Cell-Specific PEAR1 Methylation Studies Reveal a Locus that Coordinates Expression of Multiple Genes
Abstract
:1. Introduction
2. Results
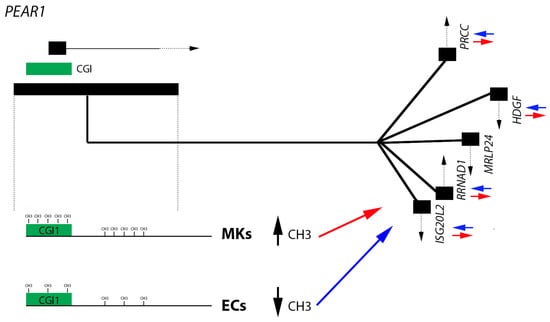
2.1. PEAR1 Hypermethylation in Megakaryocytes but Not Endothelial Cells
2.2. PEAR1 Associated Long-Distance Chromosomal Interactions in Endothelial Cells and Megakaryocytes
2.3. Gene Methylation and Expression Profile of PEAR1-Interacting Genes in Endothelial Cells and Megakaryocytes
3. Discussion
4. Material and Methods
4.1. Human HUVECs, BOECs and CD34+ Stem Cell Isolation and Differentiation In Vitro
4.2. DNA Methylation Analysis
4.3. CHiCP Analysis
4.4. Gene Expression Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bulger, M.; Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 2011, 144, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Lopes, S.; Lewis, A.; Hajkova, P.; Dean, W.; Oswald, J.; Forne, T.; Murrell, A.; Constancia, M.; Bartolomei, M.; Walter, J.; et al. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum. Mol. Genet. 2003, 12, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Ishihara, K.; Kato, R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J. Biochem. 2000, 127, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Hagege, H.; Murrell, A.; Brunel, C.; Reik, W.; Cathala, G.; Forne, T. Genomic imprinting controls matrix attachment regions in the Igf2 gene. Mol. Cell. Biol. 2003, 23, 8953–8959. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, J.J.; Cox, D.; Summerbell, D.; Rigby, P.W. A BAC transgenic analysis of the Mrf4/Myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Development 2001, 128, 1857–1868. [Google Scholar] [PubMed]
- Carter, D.; Chakalova, L.; Osborne, C.S.; Dai, Y.F.; Fraser, P. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 2002, 32, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Spitz, F.; Gonzalez, F.; Duboule, D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 2003, 113, 405–417. [Google Scholar] [CrossRef]
- Sagai, T.; Hosoya, M.; Mizushina, Y.; Tamura, M.; Shiroishi, T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development 2005, 132, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.; El-Jaick, K.; Roessler, E.; Muenke, M.; Epstein, D.J. A functional screen for sonic hedgehog regulatory elements across a 1 Mb interval identifies long-range ventral forebrain enhancers. Development 2006, 133, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, L.A.; Ahituv, N.; Moses, A.M.; Prabhakar, S.; Nobrega, M.A.; Shoukry, M.; Minovitsky, S.; Dubchak, I.; Holt, A.; Lewis, K.D.; et al. In Vivo enhancer analysis of human conserved non-coding sequences. Nature 2006, 444, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Ragoczy, T.; Telling, A.; Sawado, T.; Groudine, M.; Kosak, S.T. A genetic analysis of chromosome territory looping: Diverse roles for distal regulatory elements. Chromosome Res. 2003, 11, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Francastel, C.; Walters, M.C.; Groudine, M.; Martin, D.I. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell 1999, 99, 259–269. [Google Scholar] [CrossRef]
- Schubeler, D.; Francastel, C.; Cimbora, D.M.; Reik, A.; Martin, D.I.; Groudine, M. Nuclear localization and histone acetylation: A pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000, 14, 940–950. [Google Scholar] [PubMed]
- Gerasimova, T.I.; Byrd, K.; Corces, V.G. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 2000, 6, 1025–1035. [Google Scholar] [CrossRef]
- Carmo-Fonseca, M.; Platani, M.; Swedlow, J.R. Macromolecular mobility inside the cell nucleus. Trends Cell Biol. 2002, 12, 491–495. [Google Scholar] [CrossRef]
- Carmo-Fonseca, M. The contribution of nuclear compartmentalization to gene regulation. Cell 2002, 108, 513–521. [Google Scholar] [CrossRef]
- Phillips-Cremins, J.E.; Sauria, M.E.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.; Ong, C.T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Hadjur, S.; Williams, L.M.; Ryan, N.K.; Cobb, B.S.; Sexton, T.; Fraser, P.; Fisher, A.G.; Merkenschlager, M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 2009, 460, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Mishiro, T.; Ishihara, K.; Hino, S.; Tsutsumi, S.; Aburatani, H.; Shirahige, K.; Kinoshita, Y.; Nakao, M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009, 28, 1234–1245. [Google Scholar] [CrossRef] [PubMed]
- Nativio, R.; Wendt, K.S.; Ito, Y.; Huddleston, J.E.; Uribe-Lewis, S.; Woodfine, K.; Krueger, C.; Reik, W.; Peters, J.M.; Murrell, A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009, 5, e1000739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Handoko, L.; Xu, H.; Li, G.; Ngan, C.Y.; Chew, E.; Schnapp, M.; Lee, C.W.; Ye, C.; Ping, J.L.; Mulawadi, F.; et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011, 43, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Programming of DNA methylation patterns. Annu. Rev. Biochem. 2012, 81, 97–117. [Google Scholar] [CrossRef] [PubMed]
- Unoki, M.; Nakamura, Y. Methylation at CpG islands in intron 1 of EGR2 confers enhancer-like activity. FEBS Lett. 2003, 554, 67–72. [Google Scholar] [CrossRef]
- Shen, Y.; Takahashi, M.; Byun, H.M.; Link, A.; Sharma, N.; Balaguer, F.; Leung, H.C.; Boland, C.R.; Goel, A. Boswellic acid induces epigenetic alterations by modulating DNA methylation in colorectal cancer cells. Cancer Biol. Ther. 2012, 13, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Pope, S.D.; Jazirehi, A.R.; Attema, J.L.; Papathanasiou, P.; Watts, J.A.; Zaret, K.S.; Weissman, I.L.; Smale, S.T. Pioneer factor interactions and unmethylated CpG dinucleotides mark silent tissue-specific enhancers in embryonic stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 12377–12382. [Google Scholar] [CrossRef] [PubMed]
- Uhm, T.G.; Lee, S.K.; Kim, B.S.; Kang, J.H.; Park, C.S.; Rhim, T.Y.; Chang, H.S.; Kim do, J.; Chung, I.Y. CpG methylation at GATA elements in the regulatory region of CCR3 positively correlates with CCR3 transcription. Exp. Mol. Med. 2012, 44, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Kurukuti, S.; Tiwari, V.K.; Tavoosidana, G.; Pugacheva, E.; Murrell, A.; Zhao, Z.; Lobanenkov, V.; Reik, W.; Ohlsson, R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA 2006, 103, 10684–10689. [Google Scholar] [CrossRef] [PubMed]
- Court, F.; Camprubi, C.; Garcia, C.V.; Guillaumet-Adkins, A.; Sparago, A.; Seruggia, D.; Sandoval, J.; Esteller, M.; Martin-Trujillo, A.; Riccio, A.; et al. The PEG13-DMR and brain-specific enhancers dictate imprinted expression within the 8q24 intellectual disability risk locus. Epigenet. Chromatin 2014, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Sabato, S.; Hellman, A. DNA methylation of distal regulatory sites characterizes dysregulation of cancer genes. Genome Biol. 2013, 14, R21. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ge, B.; Casale, F.P.; Vasquez, L.; Kwan, T.; Garrido-Martin, D.; Watt, S.; Yan, Y.; Kundu, K.; Ecker, S.; et al. Genetic Drivers of Epigenetic and Transcriptional Variation in Human Immune Cells. Cell 2016, 167, 1398.e24–1414.e24. [Google Scholar] [CrossRef] [PubMed]
- Javierre, B.M.; Burren, O.S.; Wilder, S.P.; Kreuzhuber, R.; Hill, S.M.; Sewitz, S.; Cairns, J.; Wingett, S.W.; Varnai, C.; Thiecke, M.J.; et al. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell 2016, 167, 1369.e19–1384.e19. [Google Scholar] [CrossRef] [PubMed]
- The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Stunnenberg, H.G.; International Human Epigenome, C.; Hirst, M. The International Human Epigenome Consortium: A Blueprint for Scientific Collaboration and Discovery. Cell 2016, 167, 1145–1149. [Google Scholar] [CrossRef] [PubMed]
- Davison, L.J.; Wallace, C.; Cooper, J.D.; Cope, N.F.; Wilson, N.K.; Smyth, D.J.; Howson, J.M.; Saleh, N.; Al-Jeffery, A.; Angus, K.L.; et al. Long-range DNA looping and gene expression analyses identify DEXI as an autoimmune disease candidate gene. Hum. Mol. Genet. 2012, 21, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Dryden, N.H.; Broome, L.R.; Dudbridge, F.; Johnson, N.; Orr, N.; Schoenfelder, S.; Nagano, T.; Andrews, S.; Wingett, S.; Kozarewa, I.; et al. Unbiased analysis of potential targets of breast cancer susceptibility loci by Capture Hi-C. Genome Res. 2014, 24, 1854–1868. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; McGovern, A.; Orozco, G.; Duffus, K.; Yarwood, A.; Schoenfelder, S.; Cooper, N.J.; Barton, A.; Wallace, C.; Fraser, P.; et al. Capture Hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci. Nat. Commun. 2015, 6, 10069. [Google Scholar] [CrossRef] [PubMed]
- Mifsud, B.; Tavares-Cadete, F.; Young, A.N.; Sugar, R.; Schoenfelder, S.; Ferreira, L.; Wingett, S.W.; Andrews, S.; Grey, W.; Ewels, P.A.; et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015, 47, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Smemo, S.; Tena, J.J.; Kim, K.H.; Gamazon, E.R.; Sakabe, N.J.; Gomez-Marin, C.; Aneas, I.; Credidio, F.L.; Sobreira, D.R.; Wasserman, N.F.; et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 2014, 507, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Stadhouders, R.; Aktuna, S.; Thongjuea, S.; Aghajanirefah, A.; Pourfarzad, F.; van Ijcken, W.; Lenhard, B.; Rooks, H.; Best, S.; Menzel, S.; et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Investig. 2014, 124, 1699–1710. [Google Scholar] [CrossRef] [PubMed]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–5119. [Google Scholar] [CrossRef] [PubMed]
- Kauskot, A.; Di Michele, M.; Loyen, S.; Freson, K.; Verhamme, P.; Hoylaerts, M.F. A novel mechanism of sustained platelet alphaIIbbeta3 activation via PEAR1. Blood 2012, 119, 4056–4065. [Google Scholar] [CrossRef] [PubMed]
- Kauskot, A.; Vandenbriele, C.; Louwette, S.; Gijsbers, R.; Tousseyn, T.; Freson, K.; Verhamme, P.; Hoylaerts, M.F. PEAR1 attenuates megakaryopoiesis via control of the PI3K/PTEN pathway. Blood 2013, 121, 5208–5217. [Google Scholar] [CrossRef] [PubMed]
- Vandenbriele, C.; Kauskot, A.; Vandersmissen, I.; Criel, M.; Geenens, R.; Craps, S.; Luttun, A.; Janssens, S.; Hoylaerts, M.F.; Verhamme, P. Platelet endothelial aggregation receptor-1: A novel modifier of neoangiogenesis. Cardiovasc. Res. 2015. [Google Scholar] [CrossRef] [PubMed]
- Faraday, N.; Yanek, L.R.; Yang, X.P.; Mathias, R.; Herrera-Galeano, J.E.; Suktitipat, B.; Qayyum, R.; Johnson, A.D.; Chen, M.H.; Tofler, G.H.; et al. Identification of a specific intronic PEAR1 gene variant associated with greater platelet aggregability and protein expression. Blood 2011, 118, 3367–3375. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.P.; Ryan, K.; O’Connell, J.R.; Horenstein, R.B.; Damcott, C.M.; Gibson, Q.; Pollin, T.I.; Mitchell, B.D.; Beitelshees, A.L.; Pakzy, R.; et al. Genetic variation in PEAR1 is associated with platelet aggregation and cardiovascular outcomes. Circ. Cardiovasc. Genet. 2013, 6, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Suktitipat, B.; Yanek, L.R.; Faraday, N.; Wilson, A.F.; Becker, D.M.; Becker, L.C.; Mathias, R.A. Targeted deep resequencing identifies coding variants in the PEAR1 gene that play a role in platelet aggregation. PLoS ONE 2013, 8, e64179. [Google Scholar] [CrossRef] [PubMed]
- Wurtz, M.; Nissen, P.H.; Grove, E.L.; Kristensen, S.D.; Hvas, A.M. Genetic determinants of on-aspirin platelet reactivity: Focus on the influence of PEAR1. PLoS ONE 2014, 9, e111816. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, R.; Becker, L.C.; Becker, D.M.; Faraday, N.; Yanek, L.R.; Leal, S.M.; Shaw, C.; Mathias, R.; Suktitipat, B.; Bray, P.F. Genome-wide association study of platelet aggregation in African Americans. BMC Genet. 2015, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.L.; Zhao, Y.Q.; Zhou, Z.Y.; Jin, J.; Zhao, M.; Chen, X.M.; Chen, L.Y.; Cai, Y.F.; Li, J.L.; Huang, M. Associations of MDR1, TBXA2R, PLA2G7 and PEAR1 genetic polymorphisms with the platelet activity in Chinese ischemic stroke patients receiving aspirin therapy. Acta Pharmacol. Sin. 2016, 37, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- Backman, J.D.; Yerges-Armstrong, L.M.; Horenstein, R.B.; Newcomer, S.; Shaub, S.; Morrisey, M.; Donnelly, P.; Drolet, M.; Tanner, K.; Pavlovich, M.A.; et al. Prospective Evaluation of Genetic Variation in Platelet Endothelial Aggregation Receptor 1 Reveals Aspirin-Dependent Effects on Platelet Aggregation Pathways. Clin. Transl. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hu, Y.; Wen, Z.; Li, H.; Hu, X.; Zhang, Y.; Zhang, Z.; Xiao, J.; Tang, J.; Chen, X. Association of PEAR1 rs12041331 polymorphism and pharmacodynamics of ticagrelor in healthy Chinese volunteers. Xenobiotica 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Izzi, B.; Pistoni, M.; Cludts, K.; Akkor, P.; Lambrechts, D.; Verfaillie, C.; Verhamme, P.; Freson, K.; Hoylaerts, M.F. Allele-specific DNA methylation reinforces PEAR1 enhancer activity. Blood 2016, 128, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Eicher, J.D.; Chami, N.; Kacprowski, T.; Nomura, A.; Chen, M.H.; Yanek, L.R.; Tajuddin, S.M.; Schick, U.M.; Slater, A.J.; Pankratz, N.; et al. Platelet-Related Variants Identified by Exomechip Meta-analysis in 157,293 Individuals. Am. J. Hum. Genet. 2016, 99, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Yanek, L.R.; Backman, J.D.; Eicher, J.D.; Huffman, J.E.; Ben-Shlomo, Y.; Beswick, A.D.; Yerges-Armstrong, L.M.; Shuldiner, A.R.; O’Connell, J.R.; et al. Exome-chip meta-analysis identifies association between variation in ANKRD26 and platelet aggregation. Platelets 2017, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.D. Pairing megakaryopoiesis methylation with PEAR1. Blood 2016, 128, 890–892. [Google Scholar] [CrossRef] [PubMed]
- Calo, E.; Wysocka, J. Modification of enhancer chromatin: What, how and why? Mol. Cell 2013, 49, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed]
- Pekowska, A.; Benoukraf, T.; Zacarias-Cabeza, J.; Belhocine, M.; Koch, F.; Holota, H.; Imbert, J.; Andrau, J.C.; Ferrier, P.; Spicuglia, S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011, 30, 4198–4210. [Google Scholar] [CrossRef] [PubMed]
- Schofield, E.C.; Carver, T.; Achuthan, P.; Freire-Pritchett, P.; Spivakov, M.; Todd, J.A.; Burren, O.S. CHiCP: A web-based tool for the integrative and interactive visualization of promoter capture Hi-C datasets. Bioinformatics 2016, 32, 2511–2513. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.; Lambourne, J.J.; Javierre, B.M.; Grassi, L.; Kreuzhuber, R.; Ruklisa, D.; Rosa, I.M.; Tome, A.R.; Elding, H.; van Geffen, J.P.; et al. Platelet function is modified by common sequence variation in megakaryocyte super enhancers. Nat. Commun. 2017, 8, 16058. [Google Scholar] [CrossRef] [PubMed]
- Coute, Y.; Kindbeiter, K.; Belin, S.; Dieckmann, R.; Duret, L.; Bezin, L.; Sanchez, J.C.; Diaz, J.J. ISG20L2, a novel vertebrate nucleolar exoribonuclease involved in ribosome biogenesis. Mol. Cell. Proteomics MCP 2008, 7, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Efroni, S.; Duttagupta, R.; Cheng, J.; Dehghani, H.; Hoeppner, D.J.; Dash, C.; Bazett-Jones, D.P.; Le Grice, S.; McKay, R.D.; Buetow, K.H.; et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2008, 2, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Eckfeldt, C.E.; Mendenhall, E.M.; Verfaillie, C.M. The molecular repertoire of the ‘almighty’ stem cell. Nat. Rev. Mol. Cell Biol. 2005, 6, 726–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arney, K.L.; Fisher, A.G. Epigenetic aspects of differentiation. J. Cell Sci. 2004, 117, 4355–4363. [Google Scholar] [CrossRef] [PubMed]
- Shetty, A.; Dasari, S.; Banerjee, S.; Gheewala, T.; Zheng, G.; Chen, A.; Kajdacsy-Balla, A.; Bosland, M.C.; Munirathinam, G. Hepatoma-derived growth factor: A survival-related protein in prostate oncogenesis and a potential target for vitamin K2. Urol. Oncol. 2016, 34, 483.e1–483.e8. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.Y.; Zhang, A.Q.; Wang, J.; Li, C.H.; Wang, X.Q.; Pan, K.; Zhou, C.; Dong, J.H. Hepatoma-derived growth factor promotes growth and metastasis of hepatocellular carcinoma cells. Cell Biochem. Funct. 2016, 34, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Giri, K.; Pabelick, C.M.; Mukherjee, P.; Prakash, Y.S. Hepatoma derived growth factor (HDGF) dynamics in ovarian cancer cells. Apoptosis 2016, 21, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Chen, X.; Liu, N.; Wang, Z.; Miao, B.; Gan, W.; Li, D.; Guo, H. PRCC-TFE3 dual-fusion FISH assay: A new method for identifying PRCC-TFE3 renal cell carcinoma in paraffin-embedded tissue. PLoS ONE 2017, 12, e0185337. [Google Scholar] [CrossRef] [PubMed]
- Medendorp, K.; van Groningen, J.J.; Vreede, L.; Hetterschijt, L.; Brugmans, L.; van den Hurk, W.H.; van Kessel, A.G. The renal cell carcinoma-associated oncogenic fusion protein PRCCTFE3 provokes p21 WAF1/CIP1-mediated cell cycle delay. Exp. Cell Res. 2009, 315, 2399–2409. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, M.E.; Wang, W.; Chen, X.; Ji, Y.; Shakya, A.; Shen, C.; Zhang, C.; Gonzalez, V.; Brewer, M.; Ma, J.X.; et al. The regulatory role of hepatoma-derived growth factor as an angiogenic factor in the eye. Mol. Vis. 2016, 22, 374–386. [Google Scholar] [PubMed]
- Zhao, W.Y.; Wang, Y.; An, Z.J.; Shi, C.G.; Zhu, G.A.; Wang, B.; Lu, M.Y.; Pan, C.K.; Chen, P. Downregulation of miR-497 promotes tumor growth and angiogenesis by targeting HDGF in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2013, 435, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Z.; Zhao, Y.B.; Cao, W.D.; Qu, Y.; Luo, P.; Zhen, H.N.; Chen, X.Y.; Yan, Z.F.; Fei, Z. The expression of hepatoma-derived growth factor in primary central nervous system lymphoma and its correlation with angiogenesis, proliferation and clinical outcome. Med. Oncol. 2013, 30, 622. [Google Scholar] [CrossRef] [PubMed]
- Shih, T.C.; Tien, Y.J.; Wen, C.J.; Yeh, T.S.; Yu, M.C.; Huang, C.H.; Lee, Y.S.; Yen, T.C.; Hsieh, S.Y. MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J. Hepatol. 2012, 57, 584–591. [Google Scholar] [CrossRef] [PubMed]
- Thirant, C.; Galan-Moya, E.M.; Dubois, L.G.; Pinte, S.; Chafey, P.; Broussard, C.; Varlet, P.; Devaux, B.; Soncin, F.; Gavard, J.; et al. Differential proteomic analysis of human glioblastoma and neural stem cells reveals HDGF as a novel angiogenic secreted factor. Stem Cells 2012, 30, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Fisch, A.S.; Yerges-Armstrong, L.M.; Backman, J.D.; Wang, H.; Donnelly, P.; Ryan, K.A.; Parihar, A.; Pavlovich, M.A.; Mitchell, B.D.; O’Connell, J.R.; et al. Genetic Variation in the Platelet Endothelial Aggregation Receptor 1 Gene Results in Endothelial Dysfunction. PLoS ONE 2015, 10, e0138795. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Vandenbriele, C.; Kauskot, A.; Verhamme, P.; Hoylaerts, M.F.; Wright, G.J. A human platelet receptor protein microarray identifies FcepsilonR1alpha as an activating PEAR1 ligand. Mol. Cell. Proteomics MCP 2015. [Google Scholar] [CrossRef] [PubMed]
- Vandenbriele, C.; Sun, Y.; Criel, M.; Cludts, K.; Van Kerckhoven, S.; Izzi, B.; Vanassche, T.; Verhamme, P.; Hoylaerts, M.F. Dextran sulfate triggers platelet aggregation via direct activation of PEAR1. Platelets 2016, 27, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Nanda, N.; Bao, M.; Lin, H.; Clauser, K.; Komuves, L.; Quertermous, T.; Conley, P.B.; Phillips, D.R.; Hart, M.J. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J. Biol. Chem. 2005, 280, 24680–24689. [Google Scholar] [CrossRef] [PubMed]
- Dauwe, D.; Pelacho, B.; Wibowo, A.; Walravens, A.S.; Verdonck, K.; Gillijns, H.; Caluwe, E.; Pokreisz, P.; van Gastel, N.; Carmeliet, G.; et al. Neovascularization Potential of Blood Outgrowth Endothelial Cells From Patients With Stable Ischemic Heart Failure Is Preserved. J. Am. Heart Assoc. 2016, 5, e002288. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, E.A.; Nachman, R.L.; Becker, C.G.; Minick, C.R. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J. Clin. Investig. 1973, 52, 2745–2756. [Google Scholar] [CrossRef] [PubMed]
- Van Beem, R.T.; Verloop, R.E.; Kleijer, M.; Noort, W.A.; Loof, N.; Koolwijk, P.; van der Schoot, C.E.; van Hinsbergh, V.W.; Zwaginga, J.J. Blood outgrowth endothelial cells from cord blood and peripheral blood: Angiogenesis-related characteristics in vitro. J. Thromb. Haemost. JTH 2009, 7, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Izzi, B.; Binder, A.M.; Michels, K.B. Pyrosequencing Evaluation of Widely Available Bisulfite Conversion Methods: Considerations for Application. Med. Epigenet. 2014, 2, 28–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izzi, B.; Decallonne, B.; Devriendt, K.; Bouillon, R.; Vanderschueren, D.; Levtchenko, E.; de Zegher, F.; Van den Bruel, A.; Lambrechts, D.; Van Geet, C.; et al. A new approach to imprinting mutation detection in GNAS by Sequenom EpiTYPER system. Clin. Chim. Acta 2010, 411, 2033–2039. [Google Scholar] [CrossRef] [PubMed]
- Izzi, B.; Francois, I.; Labarque, V.; Thys, C.; Wittevrongel, C.; Devriendt, K.; Legius, E.; Van den Bruel, A.; D’Hooghe, M.; Lambrechts, D.; et al. Methylation defect in imprinted genes detected in patients with an Albright’s hereditary osteodystrophy like phenotype and platelet Gs hypofunction. PLoS ONE 2012, 7, e38579. [Google Scholar] [CrossRef] [PubMed]






| PEAR1 Bait | Endothelial Precursors | Location of Target | Size Target | Name Target | Score | Genomic Context |
| chr1:156,861,611–156,869,031 (7.42 KB) | chr1:156,696,068–156,699,449 | 3.38 KB | ISG20L2 and RRNAD1 | 30.34 | protein coding | |
| chr1:156,682,729–156,685,224 | 2.50 KB | RP11-66D17.5 | 17.66 | antisense | ||
| chr1:156,707,018–156,715,899 | 8.88 KB | MRPL24 and HDGF | 13.62 | protein coding | ||
| chr1:156,715,900–156,723,106 | 7.21 KB | PRCC and HDGF | 10.71 | protein coding | ||
| chr1:156,736,011–156,736,869 | 0.86 KB | PRCC and HDGF | 10.15 | protein coding | ||
| chr1:156,557,122–156,562,660 | 5.54 KB | APOA1BP and AL365181.1 | 7.8 | protein coding and miRNA | ||
| chr1:156,582,371–156,589,766 | 7.39 KB | RP11-284F21.11 and HAPLN2 | 6.65 | lincRNA and protein coding | ||
| chr1:156,622,326–156,650,015 | 27.69 KB | RP11-284F21.7 and BCAN and NES | 6.18 | antisense and protein coding | ||
| Megakaryocytes | Location of Target | Size Target | Name Target | Score | Genomic Context | |
| chr1:156,696,068–156,699,449 | 3.38 KB | ISG20L2 and RRNAD1 | 22.64 | protein coding | ||
| chr1:156,682,729–156,685,224 | 2.50 KB | RP11-66D17.5 | 11.97 | antisense | ||
| chr1:156,736,011–156,736,869 | 0.86 KB | PRCC and HDGF | 7.86 | protein coding | ||
| chr1:156,715,900–156,723,106 | 7.21 KB | PRCC and HDGF | 6.18 | protein coding | ||
| chr1:156,707,018–156,715,899 | 8.88 KB | MRPL24 and HDGF | 5.81 | protein coding |
| Gene Name | Protein Name | Gene Ontology (GO)—Biological Processes |
|---|---|---|
| ISG20L2 | Interferon Stimulated Exonuclease Gene 20 Like 2 | rRNA processing (GO:0006364); ribosome biogenesis (GO:0042254); nucleic acid phosphodiester bond hydrolysis (GO:0090305); RNA phosphodiester bond hydrolysis, exonucelolytic (GO:0090503). |
| RRNAD1 | Ribosomal RNA Adenine Dimethylase Domain Containing 1 | rRNA methylation (GO:0031167); methylation (GO:0032259). |
| PRCC | Papillary Renal Cell Carcinoma (Translocation-Associated) | mRNA splicing, via spliceosome (GO:0000398); cell cycle (GO:0007049); mitotic cell cycle checkpoint (GO:0007093). |
| HDGF | Heparin Binding Growth Factor | negative regulation of transcription from RNA polymerase II promoter (GO:0000122); transcription, DNA templated (GO:00006351); regulation of transcription (GO:0006355); signal transduction (GO:0007165); cell proliferation (GO:0008283). |
| MRPL24 | Mitochondrial Ribosomal Protein L24 | translation (GO:0070126); Mitochondrial translation elongation(GO:0070125); mitochondrial translation termination (GO:0070126) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Izzi, B.; Noro, F.; Cludts, K.; Freson, K.; Hoylaerts, M.F. Cell-Specific PEAR1 Methylation Studies Reveal a Locus that Coordinates Expression of Multiple Genes. Int. J. Mol. Sci. 2018, 19, 1069. https://doi.org/10.3390/ijms19041069
Izzi B, Noro F, Cludts K, Freson K, Hoylaerts MF. Cell-Specific PEAR1 Methylation Studies Reveal a Locus that Coordinates Expression of Multiple Genes. International Journal of Molecular Sciences. 2018; 19(4):1069. https://doi.org/10.3390/ijms19041069
Chicago/Turabian StyleIzzi, Benedetta, Fabrizia Noro, Katrien Cludts, Kathleen Freson, and Marc F. Hoylaerts. 2018. "Cell-Specific PEAR1 Methylation Studies Reveal a Locus that Coordinates Expression of Multiple Genes" International Journal of Molecular Sciences 19, no. 4: 1069. https://doi.org/10.3390/ijms19041069