d-Amino Acids Are Exuded by Arabidopsis thaliana Roots to the Rhizosphere
Abstract
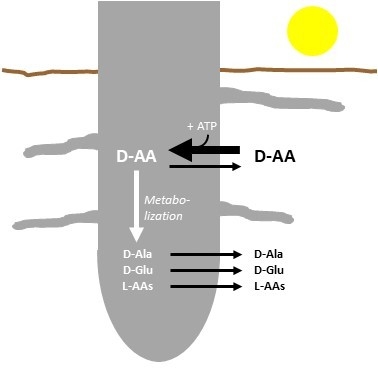
:1. Introduction
2. Results
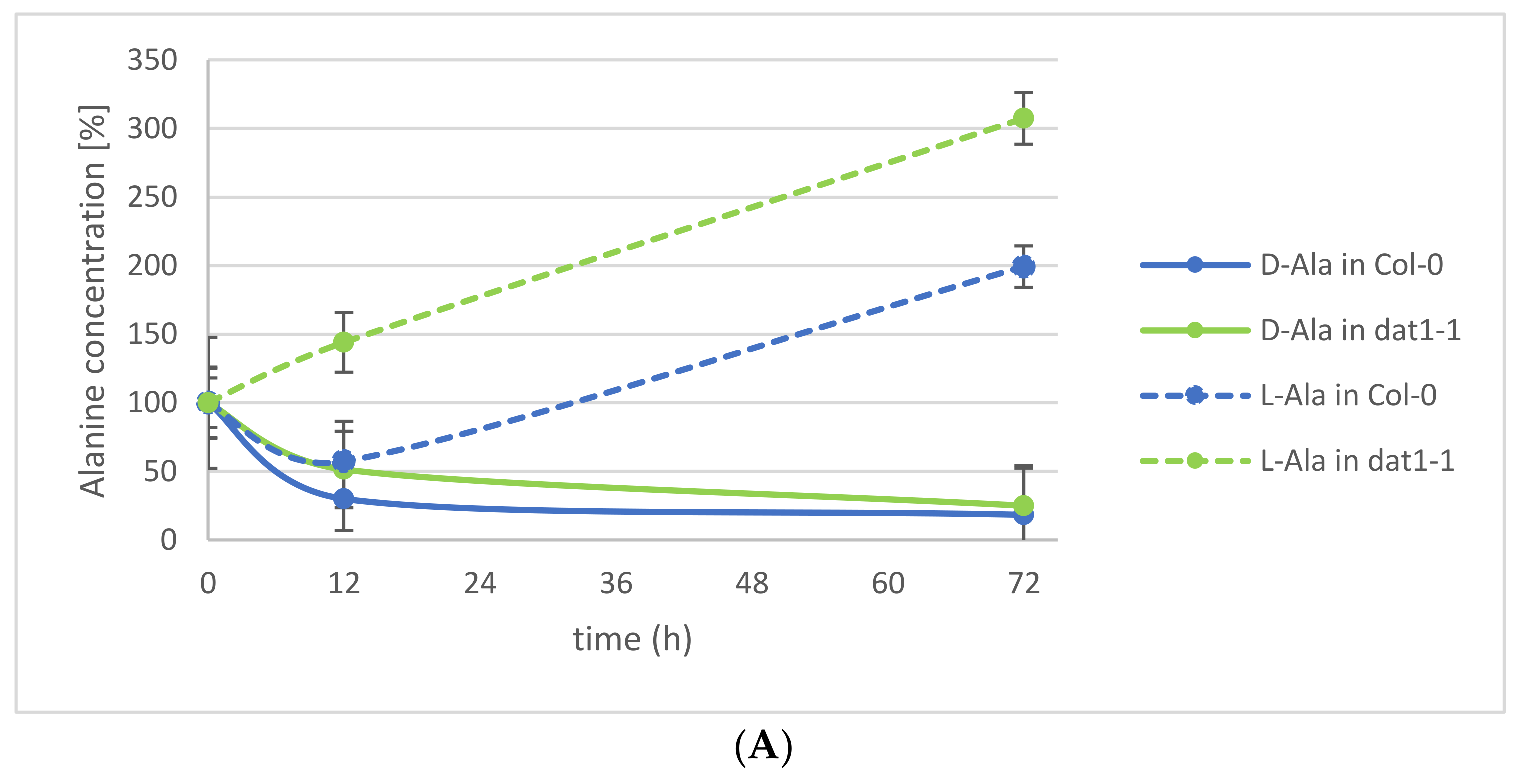
2.1. d-Ala and d-Glu Are Degraded Rapidly in Seedlings
2.2. d-AAs Are Exuded by Roots into the Medium
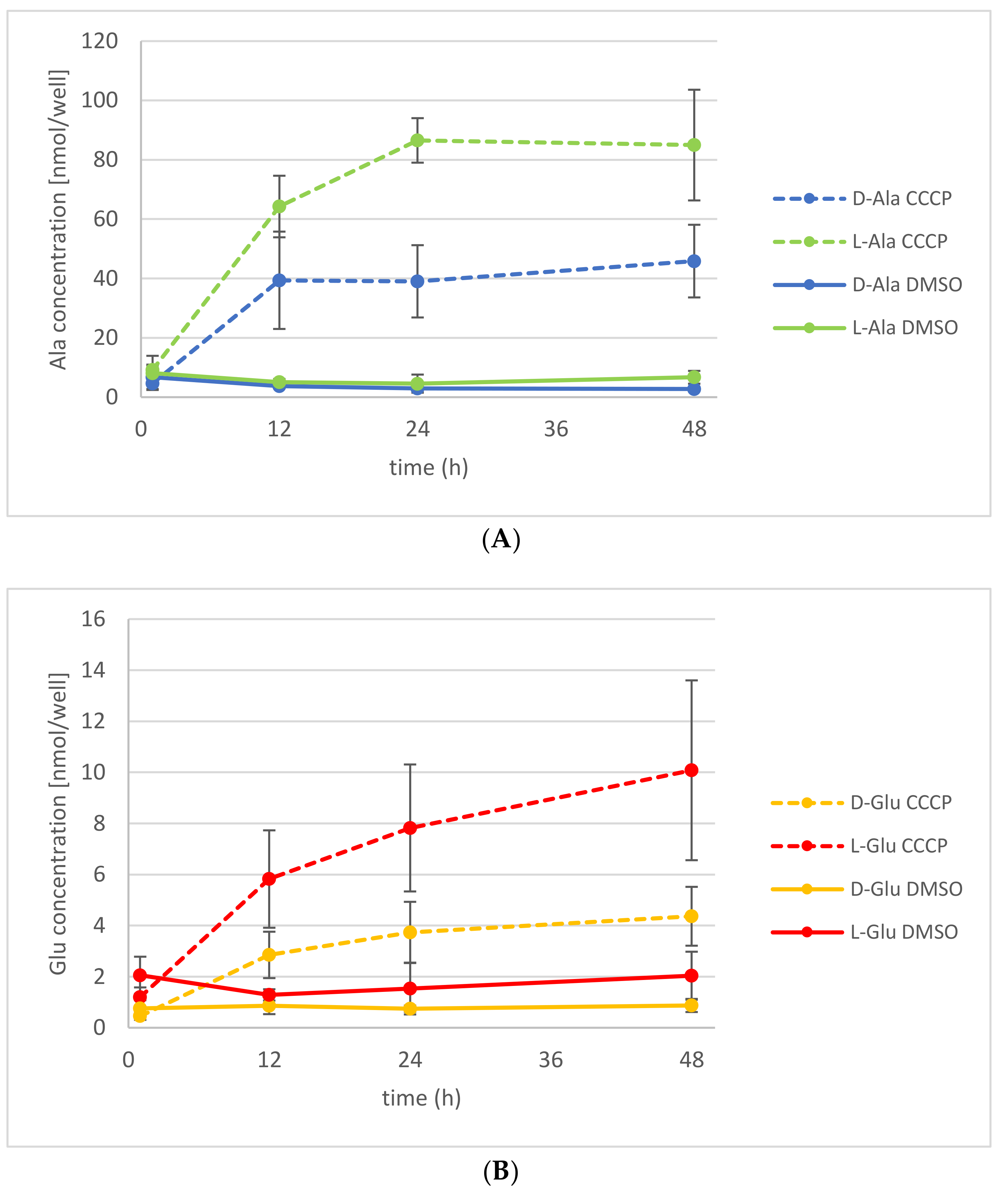
2.3. The Energetization of d-Ala Exudation
2.4. Root Exudation of Other d-AAs
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Plant Material and Growth Conditions
4.3. Extraction of Amino Acids from Seedlings and Media and Their Derivatization
4.4. LC/MS Determination of d- and l-AAs
Supplementary Materials
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2011, 10, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Ollivaux, C.; Soyez, D.; Toullec, J.Y. Biogenesis of d-amino acid containing peptides/proteins: Where, when and how? J. Pept. Sci. 2014, 20, 595–612. [Google Scholar] [CrossRef] [PubMed]
- Balu, D.T.; Coyle, J.T. The NMDA receptor ‘glycine modulatory site’ in schizophrenia: d-serine, glycine, and beyond. Curr. Opin. Pharmacol. 2015, 20, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, S.B.; Cava, F. Environmental roles of microbial amino acid racemases. Environ. Microbiol. 2016, 18, 1673–1685. [Google Scholar] [CrossRef] [PubMed]
- Sacchi, S. d-Serine metabolism: New insights into the modulation of d-amino acid oxidase activity. Biochem. Soc. Trans. 2013, 41, 1551–1556. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, M.M.; Santillo, A.; Chieffi Baccari, G. Current knowledge of d-aspartate in glandular tissues. Amino Acids 2014, 46, 1805–1818. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M. Origin, microbiology, nutrition, and pharmacology of d-amino acids. Chem. Biodivers. 2010, 7, 1491–1530. [Google Scholar] [CrossRef] [PubMed]
- Gordes, D.; Kolukisaoglu, U.; Thurow, K. Uptake and conversion of d-amino acids in Arabidopsis thaliana. Amino Acids 2011, 40, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Erikson, O.; Hertzberg, M.; Nasholm, T. A conditional marker gene allowing both positive and negative selection in plants. Nat. Biotechnol. 2004, 22, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Abe, K.; Kera, Y. Bacterial d-amino acid oxidases: Recent findings and future perspectives. Bioengineered 2015, 6, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Molla, G. Competitive Inhibitors Unveil Structure/Function Relationships in Human d-Amino Acid Oxidase. Front. Mol. Biosci. 2017, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G. An overview on d-amino acids. Amino Acids 2017, 9, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Nasholm, T.; Kielland, K.; Ganeteg, U. Uptake of organic nitrogen by plants. New Phytol. 2009, 182, 31–48. [Google Scholar] [CrossRef] [PubMed]
- Hill, P.W.; Quilliam, R.S.; DeLuca, T.H.; Farrar, J.; Farrell, M.; Roberts, P.; Newsham, K.K.; Hopkins, D.W.; Bardgett, R.D.; Jones, D.L. Acquisition and assimilation of nitrogen as peptide-bound and d-enantiomers of amino acids by wheat. PLoS ONE 2011, 6, e19220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michard, E.; Lima, P.T.; Borges, F.; Silva, A.C.; Portes, M.T.; Carvalho, J.E.; Gilliham, M.; Liu, L.H.; Obermeyer, G.; Feijo, J.A. Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil d-serine. Science 2011, 332, 434–437. [Google Scholar] [CrossRef] [PubMed]
- Monselise, E.B.; Levkovitz, A.; Kost, D. Ultraviolet radiation induces stress in etiolated Landoltia punctata, as evidenced by the presence of alanine, a universal stress signal: A 15N NMR study. Plant. Biol. (Stuttg.) 2015, 17 (Suppl. 1), 101–107. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Tanidokoro, K.; Shimizu, Y.; Kawarabayasi, Y.; Ohshima, T.; Sato, M.; Tadano, S.; Ishikawa, H.; Takio, S.; Takechi, K.; et al. Moss Chloroplasts Are Surrounded by a Peptidoglycan Wall Containing d-Amino Acids. Plant Cell 2016, 28, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Vranova, V.; Zahradnickova, H.; Janous, D.; Skene, K.R.; Matharu, A.S.; Rejsek, K.; Formanek, P. The significance of d-amino acids in soil, fate and utilization by microbes and plants: Review and identification of knowledge gaps. Plant Soil 2012, 354, 21–39. [Google Scholar] [CrossRef]
- Radkov, A.D.; McNeill, K.; Uda, K.; Moe, L.A. d-Amino acid catabolism is common among soil-dwelling bacteria. Microbes Environ. 2016, 31, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Gordes, D.; Koch, G.; Thurow, K.; Kolukisaoglu, U. Analyses of Arabidopsis ecotypes reveal metabolic diversity to convert d-amino acids. Springerplus 2013, 2, 559. [Google Scholar] [CrossRef] [PubMed]
- Svennerstam, H.; Ganeteg, U.; Bellini, C.; Nasholm, T. Comprehensive screening of Arabidopsis mutants suggests the lysine histidine transporter 1 to be involved in plant uptake of amino acids. Plant Physiol. 2007, 143, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, S.; Gumy, C.; Blatter, E.; Boeffel, S.; Fricke, W.; Rentsch, D. In planta function of compatible solute transporters of the AtProT family. J. Exp. Bot. 2011, 62, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Bruckner, H.; Westhauser, T. Chromatographic determination of l- and d-amino acids in plants. Amino Acids 2003, 24, 43–55. [Google Scholar] [PubMed]
- Fujitani, Y.; Nakajima, N.; Ishihara, K.; Oikawa, T.; Ito, K.; Sugimoto, M. Molecular and biochemical characterization of a serine racemase from Arabidopsis thaliana. Phytochemistry 2006, 67, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Suarez, J.; Hummel, S.; Hener, C.; Stahl, M.; Kolukisaoglu, Ü. AtDAT1 plays a major role in d-amino acid metabolism of Arabidopsis thaliana. New Phytol. 2018. under review. [Google Scholar]
- Kolukisaoglu, Ü.; Suarez, J. d-Amino Acids in Plants: New Insights and Aspects, but also More Open Questions. In Amino Acid-New Insights and Roles in Plant and Animal; InTech: London, UK, 2017. [Google Scholar]
- Funakoshi, M.; Sekine, M.; Katane, M.; Furuchi, T.; Yohda, M.; Yoshikawa, T.; Homma, H. Cloning and functional characterization of Arabidopsis thaliana d-amino acid aminotransferase—d-aspartate behavior during germination. FEBS J. 2008, 275, 1188–1200. [Google Scholar] [CrossRef] [PubMed]
- Badri, D.V.; Vivanco, J.M. Regulation and function of root exudates. Plant Cell Environ. 2009, 32, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Badri, D.V.; Quintana, N.; El Kassis, E.G.; Kim, H.K.; Choi, Y.H.; Sugiyama, A.; Verpoorte, R.; Martinoia, E.; Manter, D.K.; Vivanco, J.M. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 2009, 151, 2006–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Huang, X.-F.; Chaparro, J.M.; Badri, D.V.; Manter, D.K.; Vivanco, J.M.; Guo, J. Root and bacterial secretions regulate the interaction between plants and PGPR leading to distinct plant growth promotion effects. Plant Soil 2016, 401, 259–272. [Google Scholar] [CrossRef]
- Nishimura, K.; Tomoda, Y.; Nakamoto, Y.; Kawada, T.; Ishii, Y.; Nagata, Y. Alanine racemase from the green alga Chlamydomonas reinhardtii. Amino Acids 2007, 32, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Yanagida, K.; Oikawa, T.; Ogawa, T.; Soda, K. Alanine racemase of alfalfa seedlings (Medicago sativa L.): First evidence for the presence of an amino acid racemase in plants. Phytochemistry 2006, 67, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, A.; Kohnehrouz, B. Molecular cloning and expression in Escherichia coli of an active fused Zea mays L. d-amino acid oxidase. Biochemistry (Moscow) 2009, 74, 137–144. [Google Scholar] [CrossRef]
- Baetz, U.; Martinoia, E. Root exudates: The hidden part of plant defense. Trends Plant Sci. 2014, 19, 90–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, A.; Shitan, N.; Yazaki, K. Involvement of a soybean ATP-binding cassette-type transporter in the secretion of genistein, a signal flavonoid in legume-Rhizobium symbiosis. Plant Physiol. 2007, 144, 2000–2008. [Google Scholar] [CrossRef] [PubMed]
- Lesuffleur, F.; Cliquet, J.B. Characterisation of root amino acid exudation in white clover (Trifolium repens L.). Plant Soil 2010, 333, 191–201. [Google Scholar] [CrossRef]
- Hirner, A.; Ladwig, F.; Stransky, H.; Okumoto, S.; Keinath, M.; Harms, A.; Frommer, W.B.; Koch, W. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. Plant Cell 2006, 18, 1931–1946. [Google Scholar] [CrossRef] [PubMed]
- Okumoto, S.; Pilot, G. Amino Acid Export in Plants: A Missing Link in Nitrogen Cycling. Mol. Plant 2011, 4, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Dinkeloo, K.; Boyd, S.; Pilot, G. Update on amino acid transporter functions and on possible amino acid sensing mechanisms in plants. Semin. Cell Dev. Biol. 2018, 74, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interations with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.S.; Sun, H.J. Racemization in Reverse: Evidence that d-Amino Acid Toxicity on Earth Is Controlled by Bacteria with Racemases. PLoS ONE 2014, 9, e92101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Zheng, Q.; Jiao, N.Z. Microbial d-amino acids and marine carbon storage. Sci. China Earth Sci. 2016, 59, 17–24. [Google Scholar] [CrossRef]
- Semchenko, M.; Saar, S.; Lepik, A. Plant root exudates mediate neighbour recognition and trigger complex behavioural changes. New Phytol. 2014, 204, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Bertin, C.; Weston, L.A.; Huang, T.; Jander, G.; Owens, T.; Meinwald, J.; Schroeder, F.C. Grass roots chemistry: Meta-tyrosine, an herbicidal nonprotein amino acid. Proc. Natl. Acad. Sci. USA 2007, 104, 16964–16969. [Google Scholar] [CrossRef] [PubMed]




| d-AA Treated Plants | Control Plants | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| d | l | d | l | ||||||
| mean | (±SD) | mean | (±SD) | mean | (±SD) | mean | (±SD) | ||
| 12 h | Aspartate | 1.38 | (±0.30) | 2.15 | (±0.35) | 0.01 | (±0.01) | 1.85 | (±0.39) |
| Leucine | 0.32 | (±0.03) | 0.28 | (±0.21) | 0.01 | (±0.01) | 0.14 | (±0.13) | |
| Lysine | 1.46 | (±1.03) | 0.03 | (±0.02) | 0.00 | (±0.00) | 0.00 | (±0.00) | |
| Phenylalanine | 0.46 | (±0.10) | 0.00 | (±0.00) | 0.00 | (±0.00) | 0.00 | (±0.00) | |
| Proline | 2.44 | (±0.62) | 0.18 | (±0.02) | 0.76 | (±0.16) | 0.29 | (±0.06) | |
| 72 h | Aspartate | 0.77 | (±0.08) | 5.54 | (±1.51) | 0.03 | (±0.01) | 5.90 | (±0.53) |
| Leucine | 0.12 | (±0.09) | 0.00 | (±0.00) | 0.00 | (±0.00) | 0.00 | (±0.00) | |
| Lysine | 0.20 | (±0.16) | 0.00 | (±0.00) | 0.00 | (±0.00) | 0.00 | (±0.00) | |
| Phenylalanine | 0.06 | (±0.06) | 0.00 | (±0.00) | 0.00 | (±0.00) | 0.00 | (±0.00) | |
| Proline | 2.48 | (±0.27) | 0.01 | (±0.01) | 1.27 | (±0.09) | 0.09 | (±0.06) | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hener, C.; Hummel, S.; Suarez, J.; Stahl, M.; Kolukisaoglu, Ü. d-Amino Acids Are Exuded by Arabidopsis thaliana Roots to the Rhizosphere. Int. J. Mol. Sci. 2018, 19, 1109. https://doi.org/10.3390/ijms19041109
Hener C, Hummel S, Suarez J, Stahl M, Kolukisaoglu Ü. d-Amino Acids Are Exuded by Arabidopsis thaliana Roots to the Rhizosphere. International Journal of Molecular Sciences. 2018; 19(4):1109. https://doi.org/10.3390/ijms19041109
Chicago/Turabian StyleHener, Claudia, Sabine Hummel, Juan Suarez, Mark Stahl, and Üner Kolukisaoglu. 2018. "d-Amino Acids Are Exuded by Arabidopsis thaliana Roots to the Rhizosphere" International Journal of Molecular Sciences 19, no. 4: 1109. https://doi.org/10.3390/ijms19041109