with environment cues, DNA gives rise to phenotype. Accordingly, its integrity is essential for life [2]. Genomic stability is maintained by the accurate replication and adequate repair of DNA; failure of these crucial processes results in DNA damage and the inability to ensure continuation of a given species [3]. The occurrence of DNA damage is more likely to occur at genomic loci which have increased transcriptional activity [4]. Failure to maintain DNA integrity as a result of inadequate repair leads to mutations inducing structural, biochemical, and/or functional aberrations which are the cause of several diseases [2].
2. Cell Growth
The purpose of the cell cycle is to generate two genetically identical daughter cells from a single parent cell [5]. This is achieved by the coordination of cell growth, DNA replication, and cell division [5]. The cell cycle is responsive to a variety of cues and signals: internal cellular cues involving DNA damage, external cellular cues, or molecular signals that contribute to the regulation of its progression [5,6]. Cellular cues include hormones and growth factors such as insulin and insulin-like growth factor, nutrients such as amino acids and glucose, and cellular stressors such as hypoxia and osmotic stress [5,6]. The mammalian target of rapamycin (mTOR) protein kinase acts as an environmental sensor to these cues and promotes critical processes of the cell cycle [6].
The cell cycle is divided into interphase and mitosis [7]. During interphase, cell growth and DNA synthesis occur to prepare the cell for mitosis [7]. Interphase consists of the growth 1/gap 1 (G1) phase, the DNA synthesis (S) phase, and the pre-mitotic/gap 2 (G2) phase, while mitosis comprises the mitotic (M) phase [7]. In the M phase, mitosis is marked by nuclear division and cytokinesis (cytoplasmic division) [8,9]. In the G1 phase, cells are metabolically active and grow continuously [8,9]. DNA synthesis and replication occur during the S phase [8,9]. During the G2 phase, cells continue to grow and specific proteins are synthesized in preparation for mitosis [8,9]. The resting (G0) phase signifies quiescence in which non-dividing cells exit the cell cycle [8,9].
3. Cell Cycle Control and Checkpoints
Cell cycle checkpoints are important regulatory mechanisms through which DNA integrity is maintained [10,11]. They only allow cells with stable DNA to undergo DNA replication in the S phase, and only cells with correctly replicated DNA enter the M phase for cell division [10,11]. Any failure of cell cycle control mechanisms leads to a range of mutations resulting from the replication and preservation of damaged and unrepaired DNA [10,11].
Cell cycle control may be described as a three-step process [12,13]. First, DNA synthesis (S phase) and chromosome segregation (M phase) are qualitatively controlled by phosphorylation of various proteins by specific kinases [12,13]. Second, the activity of cyclin-dependent kinases (CDKs) determines the progression of cells through the cell cycle [12,13]. CDKs stimulate the transition between cell cycle phases via phosphorylation of effector protein substrates [5]. CDKs are activated by cyclins and inhibited by CDK inhibitors (CDKIs) [5]. Third, cell cycle-related regulators including cyclins and CDKIs are quantitatively controlled by ubiquitination, an important post-translational modification [12,13]. Ubiquitination results from an enzymatic cascade that involves the attachment of ubiquitin to a lysine residue of the target protein [14]. Target proteins are defined as polypeptides enriched in proline, glutamic acid, serine, and threonine residues which serve as intramolecular signals for proteolytic degradation [15]. This post-translational modification regulates vital cellular activities such as cell growth and death, chromatin organization and dynamics, gene expression, and the DNA damage response (DDR) [14].
CDK regulation is controlled by the nuclear availability of cyclins throughout the cell cycle, phosphorylation by CDK activating kinases (CAKs), and the activity of CDKI peptide inhibitors [16]. Cyclins are a family of proteins that control the progression of the cell cycle by forming complexes with CDKs thereby modulating CDK activation and activity [16,17]. Cyclin D-CDK4 and -D-CDK6 regulate the G1 phase, cyclin E-CDK2 is responsible for the G1/S phase transition, cyclin A-CDK2 regulates the S phase, cyclin A-CDK1 regulates the G2 phase, and cyclin B-CDK1 is involved in the regulation of the M phase [17]. CAKs activate all CDKs, whereas only a few inhibited by Wee1- and myelin transcription factor 1 (Myt1) kinases and promoted by cell division cycle 25 (cdc25) phosphatases [16]. Active cyclin-CDK complexes are inactivated by the binding of CDKIs from either the CDK4 inhibitor (INK) family (p15, p16, p18 and p19) or the CDK inhibitor (KIP) family (p25, p27 and p57) [16]. The INK family of CDKIs is capable of inhibiting all CDKs, whereas the KIP family of CDKIs can only inhibit CDKs involved in the G1 phase (Figure 1) [5].
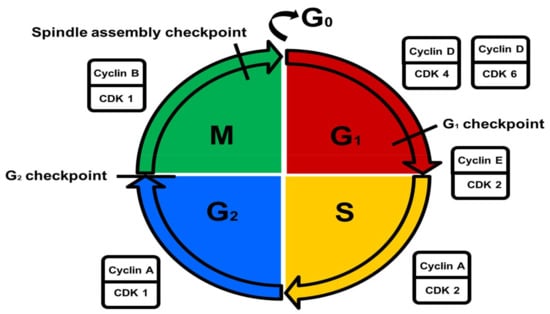
Figure 1.
Control of the cell cycle. Metabolically active growing cells are present in the growth 1/gap 1 (G1) phase. DNA replication occurs in the DNA synthesis (S) phase. Cells prepare for mitosis in the pre-mitotic/gap 2 (G2) phase. Cells undergo nuclear- and cytoplasmic division in the mitotic (M) phase. Non-dividing cells exit the cell cycle in the resting (G0) phase. The different cell cycle phases are regulated by specific cyclin/cyclin-dependent kinase (CDK) complexes.
The G1 checkpoint ensures that cell size is adequate, that nutrient supply is sufficient, that growth factors are present, and that there is no DNA damage [10,18]. The G2 checkpoint ensures that error-free DNA replication occurs by activating DNA repair mechanisms during an induced pause in the cycle if required [10,18]. Near the end of the M phase, the spindle assembly checkpoint ensures that chromosomes are stably attached to the mitotic spindle to facilitate chromosome separation [10,18].
4. Disruption of Genome Integrity
DNA damage is defined as chemical (dynamic) and physical (structural) alterations to the DNA double helix that are derived from endogenous or exogenous origins and impair the function and integrity of DNA [19,20].
If the damaged DNA is repairable, the necessary cell cycle checkpoints are activated, the DNA is repaired, genome integrity is restored, and the cell survives [21]. If the extent of DNA damage is irreparable, cells containing damaged DNA are directed to undergo senescence or programmed cell death to prevent the proliferation of mutant cells and the replication of erroneous DNA [21]. Should DNA repair mechanisms and DNA damage elimination processes fail, mutations and chromosomal aberrations arise which may lead to malignant and pathological transformation of the cell [21,22].
5. Endogenous Deoxyribonucleic Acid Damage
Endogenous DNA damage, originating from internal metabolic processes, includes damage caused by reactive oxygen species (ROS) and reactive nitrogen species (RNS) [5,20]. These products are formed during oxidative stress, metabolic processes, and the inflammatory response [5,20]. Endogenous DNA damage also includes depurination and depyrimidination at certain loci [19,20]. This occurs through the hydrolysis of N-glycosidic bonds between nitrogenous bases and deoxyribose residues, resulting in apurinic and apyrimidinic site formation [19,20,23]. In addition, the spontaneous hydrolytic deamination of cytosine bases can alter DNA, resulting in a non-native uracil base [19,20]. Replication stress represents another form of spontaneous endogenous DNA damage which occurs during the S phase and causes the stalling of replication forks [24]. The intra-S phase checkpoint is responsible for slowing replication forks to allow DNA damage to be repaired and to prevent genetically aberrant cells from progressing to the next phase of the cell cycle [25]. Furthermore, a complex interaction between checkpoint kinase 1 (Chk1), Claspin, and the Timeless (Tim)-Tim-interacting protein (Tipin) complex mediates the intra-S phase checkpoint [26,27].
6. Exogenous Deoxyribonucleic Acid Damage
Exogenous DNA damage, originating from external environmental processes, includes ionizing and solar ultraviolet radiation [19,20]. Ionizing radiation generates a wide variety of DNA lesions [19,20]. These include single and dsDNA breaks as well as oxidative modifications of nucleobases and deoxyribose moieties [19,20]. Solar ultraviolet radiation forms cyclobutane pyrimidine dimers which are strongly linked to the aetiology of skin cancer [19,20,23]. Exogenous DNA damage also includes environmental pollutants present in air, water, and food [20]. Harmful chemicals such as second-hand smoke, pesticides (e.g., organophosphates), and toxic metals (e.g., mercury) are metabolised into highly reactive metabolites that chemically react with nitrogenous bases [20]. Ultimately, these chemicals lead to deleterious DNA strand breaks and DNA adducts [20].
7. Deoxyribonucleic Acid Damage Response Pathway
The DDR is an integrated signaling and genomic maintenance network which enables cells to withstand threats posed by DNA damage [28,29,30]. The DDR is involved in signaling the presence of DNA damage to DNA repair machinery [28,29,30]. Sensor proteins recognize DNA lesions and prevent replication fork stalling by mediating the amplification of signaling pathways and stimulating transducers and effectors to impact various cellular processes [31]. These cellular processes include stabilizing replisomes (protein complexes responsible for DNA replication), regulating transcription, monitoring the cell cycle, providing energy through autophagy, remodeling chromatin, repairing damaged DNA, processing ribonucleic acid (RNA), and inducing apoptosis [31].
The DNA damage checkpoint complex is composed of sensors, signal transducers, and effector pathways [32]. The fundamental components are the phosphoinositide 3 kinase-related kinases (PIKKs), namely ataxia telangiectasia mutated (ATM), ataxia telangiectasia, and rad3-related (ATR) and DNA-dependent protein kinase (DNA-PK) (Figure 2) [32,33]. These proteins play vital roles in telomere-length regulation to protect chromosome ends from deterioration and in the prevention of their ends fusing with other chromosomes [31,32]. Additionally, the substrates of these proteins, such as Chk1 and checkpoint kinase 2 (Chk2), mediate cell cycle arrest in the G1, S, and G2 phases of the cell cycle, thus mediating DNA repair and cell death [31,32].
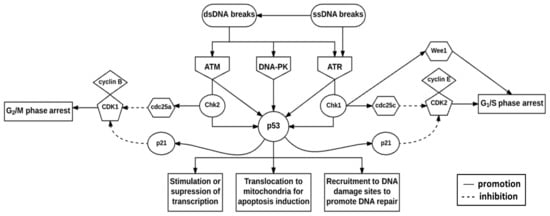
Figure 2.
Deoxyribonucleic acid damage checkpoint complex. DNA damage presenting as double-strand DNA (dsDNA)- or single-strand DNA (ssDNA) breaks initiate the DNA damage response (DDR) via ataxia telangiectasia mutated (ATM), DNA-dependent protein kinase (DNA-PK) and ataxia telangiectasia and rad3-related protein (ATR). Checkpoint kinase 2 (Chk2) is expressed throughout the cell cycle and is activated by ATM, whereas checkpoint kinase 1 (Chk1) expression is restricted to the G1- and S phases and is activated by ATR. These checkpoint kinases phosphorylate and subsequently activate p53 which integrates stress signals to determine the fate of the cell.
ATM is an important protein involved in the activation of cell cycle checkpoints [34,35]. ATM is recruited to double-strand DNA (dsDNA) breaks by the dsDNA break repair nuclease MRE11-dsDNA break repair protein RAD50 (RAD50)-nibrin (NBS1) (MRN) complex [36]. Upon recruitment, ATM is activated by autophosphorylation at three serine (Ser) sites namely Ser367, Ser1893, and Ser1981 [37]. In addition, ATM is acetylated at lysine (Lys)3016 [37]. As a result, ATM phosphorylates the MRN complex and downstream effector proteins such as Chk1 and -2 to initiate cell cycle checkpoints [38,39]. Cell cycle checkpoints allow for increased time to repair DNA damage before the cells enter either the S phase for DNA replication or the M phase for cell division [10,11].
The ATM protein is activated by dsDNA breaks, whereas the ATR protein responds to single-strand DNA (ssDNA) breaks [32,40]. ATM and ATR activate checkpoint regulator substrates Chk2 and Chk1 respectively [31,41]. These checkpoint regulator substrates are responsible for regulating CDK activity [31,32]. Chk1 activates cdc25c phosphatase by phosphorylation which subsequently inhibits CDK2 activity [41,42]. Inhibition of the cyclin E-CDK2 complex results in G1/S phase arrest [41]. Chk2 activates cdc25a phosphatase by phosphorylation which subsequently inhibits CDK1 activity [41,42]. Inhibition of the cyclin B-CDK1 complex results in G2/M phase arrest [41]. Chk1 also phosphorylates and activates Wee1 kinase to inhibit the G2/M phase transition [41,42]. ATM, ATR, DNA-PK, Chk1, and Chk2 are all capable of phosphorylating p53 which regulates the transcriptional activation of p21 to contribute to CDK1- and CDK2-mediated inhibition [40,41]. p53 phosphoprotein mediates various cellular responses to DNA damage, including the regulation of transcription, induction of cell death, and promotion of DNA repair, and is stabilized by post-translational modifications (Figure 2) [31,32]. Cell cycle arrest is promoted for the transcriptional- or post-transcriptional activation of DNA repair proteins [31,32].
8. Preservation of Genome Integrity
Cell cycle checkpoint prolongation and DDR protein recruitment is highly dependent on the characteristics and complexity of the DNA damage sustained [2,43]. Therefore, specific DNA repair mechanisms and DNA repair genes are responsible for correcting particular types of DNA lesions [2,43]. The prominent DNA repair mechanisms include mismatch repair, base-excision repair, nucleotide-excision repair, and dsDNA break repair [43,44]. dsDNA break repair can be further divided into three subtypes, namely non-homologous end-joining (NHEJ), homologous recombination (HR), and microhomology-mediated end joining (MMEJ) (Figure 3) [43,44].
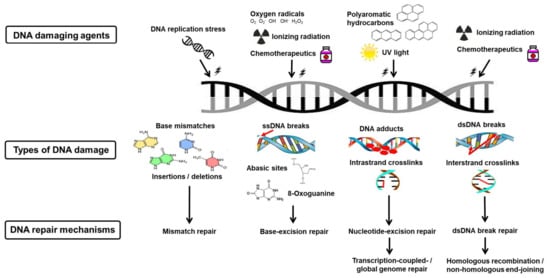
Figure 3.
Deoxyribonucleic acid damage and repair mechanisms. Various DNA damaging agents cause a range of DNA lesions. Each are corrected by a specific DNA repair mechanism, namely mismatch repair, base-excision repair, transcription-coupled/global genome repair, or homologous recombination (HR)/non-homologous end-joining (NHEJ).
9. Mismatch Repair
Mismatch repair is responsible for correcting base pair mismatches which occur when adenosine-guanine and cytosine-thymidine do not pair correctly [45]. The specific pathway that detects and removes misincorporated bases was discovered by Paul Modrich (Nobel Prize in Chemistry, 2015) [45]. Mismatch repair also corrects DNA insertions and deletions resulting from erroneous DNA replication or DNA polymerase misincorporation errors (Figure 3) [44,46,47]. Mismatch repair involves three sequential processes: recognition of the mismatch, excision of the incorrect DNA sequence, and resynthesis of the correct DNA sequence [48].
In initiating mismatch repair, the Mutator S (MutS) complex is responsible for detecting DNA mismatches [48,49,50]. The excision of the ssDNA mismatch lesion occurs upon detection of base–base mismatches and insertion/deletion loops (dsDNA with one or more unpaired nucleotides) [31,43,51]. Nuclease, polymerase, and ligase enzymes act on the subsequent ssDNA excision to ensure the new DNA strand is inserted correctly [31]. As mismatch repair is an immediate post-replicative correction mechanism, proteins involved in the repair process are regulated by the cell cycle [43,52]. These DNA mismatch repair proteins include MutS homolog 1 (MSH1) and Mutator L (MutL) homolog 1 (MLH1) which recognise base-base mismatches as well as insertion/deletion loops [43,52].
In the recognition step, MutS is recruited ahead of proliferating cell nuclear antigen (PCNA), an essential DNA replication accessory protein [53]. MutSα (MutS homolog 2 (MSH2)-MutS homolog 6 (MSH6) heteroduplex) recognizes base-base mismatches whereas MutSβ (MSH2-MutS homolog 3 (MSH3) heteroduplex) recognizes insertion/deletion loops [48,49,50,54]. Upon MutS recruitment, the DNA mismatch repair protein MutL is recruited to MutS [48,49,50]. In the excision step, MutL activates Mutator H (MutH) endonuclease to generate a ssDNA break (DNA nick) containing the mismatch which allows for the attachment of exonuclease 1 (Exo1) [48,49,50]. Exo1 excises the mismatched DNA strand. This is proceeded by the recruitment of replication protein A (RPA) to protect the resulting ssDNA [48,49,50]. In the resynthesis step, DNA polymerase synthesizes a new complementary DNA strand to correct the mismatch and DNA ligase repairs the DNA nick and restores the DNA double helix [48,49,50].
10. Base-Excision Repair
Base-excision repair is involved in the removal and replacement of damaged DNA bases [55]. Tomas Lindahl described the pathway in which these modified bases are repaired, for which he was awarded the Nobel Prize in Chemistry (2015) [55]. Glycosylases are responsible for the detection and removal of the damaged DNA base(s) and the subsequent forming of an abasic site [56]. DNA damage-causing agents that specifically induce the base-excision repair pathway include ROS (e.g., superoxide (O2−) and hydrogen peroxide (H2O2)), X-rays (e.g., computed axial tomography scans), alkylating agents (e.g., cisplatin), and spontaneous reactions (e.g., replication fork stalling) [44,57]. DNA lesions arise from these mutagens via alkylation, deamination, and oxidation reactions, and as a result of ROS-induced oxidative damage to guanine and the subsequent formation of 8-oxoguanine (Figure 3) [43,58].
ROS are generated during normal cellular respiration [23,59]. Electron transfer occurs between various metabolic intermediates and a terminal electron acceptor, namely molecular oxygen (O2) during aerobic respiration [23]. ROS have the potential to cause oxidative damage to DNA as a result of their unpaired electrons which make them highly reactive [23]. H2O2 is a by-product of numerous biochemical reactions such as uric acid formation, but may also be generated by ionizing radiation [23]. H2O2 produces two oxidized base products, namely 8-oxoguanine, which binds to adenine or cytosine to form a transversion mutation (conversion from a purine to a pyrimidine and vice versa), and thymine glycol which inhibits DNA replication [23,59].
In base-excision repair, a substrate-specific DNA glycosylase enzyme detects a damaged DNA base and removes it by cleaving the N-glycosidic bond between deoxyribose and the damaged base [43,56,58,60,61,62]. Nuclease, polymerase, and ligase enzymes are subsequently recruited in order to complete DNA repair in a similar manner as in ssDNA break repair [56,60,61,62]. Upon recognition and excision of a damaged DNA base by a substrate-specific DNA glycosylase enzyme, an abasic site is formed and is cleaved by the apurinic-apyrimidinic endonuclease [56,58,60,61,62]. Scaffolding proteins, namely poly (adenosine diphosphate (ADP)-ribose) polymerase 1 (PARP1) and X-ray repair cross-complementation protein 1 (XRCC1), protect the resulting ssDNA and recruit downstream base-excision repair proteins [56,60,61,62]. In short-patch base excision repair, DNA polymerase β inserts the modified base and DNA ligase I or -III seal the remaining DNA nick [56,58,60,61,62]. In long-patch base excision repair, DNA polymerases δ and -ε insert the correct bases past the gap, while flap endonuclease 1 (FEN1) cleaves the displaced DNA and DNA ligase 1 along with PCNA which seals the nick [56,58,60,61,62].
Base excision repair corrects large numbers of small DNA base lesions caused by alkylation, deamination, and oxidation reactions [43,44]. Components of base excision repair machinery such as glycosylases are regulated in a cell cycle-specific manner [43]. The expression of uracil-DNA glycosylase, encoded by the UNG gene, peaks in the late G1 phase continuing throughout the S phase [43,58]. The expression of thymine/uracil mismatch glycosylase, encoded by the TDG gene, peaks in the G1 phase and declines in the S phase [43,58].
11. Nucleotide-Excision Repair
Nucleotide-excision repair controls the removal of DNA adducts (segments of DNA covalently bound to carcinogenic chemicals) from DNA by excising an oligonucleotide containing the lesion to replace it with newly synthesised DNA [63]. The discovery of the mechanism by which this is achieved is attributed to Aziz Sancar (Nobel Prize in Chemistry, 2015) [63]. DNA-damaging agents that induce the nucleotide-excision repair pathway include ultraviolet light and polycyclic aromatic hydrocarbons which contribute to destabilization of the DNA double helix [44,64]. These agents may cause DNA adducts, such as etheno-DNA adducts (e.g., 1,N6-ethenodeoxyadenosine and 3,N4-ethenodeoxycytidine), which are generated from exogenous carcinogen metabolism and endogenous lipid peroxidation, as well as intrastrand crosslinks, characterised by the covalent binding of nucleotides within the same DNA strand (Figure 3) [43,65,66].
DNA double helix-distorting lesions are recognized by the xeroderma pigmentosum group A (XPA) protein and undergo repair in one of two pathways depending on the type of lesion i.e., transcription-coupled nucleotide-excision repair or global genome nucleotide excision repair [31,67]. DNA double helix distortion is most commonly caused by pyrimidine dimers formed by ultraviolet light [43,44]. Transcription-coupled nucleotide excision repair targets lesions blocking transcription while global genome nucleotide-excision repair targets lesions in both transcribed- and non-transcribed DNA [31,67]. Nucleotide excision repair is characterised by the excision of the 25–30 base oligonucleotide segments containing the adduct, resulting in ssDNA on which DNA polymerases act before ligation occurs [68].
Global genome nucleotide excision repair is initiated by the xeroderma pigmentosum group C-RAD23 homolog B (XPC-RAD23B) complex which binds to the non-damaged DNA strand opposite to the lesion [68,69,70]. Transcription factor II human (TFIIH) interacts with XPC-RAD23B to recruit the group B subunit (XPB) to separate DNA strands and allow the group D subunit (XPD) to detect DNA damage and verify the chemical composition of the lesion [68,69,70]. The pre-incision complex is formed with the recruitment of RPA, the group A subunit (XPA) and the group G subunit (XPG) [68,69,70]. The excision repair cross-complementation group 1-xeroderma pigmentosum group F (ERCC1-XPF) complex interacts with XPA to form a 5′ DNA incision at the lesion [68,69,70]. DNA repair synthesis is initiated by polymerases δ and -κ or polymerase ε and is followed by a 3′ DNA incision at the lesion by XPG [68,69,70]. The DNA nick is sealed by DNA ligase I or the DNA ligase IIIa-XRCC1 complex [68,69,70].
12. Double-Strand Deoxyribonucleic Acid Break Repair
DsDNA breaks occur when the sugar-phosphate backbones of both DNA strands are broken at a similar position or in close proximity to one other [71]. Subsequently, physical dissociation of the DNA double helix takes place resulting in the formation of two separate single-stranded molecules [71]. Genetic information is lost as a result of the absence of a DNA template for accurate repair in the newly synthesized DNA [71]. DNA-damaging agents that cause dsDNA breaks and the repair pathway include X-rays, ionizing radiation, and anti-cancer drugs (e.g., cisplatin) [44,72]. These agents may also cause other DNA lesions such as interstrand crosslinks (covalent bonds which form between complementary strands thereby inhibiting their separation and replication) (Figure 3) [43]. Replication fork stalling may be the result of dsDNA-damaging agents or may be responsible for the formation of dsDNA breaks as a result of origin re-firing in an attempt to promote replication fork speed [25,73]. Thus, dsDNA break repair mechanisms are essential for replication fork progression and stable DNA replication [25,73].
The MRN complex detects dsDNA breaks and subsequently recruits and activates members of the DDR machinery such as those of the phosphatidylinositol 3 kinase (PI3K) family [74,75]. Activated ATM phosphorylates histone variant H2AX at Ser139 resulting in the formation of foci at sites of DNA damage lending to the recruitment of repair proteins [76,77]. Although the phosphorylated form of histone H2AX (γ-H2AX) may be regarded as a sensitive quantitative indicator of dsDNA damage, specificity is not high as it may also serve as evidence of other DNA stressors such as stalled replication forks [78].
With dsDNA breaks, two principle mechanisms may be implemented in the repair process, namely NHEJ and HR [79]. This classification depends on whether sequence homology (template DNA sequence) is used to join dsDNA break ends [79,80]. In NHEJ, sequence homology is not required for dsDNA break end-joining and it involves minimal DNA processing [79,80]. In HR, sequence homology is required in order to align dsDNA break ends prior to ligation [79,80].
The Ku70/Ku80 heterodimer recognizes dsDNA breaks by binding to both blunt or near-blunt broken DNA ends to elicit NHEJ [81,82,83]. Additionally, it binds and activates the DNA-PK catalytic subunit (DNA-PKCS) [81,82]. NHEJ is facilitated by scaffold proteins, namely X-ray repair cross-complementation protein 4 (XRCC4) and XRCC4-like factor (XLF), which bind to DNA ligase IV to seal the DNA nick [81,82,83]. DNA end-processing occurs prior to ligation to ensure compatible DNA ends by either the DNA-PKCS-interacting protein Artemis endonuclease trimming DNA ends or polymerases filling DNA ends (Figure 4) [81,82,84]. MMEJ, also known as alternative end-joining, is a Ku protein-independent NHEJ pathway that commonly results in DNA sequence deletions [71]. Both NHEJ and MMEJ can function in all phases of the cell cycle [71].
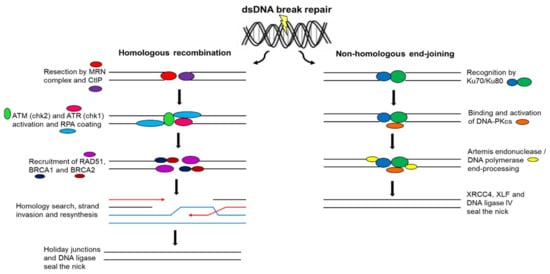
Figure 4.
Double-strand deoxyribonucleic acid break repair. DsDNA breaks are repaired by HR or NHEJ. HR involves the restoration of DNA sequences using sister chromatid sequence homology as a template and functions in all phases of the cell cycle, whereas NHEJ involves damaged DNA sequence deletions and functions in only S and G2 phases.
A paralog of XRCC4 and XLF (PAXX), a member of the XRCC4 superfamily, is recruited to dsDNA damage breaks and interacts directly with the Ku70/Ku80 heterodimer to initiate NHEJ [85,86]. Moreover, PAXX functions with structurally similar scaffold proteins XRCC4 and XLF to facilitate ligation of the DNA nick and conclude dsDNA break repair [85,86]. PAXX is a novel component of the NHEJ machinery and promotes cell survival in response to dsDNA break-inducing agents [85,86].
In HR, replicated sister chromatid DNA sequences are used as templates to restore missing DNA sequences on the damaged chromatid [87,88]. For this reason, HR can only operate in the S and G2 phases of the cell cycle when replicated sister chromatids are available [87,88]. HR is initiated by resection of broken DNA ends by the MRN complex and the C-terminal-binding protein interacting protein (CtIP) generating 3′-ssDNA tails [87,88,89]. RPA coats 3′-ssDNA tails and is replaced by RAD51 with the help of breast cancer gene 1 (BRCA1) and 2 (BRCA2) [79,87,88,89]. A nucleoprotein complex is created that detects the homologous sequence on the sister chromatid, a process referred to as strand invasion [89,90]. RAD51 catalyzes strand invasion on the homologous template to allow the restoration of lost sequence information [87,90]. Broken DNA ends are resolved by junctions called Holiday junctions that result in crossover and non-crossover products (Figure 4) [84]. As HR makes use of a non-damaged DNA template to restore chromosome integrity, it is a more precise method of DNA repair than that of NHEJ and MMEJ [71].
Two additional dsDNA break repair pathways exist that require sequence homology, namely single-strand annealing (SSA) and alternative non-homologous end-joining (alt-NHEJ) [91]. SSA is mediated by a single-strand annealing protein (SSAP) (such as RAD52) which uses a short single-stranded region to locate sequence identity and initiate HR [91,92]. In the SSA pathway, resection of the dsDNA break exposes complementary sequences in the ssDNA tails of the two ends [93]. Complementary sequences anneal, leaving non-complementary flaps of ssDNA [93]. Nucleolytic flap removal and ligation finalizes the repair of the dsDNA break [93]. In the absence of XRCC4 and DNA ligase IV, which are responsible for concluding the classical NHEJ repair pathway, alt-NHEJ resolves the dsDNA break [94]. Alt-NHEJ relies on CtIP-dependent resection as in HR but requires a unique set of repair factors, namely PARP1 together with DNA ligase I or III [95,96].
Aurora kinase A and ninein-interacting protein (AUNIP) acts as a dsDNA damage sensor and interacts with CtIP to ensure adequate accumulation of CtIP at dsDNA breaks to initiate DNA end resection and subsequently HR [97,98]. AUNIP is recruited to dsDNA damage sites through a DNA-binding motif displaying a preference for substrates that are structurally similar to those formed at replication forks during replication stalling [97]. The absence of AUNIP results in the failure of the HR pathway and is accompanied by hypersensitivity to DNA damage agents that cause replication-associated dsDNA breaks [97]. When AUNIP is overexpressed, it promotes HR and inhibits NHEJ; however, when AUNIP is inhibited, the frequency of NHEJ is increased [97].
Ubiquitin-specific protease 4 (USP4) facilitates HR by directly participating in dsDNA break end-resection through the post-translational process of autoubiquitination [99,100]. USP4 interacts with both CtIP and the MRN complex via a specific conserved region on USP4 and the catalytic domain of USP4, respectively, to promote the recruitment of the DNA end-resection factor CtIP to dsDNA break sites [100]. USP4 contains several ubiquitinated sites, mostly on cysteine residues [99]. USP4 catalytic activity is responsible for the deubiquitination of these cysteine residues to promote CtIP recruitment [99]. USP4 is a novel HR regulator whose enzymatic activity is regulated by ubiquitin adducts [99,100].
13. Pathophysiology of Deoxyribonucleic Acid Repair Failure
Rare hereditary diseases characterized by DNA repair deficiencies arise when repair machinery and mechanisms are defective [101]. Germline mutations in the relevant DNA repair genes are responsible for a range of diseases including Werner, Bloom, and Cockayne syndromes [101].
Mismatch repair deficiencies are present in adult-onset autosomal dominant Lynch syndrome and child-onset autosomal recessive constitutional mismatch repair deficiency syndrome, which include well-established colorectal and endometrial cancer syndromes [31,101,102]. Both adult- and child-onset hereditary tumour syndromes are characterized by mutated MSH and MLH genes which are responsible for sensing base-base mismatches and insertion/deletion loops (Table 1) [31,101,102]. Xeroderma pigmentosum is a non-curable genetic disease characterized by germline mutations in nucleotide-excision repair genes causing neurodegeneration, photosensitivity, and skin cancer [31,101,103]. Mutated xeroderma pigmentosum genes foster the creation of altered protein products which are responsible for the inability to repair DNA adducts and intrastrand crosslinks resulting from defective recognition and signaling of these nucleotide lesions (Table 1) [103]. Spinocerebellar ataxia with axonal neuropathy (SCAN1) results from a tyrosyl-DNA phosphodiesterase deficiency, an enzyme involved in controlling DNA winding by topoisomerase during base excision repair [104]. Ataxia with oculomotor apraxia 1 (AOA1) results from an aprataxin deficiency which is associated with scaffolding proteins which facilitate accurate base-excision repair [105]. Both SCAN1 and AOA1 are characterized by base excision repair deficiencies that produce ataxia and neurodegeneration (Table 1) [104,105]. DDR defects are present in Li-Fraumeni syndrome in which soft tissue sarcomas, breast cancer, and brain tumours are prevalent [31,101,106]. A mutation in the p53 gene inhibits the DDR and interferes with cell cycle regulation and tumour suppression (Table 1) [106]. If any component of the DDR machinery is impaired, it results in defective DNA damage sensing and signaling, potentially leading to pathological conditions that include Alzheimer’s, Parkinson’s, and Huntington’s diseases [31,101].

Table 1.
Diseases and disorders associated with defective DNA repair.
The identification of various pathologies related to inadequate DNA damage detection and DNA repair mechanisms highlights the fundamental role these processes play in maintaining genomic stability [107]. These networks are of particular importance in the prevention of neurodegeneration and malignant transformation [107]. Furthermore, they govern normal growth, neurogenesis, and immune system development [107].
14. Deoxyribonucleic Acid Repair Pathways as Therapeutic Targets and Future Directions
A defective DDR system is a hallmark of certain cancers which allows tumour cells to proliferate and acquire mutations [108]. These DNA repair defects serve as a platform for the discovery of specific treatments for selected cancers [108].
PARP1 binds to damaged DNA ends via two homologous N-terminal zinc (Zn) finger domains, Zn1 and Zn2 [109]. The carboxylic catalytic domain of PARP1 uses nicotinamide adenine dinucleotide (NAD+) as a substrate to synthesize poly (ADP-ribose) chains by an autoregulated process [109]. A structurally unique third Zn finger domain, Zn3, plays a vital role in the synthesis of these poly (ADP-ribose) chains [110]. Poly (ADP-ribose) chain synthesis at the carboxylic catalytic domain leads to the recruitment of the base-excision repair complex through interaction with condensin I and XRCC1 [110,111]. When PARP1 is bound to damaged DNA ends it prevents the conversion of ssDNA breaks into dsDNA breaks until base excision repair is completed [110].
Defects in HR proteins, specifically BRCA1 and -2, lead to the failure of dsDNA break repair and increase the likelihood of breast and ovarian cancer [112,113]. Cancer cells harbouring BRCA mutations are unable to recruit RAD51 to dsDNA break sites during HR, thus forcing cells into the more error-prone NHEJ repair pathway [114]. This HR defect promotes tumour cell sensitivity to treatments that induce ssDNA breaks [115]. One such treatment strategy is the inhibition of scaffold protein PARP1 which is involved in the repair of ssDNA lesions [114,116,117]. Furthermore, PARP inhibition leads to an accumulation of dsDNA aberrations giving rise to cell death, a process referred to as synthetic lethality [114,116,117].
ATM regulates responses associated with dsDNA break repair by phosphorylating downstream regulatory proteins and repair factors such as BRCA1, Chk2 and p53 [118]. Williamson et al. (2012) showed that mantle cell lymphoma expressing ATM and p53 mutations exhibit enhanced cytotoxicity to olaparib (PARP inhibitor) treatment both in vitro and in vivo [119]. In addition, intact DNA-PK, together with mutated ATM/p53, contribute to the induction of NHEJ as well as the synthetic lethal response consequent to PARP inhibition [119]. PARP activity is required for the detection and resumption of stalled replication forks following replication stress [120]. Following recognition by PARP, the MRN complex is recruited and the HR repair pathway repairs the damage in order to restart the replication fork [121,122]. PARP inhibition thus prevents the downstream processes required for the continuation of replication forks and subsequent DNA replication [122]. Cytogenetic aberrations involving chromosome 11q, which contains cancer-associated genes such as ATM and Chk1, have been implicated in neuroblastoma [123]. Defective DDR systems display a sensitivity to PARP inhibition, and thus PARP inhibitors are promising neuroblastoma therapeutics [123].
Olaparib was approved in 2014 by the Food and Drug Administration (FDA) as a monotherapy for women diagnosed with BRCA-deficient or -mutant ovarian cancer who had undergone three or more failed chemotherapy regimens [124]. The administration of olaparib within this patient subset resulted in progression-free survival that was significantly longer in the olaparib treatment group (48%) when compared to the placebo group (15%) [125]. Olaparib has a good oral bioavailability but myelodysplastic syndrome and acute myeloid leukaemia have been reported as more substantive unwanted effects [124,126]. Olaparib is the first clinical chemotherapeutic agent inhibiting PARP in order to target DNA repair defects in malignant cells [127].
DNA strand break bait (Dbait) molecules are DNA repair inhibitors that mimic dsDNA breaks and sequester dsDNA break repair proteins such as DNA-PK and PARP1 [128]. These large molecules are comprised of 32-base pair double helices that interfere with dsDNA break signaling by acting as bait for repair enzymes and thus inhibit HR and NHEJ [128]. Dbait molecules cause DNA-PK hyper-activation, resulting in the phosphorylation of DNA damage signaling molecules, including H2AX, Chk2, and p53, ultimately preventing the recruitment of DNA repair complexes to DNA damage sites [129].
Biau et al. (2014) conducted a preclinical study in which a cholesterol-conjugated Dbait molecule, DT01, sensitized melanoma cells to radiotherapy both in vitro and in vivo [128]. In addition, DT01 has been shown to improve the efficacy of the chemotherapeutic doxorubicin in mouse models bearing hepatocellular carcinoma [130]. Herath et al. (2016) investigated the chemosensitizing effects of DT01 in combination with a two-drug chemotherapeutic regimen (oxaliplatin and 5-fluorouracil) in an in vivo colorectal liver metastases model, and have reported significant anti-tumour effects using the combined treatment [131]. Moreover, H2AX phosphorylation by DNA-PK was exclusive to tumour cells, thus indicating sparing of surrounding non-tumourigenic tissue [131]. A signal-interfering DNA (AsiDNA), which is a cholesterol-conjugated member of the Dbait family, induces preferential toxicity towards tumourigenic tissue whilst sparing non-tumourigenic hematologic cells and preserving immune function [132]. Thierry et al. (2017) reported the induction of necrotic and apoptotic cell death by AsiDNA through p53-independent mechanisms in several lymphoma and leukaemia cell lines [132]. AsiDNA enters cells through low density lipoprotein (LDL) receptors and subsequently activates DNA-PK [132]. Dbait molecules improve the clinical outcomes of chemo- and radiotherapy by disturbing DNA repair processes in treated tumour tissue [128,132,133]. The combination of PARP inhibitor and Dbait leads to increased unrepaired dsDNA breaks, resulting in amplified tumour cell death while sparing non-tumour cells [133].
PARP inhibitors constitute a major emerging class of promising therapeutics; however, various other DNA repair pathway inhibitors are also currently being investigated [134]. Preclinical and clinical development of highly selective small molecule inhibitors of ATM and ATR is aimed at targeting the DDR and subsequently DNA repair [135,136,137]. Base excision repair inhibitors include apurinic-apyrimidinic endonuclease inhibitors that prevent abasic site cleavage and DNA polymerase β inhibitors, preventing the insertion of modified bases [134,138]. Protein-protein and protein-DNA interactions involved in nucleotide-excision repair have been identified as targets of the repair pathway and include ERCC1-XPF, ERCC1-XPA, and RPA-DNA [134,139]. Inhibition of vital proteins involved in NHEJ, such as DNA-PK and the Ku70/Ku80 heterodimer, inhibit recognition of termini and end-bridging, thus implicating dsDNA break repair [140,141]. Furthermore, inhibition of vital components of HR machinery such as RAD51 targets the alternative dsDNA break repair pathway [142].
Genetic engineering is an emergent experimental treatment strategy with potential applications in incurable genetic disorders [143]. One area which has received considerable attention is gene editing and, in particular, the use of the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease (Cas) system [143]. There are three CRISPR/Cas systems which are classified according to a specific Cas protein [143]. Type I is identifiable by the presence of Cas3, type II by Cas9, and type III by Cas10 [143]. Each system uses a unique mechanism to recognize and cleave nucleic acids [144]. Type I and III use Cas complexes to target specific DNA sites; however, type II requires only a single Cas protein [144]. For this reason, the type II CRISPR/Cas9 system has been used preferentially for genetic engineering [144,145]. CRISPR/Cas9 uses a complementary guide-RNA (gRNA) sequence and a protospacer adjacent motif (PAM) sequence to recognize a specific targeted DNA sequence [144,145]. PAM sequences, usually 5′-asparagine-glycine-glycine-3′(5′-NGG-3′), are used by both type I and -II CRISPR/Cas systems to recognize target DNA [146]. PAM sequences lie within the target DNA sequence and, if absent, Cas9-binding will not occur even if the gRNA sequence is complementary to the target DNA sequence [147]. The C-terminal of Cas9 interacts with the PAM sequence via arginine (Arg) motifs 1333 and -1335 to trigger the separation of the upstream strands of the target sequence at the first base pair position [148]. Cas9 has two catalytic nuclease domains, histidine-asparagine-histidine (HNH) and RuvC, each of which are responsible for cleaving one DNA strand in a coordinated manner [149,150]. The HNH domain cleaves the complementary strand while the non-complementary strand is cleaved by the RuvC domain [149,150]. This concurrent cleaving activity produces a dsDNA break which is repairable by either HR or NHEJ [150,151,152]. Error-prone repair pathways such as NHEJ may result in gene-silencing or insertion/deletion mutations which are generally used for gene knock-out experiments [150,151,152]. The homology-directed repair pathway ensures precise DNA repair through a ‘copy-paste’ mechanism using a donor template; however, NHEJ is the preferred repair pathway in response to Cas9 cleavage [151,153].
Although the potential clinical applications of CRISPR/Cas9 are numerous, these are in early stages of research [154]. CRISPR/Cas9 may be applicable in the treatment of cancer and genetic disorders such as Duchenne muscular dystrophy, retinitis pigmentosa, β-thalassaemia, and xeroderma pigmentosum disorder [154,155,156]. Huntington’s disease is characterized by an expansion of cytosine-adenine-guanine/glutamine repeats; a potential therapeutic target may lie in the use of CRISPR/Cas9 to silence the mutant form of the huntingtin gene (mHtt) [152].Yang et al. (2017) demonstrated successful CRISPR/Cas9 suppression of mHtt in mouse striatal neuronal cells which resulted in the alleviation of motor impairments and neurotoxicity [152]. Ou et al. (2016) combined CRISPR/Cas9 technology with that of Takahashi and Yamanaka’s (2006) induced pluripotent stem cells (iPSCs) with the aim of curing β-thalassaemia by correcting the defective β-globin gene [157,158]. Corrected iPSCs were used to generate haematopoietic stem cells that could successfully differentiate and survive in mice without exhibiting tumourigenic properties [158]. CRISPR/Cas9 is a promising tool harnessing DNA damage as well as repair pathways and mechanisms in the treatment of incurable diseases, disease mapping, drug screening, and personalized medicine [144,145].
15. Conclusions
DNA is responsible for carrying hereditary information across generations; it accomplishes this by controlling the production and function of proteins. As a result, it is essential for growth, survival, and reproduction. Should aberrations occur in DNA, genome integrity is maintained through accurate DNA replication and adequate DNA repair. DNA damaging agents may be of endogenous or exogenous origin and the resulting DNA lesions may cause morbidity and mortality if not repaired. Specific DNA repair mechanisms that are closely associated with the cell cycle exist to correct the different types of DNA lesion that occur. Failure of these vital repair processes may lead to a variety of mutations and, consequently, diseases. Through an understanding of the causes of DNA damage and the corresponding repair mechanisms, it is possible to design strategies to create or improve methods for the prevention, diagnosis, and treatment of pathologies related to deficiencies in the mechanisms of DNA repair.
Acknowledgments
Funding and support of this research is gratefully acknowledged from the Cancer Association of South Africa (CANSA) (A0V741) (A0W228), the National Research Foundation (NRF) (105992) (90523) (85818), the South African Medical Research Council (SAMRC) (A0W110) (University Flagship and Stem Cell Extramural Unit awards to MSP), Struwig-Germeshuysen Trust (A0N074), the School of Medicine Research Committee of the University of Pretoria (RESCOM) (A0R984), and the Research Development Programme of the University of Pretoria (RDP-UP). All images were created using Microsoft PowerPoint 2016.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ADP | adenosine diphosphate |
| alt-NHEJ | alternative non-homologous end-joining |
| AOA1 | ataxia with oculomotor apraxia 1 |
| Arg | arginine |
| AsiDNA | a signal interfering DNA |
| ATM | ataxia telangiectasia mutated |
| ATR | ataxia telangiectasia and rad3-related |
| AUNIP | aurora kinase A and ninein interacting protein |
| BRCA1 | breast cancer gene 1 |
| BRCA2 | breast cancer gene 2 |
| CAKs | CDK activating kinases |
| Cas | CRISPR-associated nuclease |
| cdc25 | cell division cycle 25 |
| CDKs | cyclin-dependent kinases |
| CDKIs | CDK inhibitors |
| Chk1 | checkpoint kinase 1 |
| Chk2 | checkpoint kinase 2 |
| CRISPR | clustered regularly interspaced short palindromic repeats |
| CtIP | C-terminal-binding protein interacting protein |
| Dbait | DNA strand break bait |
| DDR | DNA damage response |
| DNA | deoxyribonucleic acid |
| DNA-PK | DNA-dependent protein kinase |
| DNA-PKCS | DNA-PK catalytic subunit |
| dsDNA | double-strand DNA |
| ERCC1 | excision-repair cross-complementation group 1 |
| Exo1 | exonuclease 1 |
| FDA | Food and Drug Administration |
| FEN1 | flap endonuclease 1 |
| G | glycine |
| G0 | resting |
| G1 | growth 1/gap 1 |
| G2 | pre-mitotic/gap 2 |
| gRNA | guide RNA |
| H | histidine |
| H2O2 | hydrogen peroxide |
| HR | homologous recombination |
| INK | CDK4 inhibitor |
| iPSCs | induced pluripotent stem cells |
| KIP | CDK inhibitor |
| LDL | low density lipoprotein |
| Lys | lysine |
| M | mitotic |
| mHtt | mutant huntingtin gene |
| MLH1 | MutL homolog 1 |
| MMEJ | microhomology-mediated end-joining |
| MRE11 | dsDNA break repair nuclease MRE11 |
| MRN | MRE11-RAD50-NBS1 |
| MSH1 | MutS homolog 1 |
| MSH2 | MutS homolog 2 |
| MSH3 | MutS homolog 3 |
| MSH6 | MutS homolog 6 |
| mTOR | mammalian target of rapamycin |
| MutH | Mutator H |
| MutL | Mutator L |
| MutS | Mutator S |
| MutSα | MSH2-MSH6 heteroduplex |
| MutSβ | MSH2-MSH3 heteroduplex |
| Myt1 | myelin transcription factor 1 |
| N | asparagine |
| NAD+ | nicotinamide adenine dinucleotide |
| NBS1 | nibrin |
| NHEJ | non-homologous end-joining |
| O2 | oxygen |
| O2− | superoxide |
| PAM | protospacer adjacent motif |
| PARP1 | poly (ADP-ribose) polymerase 1 |
| PAXX | paralog of XRCC4 and XLF |
| PCNA | proliferating cell nuclear antigen |
| PI3K | phosphatidylinositol 3 kinase |
| PIKKs | phosphoinositide 3 kinase-related kinases |
| RAD50 | dsDNA break repair protein RAD50 |
| RNA | ribonucleic acid |
| RNS | reactive nitrogen species |
| ROS | reactive oxygen species |
| RPA | replication protein A |
| S | DNA synthesis |
| SCAN1 | spinocerebellar ataxia with axonal neuropathy |
| Ser | serine |
| SSA | single-strand annealing |
| SSAP | single-strand annealing protein |
| ssDNA | single-strand DNA |
| TFIIH | transcription factor II human |
| Tim | Timeless |
| Tipin | Tim-interacting protein |
| USP4 | ubiquitin-specific protease 4 |
| XLF | XRCC4-like factor |
| XPA | xeroderma pigmentosum group A |
| XPB | xeroderma pigmentosum group B |
| XPC-RAD23B | xeroderma pigmentosum group C-RAD23 homolog B |
| XPD | xeroderma pigmentosum group D |
| XPF | xeroderma pigmentosum group F |
| XPG | xeroderma pigmentosum group G |
| XRCC1 | X-ray repair cross-complementation protein 1 |
| XRCC4 | X-ray repair cross-complementation protein 4 |
| Zn | zinc |
References
- Watson, J.D.; Crick, F.H. Genetical implications of the structure of deoxyribonucleic acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Clancy, S. DNA damage and repair: Mechanisms for maintaining DNA integrity. Nat. Educ. 2008, 1, 103. [Google Scholar]
- Wang, J.; Lindahl, T. Maintenance of genome stability. Genom. Proteom. Bioinform. 2016, 14, 119–121. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Abdulovic, A.L.; Gealy, R.; Lippert, M.J.; Jinks-Robertson, S. Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair 2007, 6, 1285–1296. [Google Scholar] [CrossRef] [PubMed]
- Khanna, K.K.; Shiloh, Y. The DNA Damage Response: Implications on Cancer Formation and Treatment; Springer: Dordrecht, The Netherlands, 2009. [Google Scholar]
- Ekim, B.; Magnuson, B.; Acosta-Jaquez, H.A.; Keller, J.A.; Feener, E.P.; Fingar, D.C. mTOR kinase domain phosphorylation promotes mTORC1 signaling, Cell Growth, and Cell Cycle Progression. Mol. Cell. Biol. 2011, 31, 2787–2801. [Google Scholar] [CrossRef] [PubMed]
- Ly, T.; Ahmad, Y.; Shlien, A.; Soroka, D.; Mills, A.; Emanuele, M.J.; Stratton, M.R.; Lamond, A.I. A proteomic chronology of gene expression through the cell cycle in human myeloid leukemia cells. eLife 2014, 3, e01630. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, E.; Gadaleta, M.C. Cell Cycle Control: Mechanisms and Protocols; Springer: New York, NY, USA, 2014. [Google Scholar]
- Cooper, G.M. The Cell: A Molecular Approach, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Gabrielli, B.; Brooks, K.; Pavey, S. Defective cell cycle checkpoints as targets for anti-cancer therapies. Front. Pharmacol. 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Barnum, K.; O’Connell, M. Cell cycle regulation by checkpoints. In Cell Cycle Control. Methods in Molecular Biology; Noguchi, E., Gadaleta, M.C., Eds.; Springer: New York, NY, USA, 2014; pp. 29–40. [Google Scholar]
- Robert, J. Cell Cycle Control. Textbook of Cell Signalling in Cancer; Springer: Cham, Switzerland, 2015; pp. 203–219. [Google Scholar]
- Morgan, D.O. The Cell Cycle: Principles of Control; New Science Press: London, UK, 2007. [Google Scholar]
- Levy-Cohen, G.; Blank, M. Functional analysis of protein ubiquitination. Anal. Biochem. 2015, 484, 37–39. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.; Gomes, A. PEST sequences in calmodulin-binding proteins. In Signal Transduction Mechanisms. Developments in Molecular and Cellular Biochemistry; Barnes, J., Coore, H., Mohammed, A., Sharma, R., Eds.; Springer: New York, NY, USA, 1995; pp. 17–27. [Google Scholar]
- Weis, M.C.; Avva, J.; Jacobberger, J.W.; Sreenath, S.N. A data-driven, mathematical model of mammalian cell cycle regulation. PLoS ONE 2014, 9, e97130. [Google Scholar] [CrossRef] [PubMed]
- Goodman, S.R. Medical Cell Biology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Zachos, G.; Black, E.J.; Walker, M.; Scott, M.T.; Vagnarelli, P.; Earnshaw, W.C.; Gillespie, D.A. Chk1 is required for spindle checkpoint function. Dev. Cell 2007, 12, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Siede, W.; Doetsch, P.W. DNA Damage Recognition; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Geacintov, N.E.; Broyde, S. The Chemical Biology of DNA Damage; Wiley: Hoboken, NJ, USA, 2011. [Google Scholar]
- Speidel, D. The role of DNA damage responses in p53 biology. Arch. Toxicol. 2015, 89, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Jeggo, P.A.; Pearl, L.H.; Carr, A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer 2016, 16, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Tropp, B.E. Molecular Biology: Genes to Proteins, 3rd ed.; Jones and Bartlett Publishers: Burlington, MA, USA, 2008. [Google Scholar]
- Henriksson, S.; Groth, P.; Gustafsson, N.; Helleday, T. Distinct mechanistic responses to replication fork stalling induced by either nucleotide or protein deprivation. Cell Cycle 2017. [Google Scholar] [CrossRef] [PubMed]
- Iyer, D.R.; Rhind, N. Replication fork slowing and stalling are distinct, checkpoint-independent consequences of replicating damaged DNA. PLoS Genet. 2017, 13, e1006958. [Google Scholar] [CrossRef] [PubMed]
- Ünsal-Kaçmaz, K.; Chastain, P.D.; Qu, P.-P.; Minoo, P.; Cordeiro-Stone, M.; Sancar, A.; Kaufmann, W.K. The human Tim/Tipin complex coordinates an intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell. Biol. 2007, 27, 3131–3142. [Google Scholar] [CrossRef] [PubMed]
- Gagou, M.E.; Zuazua-Villar, P.; Meuth, M. Enhanced H2AX phosphorylation, DNA replication fork arrest, and cell death in the absence of Chk1. Mol. Biol. Cell 2010, 21, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Karl, S.; Pritschow, Y.; Volcic, M.; Häcker, S.; Baumann, B.; Wiesmüller, L.; Debatin, K.M.; Fulda, S. Identification of a novel pro-apopotic function of NF-κB in the DNA damage response. J. Cell. Mol. Med. 2009, 13, 4239–4256. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Musich, P.R.; Zou, Y. Differential DNA damage responses in p53 proficient and deficient cells: Cisplatin-induced nuclear import of XPA is independent of ATR checkpoint in p53-deficient lung cancer cells. Int. J. Biochem. Mol. Biol. 2011, 2, 138–145. [Google Scholar] [PubMed]
- Pabla, N.; Huang, S.; Mi, Q.-S.; Daniel, R.; Dong, Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J. Biol. Chem. 2008, 283, 6572–6583. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Lukas, J. DNA damage checkpoints: From initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007, 19, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Rodier, F.; Campisi, J.; Bhaumik, D. Two faces of p53: Aging and tumor suppression. Nucleic Acids Res. 2007, 35, 7475–7484. [Google Scholar] [CrossRef] [PubMed]
- Bakkenist, C.J.; Kastan, M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003, 421, 499. [Google Scholar] [CrossRef] [PubMed]
- Falck, J.; Coates, J.; Jackson, S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005, 434, 605. [Google Scholar] [CrossRef] [PubMed]
- Uziel, T.; Lerenthal, Y.; Moyal, L.; Andegeko, Y.; Mittelman, L.; Shiloh, Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003, 22, 5612–5621. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, S.V.; Graham, M.E.; Jakob, B.; Tobias, F.; Kijas, A.W.; Tanuji, M.; Chen, P.; Robinson, P.J.; Taucher-Scholz, G.; Suzuki, K.; et al. Autophosphorylation and ATM activation: Additional sites add to the complexity. J. Biol. Chem. 2011, 286, 9107–9119. [Google Scholar] [CrossRef] [PubMed]
- Riballo, E.; Kühne, M.; Rief, N.; Doherty, A.; Smith, G.C.; Recio, M.J.; Reis, C.; Dahm, K.; Fricke, A.; Krempler, A.; et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell 2004, 16, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Beucher, A.; Birraux, J.; Tchouandong, L.; Barton, O.; Shibata, A.; Conrad, S.; Goodarzi, A.A.; Krempler, A.; Jeggo, P.A.; Löbrich, M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009, 28, 3413–3427. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef] [PubMed]
- Bartek, J.; Lukas, J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 2003, 3, 421–429. [Google Scholar] [CrossRef]
- Busino, L.; Donzelli, M.; Chiesa, M.; Guardavaccaro, D.; Ganoth, D.; Dorrello, N.V.; Hershko, A.; Pagano, M.; Draetta, G.F. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature 2003, 426, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Mjelle, R.; Hegre, S.A.; Aas, P.A.; Slupphaug, G.; Drabløs, F.; Sætrom, P.; Krokan, H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair 2015, 30, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Torgovnick, A.; Schumacher, B. DNA repair mechanisms in cancer development and therapy. Front. Genet. 2015, 6, 157. [Google Scholar] [CrossRef] [PubMed]
- Modrich, P. Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 1991, 25, 229–253. [Google Scholar] [CrossRef] [PubMed]
- Tropp, B.E. Molecular Biology: Genes to Proteins, 4th ed.; Jones & Bartlett Learning: Burlington, MA, USA, 2012. [Google Scholar]
- Spies, M.; Fishel, R. Mismatch Repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol. 2015, 7, a022657. [Google Scholar] [CrossRef] [PubMed]
- Constantin, N.; Dzantiev, L.; Kadyrov, F.A.; Modrich, P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005, 280, 39752–39761. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Clark, A.B.; Wong, E.; Kane, M.F.; Mazur, D.J.; Parris, T.; Kolas, N.K.; Russel, R.; Hou, H.; Kneitz, B.; et al. Inactivation of exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003, 17, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Dherin, C.; Gueneau, E.; Francin, M.; Nunez, M.; Miron, S.; Liberti, S.E.; Rasmussen, L.J.; Zinn-Justin, S.; Gilquin, B.; Charbonnier, J.B.; et al. Characterization of a highly conserved binding site of Mlh1 required for exonuclease I-dependent mismatch repair. Mol. Cell. Biol. 2009, 29, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Ganten, D.; Ruckpaul, K. Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine; Springer: Berlin, Germany, 2006. [Google Scholar]
- Wood, R.D.; Mitchell, M.; Sgouros, J.; Lindahl, T. Human DNA repair genes. Science 2001, 291, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.; Tran, P.T.; Joshi, A.; Liskay, R.M.; Alani, E. MSH-MLH complexes formed at a DNA mismatch are disrupted by the PCNA sliding clamp. J. Mol. Biol. 2001, 306, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Predeus, A.V.; Kovacs, N.; Feig, M. Differential mismatch recognition specificities of eukaryotic MutS homologs, MutSα and MutSβ. Biophys. J. 2014, 106, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709. [Google Scholar] [CrossRef] [PubMed]
- Odell, I.D.; Barbour, J.-E.; Murphy, D.L.; Della-Maria, J.A.; Sweasy, J.B.; Tomkinson, A.E.; Wallace, S.S.; Pederson, D.S. Nucleosome disruption by DNA ligase III-XRCC1 promotes efficient base excision repair. Mol. Cell. Biol. 2011, 31, 4623–4632. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Chen, Z.; Medhurst, A.L.; Neal, J.A.; Bao, Z.; Mortusewicz, O.; McGouran, J.; Song, X.; Shen, H.; Hamdy, F.C.; et al. DNA-PKcs and PARP1 bind to unresected stalled DNA replication forks where they recruit XRCC1 to mediate repair. Cancer Res. 2016, 76, 1078–1088. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjørås, M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, P.H.N.; Furtado, C.; Repolês, B.M.; Ribeiro, G.A.; Mendes, I.C.; Peloso, E.F.; Gadelha, F.R.; Macedo, A.M.; Franco, G.R.; Pena, S.D.; et al. Oxidative stress and DNA lesions: The role of 8-oxoguanine lesions in trypanosoma cruzi cell viability. PLoS Negl. Trop. Dis. 2013, 7, e2279. [Google Scholar] [CrossRef] [PubMed]
- Dianova, I.I.; Sleeth, K.M.; Allinson, S.L.; Parsons, J.L.; Breslin, C.; Caldecott, K.W.; Dianov, G.L. XRCC1–DNA polymerase β interaction is required for efficient base excision repair. Nucleic Acids Res. 2004, 32, 2550–2555. [Google Scholar] [CrossRef] [PubMed]
- Asagoshi, K.; Tano, K.; Chastain, P.D.; Adachi, N.; Sonoda, E.; Kikuchi, K.; Koyama, H.; Nagata, K.; Kaufman, D.G.; Takeda, S.; et al. FEN1 functions in long patch base excision repair under conditions of oxidative stress in vertebrate cells. Mol. Cancer Res. 2010, 8, 204–215. [Google Scholar] [CrossRef] [PubMed]
- Wiederhold, L.; Leppard, J.B.; Kedar, P.; Karimi-Busheri, F.; Rasouli-Nia, A.; Weinfeld, M.; Tomkinson, A.E.; Izumi, T.; Prasad, R.; Wilson, S.H.; et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 2004, 15, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Petit, C.; Sancar, A. Nucleotide excision repair: From, E. coli to man. Biochimie 1999, 81, 15–25. [Google Scholar] [CrossRef]
- Ranes, M.; Boeing, S.; Wang, Y.; Wienholz, F.; Menoni, H.; Walker, J.; Encheva, V.; Chakravarty, P.; Mari, P.O.; Stewart, A.; et al. A ubiquitylation site in Cockayne syndrome B required for repair of oxidative DNA damage, but not for transcription-coupled nucleotide excision repair. Nucleic Acids Res. 2016, 44, 5246–5255. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Li, H.; Wang, S.; Jiang, X.; Zhang, S.; Zhang, R.; Fu, P.P.; Sun, X. Ultrasensitive UPLC-MS-MS method for the quantitation of etheno-DNA adducts in human urine. Int. J. Environ. Res. Public Health 2014, 11, 10902. [Google Scholar] [CrossRef] [PubMed]
- Chaim, I.A.; Gardner, A.; Wu, J.; Iyama, T.; Wilson, D.M.; Samson, L.D. A novel role for transcription-coupled nucleotide excision repair for the in vivo repair of 3,N4-ethenocytosine. Nucleic Acids Res. 2017, 45, 3242–3252. [Google Scholar] [PubMed]
- Bee, L.; Marini, S.; Pontarin, G.; Ferraro, P.; Costa, R.; Albrecht, U.; Celotti, L. Nucleotide excision repair efficiency in quiescent human fibroblasts is modulated by circadian clock. Nucleic Acids Res. 2015, 43, 2126–2137. [Google Scholar] [CrossRef] [PubMed]
- Hoogstraten, D.; Bergink, S.; Ng, J.M.Y.; Verbiest, V.H.M.; Luijsterburg, M.S.; Geverts, B.; Raams, A.; Dinant, C.; Hoeijmakers, J.H.; Vermeulen, W.; et al. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J. Cell Sci. 2008, 121, 2850–2859. [Google Scholar] [CrossRef] [PubMed]
- Sugasawa, K.; Okamoto, T.; Shimizu, Y.; Masutani, C.; Iwai, S.; Hanaoka, F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001, 15, 507–521. [Google Scholar] [CrossRef] [PubMed]
- Lans, H.; Marteijn, J.A.; Schumacher, B.; Hoeijmakers, J.H.J.; Jansen, G.; Vermeulen, W. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. 2010, 6, e1000941. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, T.; Baer, R.; Gautier, J. DNA double-strand break repair pathway choice and cancer. DNA Repair 2014, 19, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Weterings, E.; Gallegos, A.C.; Dominick, L.N.; Cooke, L.S.; Bartels, T.N.; Vagner, J.; Matsunaga, T.O.; Mahadevan, D. A novel small molecule inhibitor of the DNA repair protein Ku70/80. DNA Repair 2016, 43, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Barrasa, M.I.; Orr-Weaver, T.L. Replication fork progression during re-replication requires the DNA damage checkpoint and double-strand break repair. Curr. Biol. 2015, 25, 1654–1660. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.M.; Misri, S.; Meyer, B.; Raj, S.; Zobel, C.L.; Sleckman, B.P.; Hallahan, D.E.; Sharma, G.G. Unique epigenetic influence of H2AX phosphorylation and H3K56 acetylation on normal stem cell radioresponses. Mol. Biol. Cell 2016, 27, 1332–1345. [Google Scholar] [CrossRef] [PubMed]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef] [PubMed]
- Redon, C.E.; Nakamura, A.J.; Zhang, Y.-W.; Ji, J.; Bonner, W.M.; Kinders, R.J.; Parchment, R.E.; Doroshow, J.H.; Pommier, Y. Histone γH2AX and poly(ADP-Rribose) as clinical pharmacodynamic biomarkers. Clin. Cancer Res. 2010, 16, 4532–4542. [Google Scholar] [CrossRef] [PubMed]
- Löbrich, M.; Shibata, A.; Beucher, A.; Fisher, A.; Ensminger, M.; Goodarzi, A.A.; Barton, O.; Jeggo, P.A. γH2AX foci analysis for monitoring DNA double-strand break repair: Strengths, limitations and optimization. Cell Cycle 2010, 9, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Cleaver, J.E. γH2Ax: Biomarker of damage or functional participant in DNA repair “all that glitters is not gold!”. Photochem. Photobiol. 2011, 87, 1230–1239. [Google Scholar] [CrossRef] [PubMed]
- Kass, E.M.; Helgadottir, H.R.; Chen, C.-C.; Barbera, M.; Wang, R.; Westermark, U.K.; Ludwig, T.; Moynahan, M.E.; Jasin, M. Double-strand break repair by homologous recombination in primary mouse somatic cells requires BRCA1 but not the ATM kinase. Proc. Natl. Acad. Sci. USA 2013, 110, 5564–5569. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair 2008, 7, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Adachi, N.; Suzuki, H.; Iiizumi, S.; Koyama, H. Hypersensitivity of nonhomologous DNA end-joining mutants to VP-16 and ICRF-193: Implications for the repair of topoisomerase II-mediated DNA damage. J. Biol. Chem. 2003, 278, 35897–35902. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Lu, H.; Tippin, B.; Goodman, M.F.; Shimazaki, N.; Koiwai, O.; Hsieh, C.L.; Schwarz, K.; Lieber, M.R. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell 2004, 16, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Uentrop, L.; El-Awady, R.A.; Schliecker, L.; Willers, H.; Dahm-Daphi, J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008, 36, 2561–2569. [Google Scholar] [CrossRef] [PubMed]
- Brandsma, I.; van Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed]
- Ochi, T.; Blackford, A.N.; Coates, J.; Jhujh, S.; Mehmood, S.; Tamura, N.; Travers, J.; Wu, Q.; Draviam, V.M.; Robinson, C.V.; et al. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 2015, 347, 185–188. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shao, Z.; Jiang, W.; Lee, B.J.; Zha, S. PAXX promotes Ku accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice. Nat. Commun. 2017, 8, 13816. [Google Scholar] [CrossRef] [PubMed]
- Falck, J.; Forment, J.V.; Coates, J.; Mistrik, M.; Lukas, J.; Bartek, J.; Jackson, S.P. Cyclin-dependent kinase targeting of NBS1 promotes DNA-end resection, replication restart and homologous recombination. EMBO Rep. 2012, 13, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Limbo, O.; Chahwan, C.; Yamada, Y.; de Bruin, R.A.; Wittenberg, C.; Russell, P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 2007, 28, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Moiani, D.; Arvai, A.S.; Perry, J.; Harding, S.M.; Genois, M.M.; Maity, R.; van Rossum-Fikkert, S.; Kertokalio, A.; Romoli, F.; et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 2014, 53, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Stark, J.M.; Pierce, A.J.; Oh, J.; Pastink, A.; Jasin, M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004, 24, 9305–9316. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, A.J.; Mohni, K.N.; Kan, Y.; Hendrickson, E.A.; Stark, J.M.; Weller, S.K. The HSV-1 exonuclease, UL12, stimulates recombination by a single strand annealing mechanism. PLoS Pathog. 2012, 8, e1002862. [Google Scholar] [CrossRef] [PubMed]
- Ander, M.; Subramaniam, S.; Fahmy, K.; Stewart, A.F.; Schäffer, E. A single-strand annealing protein clamps DNA to detect and secure homology. PLoS Biol. 2015, 13, e1002213. [Google Scholar] [CrossRef] [PubMed]
- Morrical, S.W. DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb. Perspect. Biol. 2015, 7, a016444. [Google Scholar] [CrossRef] [PubMed]
- Nussenzweig, A.; Nussenzweig, M.C. A backup DNA repair pathway moves to the forefront. Cell 2007, 131, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Biehs, R.; Steinlage, M.; Barton, O.; Juhász, S.; Künzel, J.; Spies, J.; Shibata, A.; Jeggo, P.A.; Löbrich, M. DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol. Cell 2017, 65, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Newman, E.A.; Lu, F.; Bashllari, D.; Wang, L.; Opipari, A.W.; Castle, V.P. Alternative NHEJ pathway components are therapeutic targets in high-risk neuroblastoma. Mol. Cancer Res. 2015, 13, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Chen, H.; Han, J.; He, H.; Huen, M.S.Y.; Feng, X.; Liu, T.; Huang, J. AUNIP/C1orf135 directs DNA double-strand breaks towards the homologous recombination repair pathway. Nat. Commun. 2017, 8, 895. [Google Scholar] [CrossRef] [PubMed]
- Lieu, A.S.; Chen, T.S.; Chou, C.H.; Wu, C.H.; Hsu, C.Y.; Huang, C.Y.; Chang, L.K.; Loh, J.K.; Chang, C.S.; Hsu, C.M.; et al. Functional characterization of AIBp, a novel Aurora-A binding protein in centrosome structure and spindle formation. Int. J. Oncol. 2010, 37, 429–436. [Google Scholar] [PubMed]
- Wijnhoven, P.; Konietzny, R.; Blackford, A.N.; Travers, J.; Kessler, B.M.; Nishi, R.; Jackson, S.P. USP4 auto-deubiquitylation promotes homologous recombination. Mol. Cell 2015, 60, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, H.; Wang, X.; Tian, Q.; Hu, Z.; Peng, C.; Jiang, P.; Wang, T.; Guo, W.; Chen, Y.; et al. The deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep. 2015, 13, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, A.J.F.; Miller, J.H.; Suzuki, D.T.; Lewontin, R.C.; Gelbart, W.M. An Introduction to Genetic Analysis, 7th ed.; W.H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Maletzki, C.; Huehns, M.; Bauer, I.; Ripperger, T.; Mork, M.M.; Vilar, E.; Klöcking, S.; Zettl, H.; Prall, F.; Linnebacher, M. Frameshift mutational target gene analysis identifies similarities and differences in constitutional mismatch repair-deficiency and Lynch syndrome. Mol. Carcinog. 2017, 56, 1753–1764. [Google Scholar] [CrossRef] [PubMed]
- Bowden, N.A.; Beveridge, N.J.; Ashton, K.A.; Baines, K.J.; Scott, R.J. Understanding xeroderma pigmentosum complementation groups using gene expression profiling after UV-light exposure. Int. J. Mol. Sci. 2015, 16, 15985–15996. [Google Scholar] [CrossRef] [PubMed]
- Hirano, R.; Interthal, H.; Huang, C.; Nakamura, T.; Deguchi, K.; Choi, K.; Bhattacharjee, M.B.; Arimura, K.; Umehara, F.; Izumo, S.; et al. Spinocerebellar ataxia with axonal neuropathy: Consequence of a Tdp1 recessive neomorphic mutation? EMBO J. 2007, 26, 4732–4743. [Google Scholar] [CrossRef] [PubMed]
- Çağlayan, M.; Horton, J.K.; Prasad, R.; Wilson, S.H. Complementation of aprataxin deficiency by base excision repair enzymes. Nucleic Acids Res. 2015, 43, 2271–2281. [Google Scholar] [CrossRef] [PubMed]
- Akouchekian, M.; Hemati, S.; Jafari, D.; Jalilian, N.; Dehghan Manshadi, M. Does PTEN gene mutation play any role in Li-Fraumeni syndrome. Med. J. Islam. Repub. Iran 2016, 30, 378. [Google Scholar] [PubMed]
- Lawrence, K.S.; Chau, T.; Engebrecht, J. DNA damage response and spindle assembly checkpoint function throughout the cell cycle to ensure genomic integrity. PLoS Genet. 2015, 11, e1005150. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Steffen, J.D.; McCauley, M.M.; Pascal, J.M. Fluorescent sensors of PARP-1 structural dynamics and allosteric regulation in response to DNA damage. Nucleic Acids Res. 2016, 44, 9771–9783. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.F.; Ruhl, D.D.; Planck, J.L.; Kraus, W.L.; Pascal, J.M. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1) functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J. Biol. Chem. 2010, 285, 18877–18887. [Google Scholar] [CrossRef] [PubMed]
- Heale, J.T.; Ball, A.R.; Schmiesing, J.A.; Kim, J.-S.; Kong, X.; Zhou, S.; Hudson, D.F.; Earnshaw, W.C.; Yokomori, K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol. Cell 2006, 21, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Audeh, M.W.; Carmichael, J.; Penson, R.T.; Friedlander, M.; Powell, B.; Bell-McGuinn, K.M.; Scott, C.; Weitzel, J.N.; Oaknin, A.; Loman, N.; et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet 2010, 376, 245–251. [Google Scholar] [CrossRef]
- Tutt, A.; Robson, M.; Garber, J.E.; Domchek, S.M.; Audeh, M.W.; Weitzel, J.N.; Friedlander, M.; Arun, B.; Loman, N.; Schmutzler, R.K.; et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet 2010, 376, 235–244. [Google Scholar] [CrossRef]
- Wiener, D.; Gajardo-Meneses, P.; Ortega-Hernández, V.; Herrera-Cares, C.B.; Díaz, S.N.; Fernández, W.; Cornejo, V.; Gamboa, J.; Tapia, T.; Alvarez, C.; et al. BRCA1 and BARD1 colocalize mainly in the cytoplasm of breast cancer tumors, and their isoforms show differential expression. Breast Cancer Res. Treat. 2015, 153, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Sartori, A.A.; Lukas, C.; Coates, J.; Mistrik, M.; Fu, S.; Bartek, J.; Baer, R.; Lukas, J.; Jackson, S.P. Human CtIP promotes DNA end resection. Nature 2007, 450, 509. [Google Scholar] [CrossRef] [PubMed]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005, 434, 917. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, R.; Bakkenist, C.J.; McKinnon, P.J.; Kastan, M.B. Phosphorylation of SMC1 is a critical downstream event in the ATM–NBS1–BRCA1 pathway. Genes Dev. 2004, 18, 1423–1438. [Google Scholar] [CrossRef] [PubMed]
- Williamson, C.T.; Kubota, E.; Hamill, J.D.; Klimowicz, A.; Ye, R.; Muzik, H.; Dean, M.; Tu, L.; Gilley, D.; Magliocco, A.M.; et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol. Med. 2012, 4, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Ronson, G.; Piberger, A.L.; Higgs, M.; Olsen, A.; Stewart, G.; McHugh, P.; Petermann, E.; Lakin, N. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat. Commun. 2018, 9, 746. [Google Scholar] [CrossRef] [PubMed]
- Ray Chaudhuri, A.; Callen, E.; Ding, X.; Gogola, E.; Duarte, A.A.; Lee, J.E.; Wong, N.; Lafarga, V.; Calvo, J.A.; Panzarino, N.J.; et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 2016, 535, 382. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Petermann, E.; Schultz, N.; Jemth, A.-S.; Loseva, O.; Issaeva, N.; Johansson, F.; Fernandez, S.; McGlynn, P.; Helleday, T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009, 28, 2601–2615. [Google Scholar] [CrossRef] [PubMed]
- Takagi, M.; Yoshida, M.; Nemoto, Y.; Tamaichi, H.; Tsuchida, R.; Seki, M.; Uryu, K.; Nishii, R.; Miyamoto, S.; Saito, M.; et al. Loss of DNA damage response in neuroblastoma and utility of a PARP inhibitor. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.; Ison, G.; McKee, A.E.; Zhang, H.; Tang, S.; Gwise, T.; Sridhara, R.; Lee, E.; Tzou, A.; Philip, R.; et al. FDA approval summary: Olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin. Cancer Res. 2015, 21, 4257–4261. [Google Scholar] [CrossRef] [PubMed]
- Pujade-Lauraine, E.; Ledermann, J.A.; Selle, F.; Gebski, V.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 1274–1284. [Google Scholar] [CrossRef]
- Bundred, N.; Gardovskis, J.; Jaskiewicz, J.; Eglitis, J.; Paramonov, V.; McCormack, P.; Swaisland, H.; Cavallin, M.; Parry, T.; Carmichael, J.; et al. Evaluation of the pharmacodynamics and pharmacokinetics of the PARP inhibitor olaparib: A phase I multicentre trial in patients scheduled for elective breast cancer surgery. Investig. New Drugs 2013, 31, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Fong, P.C.; Boss, D.S.; Yap, T.A.; Tutt, A.; Wu, P.; Mergui-Roelvink, M.; Mortimer, P.; Swaisland, H.; Lau, A.; O’Connor, M.J.; et al. Inhibition of poly(ADP-Ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009, 361, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Biau, J.; Devun, F.; Jdey, W.; Kotula, E.; Quanz, M.; Chautard, E.; Sayarath, M.; Sun, J.S.; Verrelle, P.; Dutreix, M. A preclinical study combining the DNA repair inhibitor Dbait with radiotherapy for the treatment of melanoma. Neoplasia 2014, 16, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Qiu, H.; Shao, Z.; Wang, G.; Wang, J.; Yao, Y.; Xin, Y.; Zhou, M.; Wang, A.Z.; Zhang, L. Nanoparticle formulation of small DNA molecules, Dbait, improves the sensitivity of hormone-independent prostate cancer to radiotherapy. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 2261–2271. [Google Scholar] [CrossRef] [PubMed]
- Herath, N.I.; Devun, F.; Herbette, A.; Lienafa, M.C.; Sun, J.S.; Dutreix, M.; Denys, A. Potentiation of doxorubicin efficacy in hepatocellular carcinoma by the DNA repair inhibitor DT01 in preclinical models. Eur. Radiol. 2017, 27, 4435–4444. [Google Scholar] [CrossRef] [PubMed]
- Herath, N.I.; Devun, F.; Lienafa, M.C.; Herbette, A.; Denys, A.; Sun, J.S.; Dutreix, M. The DNA repair inhibitor DT01 as a novel therapeutic strategy for chemosensitization of colorectal liver metastasis. Mol. Cancer Ther. 2016, 15, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Thierry, S.; Jdey, W.; Alculumbre, S.; Soumelis, V.; Noguiez-Hellin, P.; Dutreix, M. The DNA repair inhibitor Dbait is specific for malignant hematologic cells in blood. Mol. Cancer Ther. 2017, 16, 2817–2827. [Google Scholar] [CrossRef] [PubMed]
- Jdey, W.; Thierry, S.; Russo, C.; Devun, F.; Al Abo, M.; Noguiez-Hellin, P.; Sun, J.S.; Barillot, E.; Zinovyev, A.; Kuperstein, I.; et al. Drug driven synthetic lethality: Bypassing tumor cell genetics with a combination of Dbait and PARP inhibitors. Clin. Cancer Res. 2017, 23, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Gavande, N.S.; VanderVere-Carozza, P.S.; Hinshaw, H.D.; Jalal, S.I.; Sears, C.R.; Pawelczak, K.S.; Turchi, J.J. DNA repair targeted therapy: The past or future of cancer treatment? Pharmacol. Ther. 2016, 160, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Batey, M.A.; Zhao, Y.; Kyle, S.; Richardson, C.; Slade, A.; Martin, N.M.B.; Lau, A.; Newell, D.R.; Curtin, N.J. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol. Cancer Ther. 2013, 12, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Biddlestone-Thorpe, L.; Sajjad, M.; Rosenberg, E.; Beckta, J.M.; Valerie, N.C.K.; Tokarz, M.; Adams, B.R.; Wagner, A.F.; Khalil, A.; Gilfor, D.; et al. ATM kinase inhibition preferentially sensitizes p53-mutant glioma to ionizing radiation. Clin. Cancer Res. 2013, 19, 3189–3200. [Google Scholar] [CrossRef] [PubMed]
- Foote, K.M.; Blades, K.; Cronin, A.; Fillery, S.; Guichard, S.S.; Hassall, L.; Hickson, I.; Jacq, X.; Jewsbury, P.J.; McGuire, T.M.; et al. Discovery of 4-{4-[(3R)-3-Methylmorpholin-4-yl]-6-[1-(methylsulfonyl)cyclopropyl]pyrimidin-2-yl}-1H-indole (AZ20): A potent and selective inhibitor of ATR protein kinase with monotherapy in vivo antitumor activity. J. Med. Chem. 2013, 56, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Madhusudan, S.; Smart, F.; Shrimpton, P.; Parsons, J.L.; Gardiner, L.; Houlbrook, S.; Talbot, D.C.; Hammonds, T.; Freemont, P.A.; Sternberg, M.J.; et al. Isolation of a small molecule inhibitor of DNA base excision repair. Nucleic Acids Res. 2005, 33, 4711–4724. [Google Scholar] [CrossRef] [PubMed]
- Andrews, B.J.; Turchi, J.J. Development of a high-throughput screen for inhibitors of replication protein A and its role in nucleotide excision repair. Mol. Cancer Ther. 2004, 3, 385–391. [Google Scholar] [PubMed]
- Boeckman, H.J.; Trego, K.S.; Turchi, J.J. Cisplatin sensitizes cancer cells to ionizing radiation via inhibition of nonhomologous end joining. Mol. Cancer Res. 2005, 3, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Munck, J.M.; Batey, M.A.; Zhao, Y.; Jenkins, H.; Richardson, C.J.; Cano, C.; Tavecchio, M.; Barbeau, J.; Bardos, J.; Cornell, L.; et al. Chemosensitization of cancer cells by KU-0060648, a dual inhibitor of DNA-PK and PI-3K. Mol. Cancer Ther. 2012, 11, 1789–1798. [Google Scholar] [CrossRef] [PubMed]
- Alagpulinsa, D.A.; Ayyadevara, S.; Shmookler Reis, R.J. A small-molecule inhibitor of RAD51 reduces homologous recombination and sensitizes multiple myeloma cells to doxorubicin. Front. Oncol. 2014, 4, 289. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. eLife 2013, 2, e00471. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.; Erdmann, S.; Mojica, F.J.M.; Garrett, R.A. Protospacer recognition motifs: Mixed identities and functional diversity. RNA Biol. 2013, 10, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Bothmer, A.; Phadke, T.; Barrera, L.A.; Margulies, C.M.; Lee, C.S.; Buquicchio, F.; Moss, S.; Abdulkerim, H.S.; Selleck, W.; Jayaram, H.; et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat. Commun. 2017, 8, 13905. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; González, F.; Huangfu, D. The iCRISPR platform for rapid genome editing in human Pluripotent Stem Cells. Methods Enzymol. 2014, 546, 215–250. [Google Scholar] [PubMed]
- Yang, S.; Chang, R.; Yang, H.; Zhao, T.; Hong, Y.; Kong, H.E.; Sun, X.; Qin, Z.; Jin, P.; Li, S.; et al. CRISPR/Cas9-mediated gene editing ameliorates neurotoxicity in mouse model of Huntington’s disease. J. Clin. Investig. 2017, 127, 2719–2724. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D. Genome editing: Progress and challenges for medical applications. Genome Med. 2016, 8, 120. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Xu, Y.; Zhang, X.; Ding, C.; Huang, R.; Zhang, Z.; Lv, J.; Xie, X.; Chen, Y.; Li, Y.; et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015, 6, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Castellanos Rivera, R.M.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Tong, Y.; Liu, X.Z.; Wang, T.T.; Cheng, L.; Wang, B.Y.; Lv, X.; Huang, Y.; Liu, D.P. Both TALENs and CRISPR/Cas9 directly target the HBB IVS2–654 (C > T) mutation in β-thalassemia-derived iPSCs. Sci. Rep. 2015, 5, 12065. [Google Scholar] [CrossRef] [PubMed]
- Ou, Z.; Niu, X.; He, W.; Chen, Y.; Song, B.; Xian, Y.; Fan, D.; Tang, D.; Sun, X. The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human β-thalassemia in mice. Sci. Rep. 2016, 6, 32463. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).