Fine Mapping Identifies SmFAS Encoding an Anthocyanidin Synthase as a Putative Candidate Gene for Flower Purple Color in Solanum melongena L.
Abstract
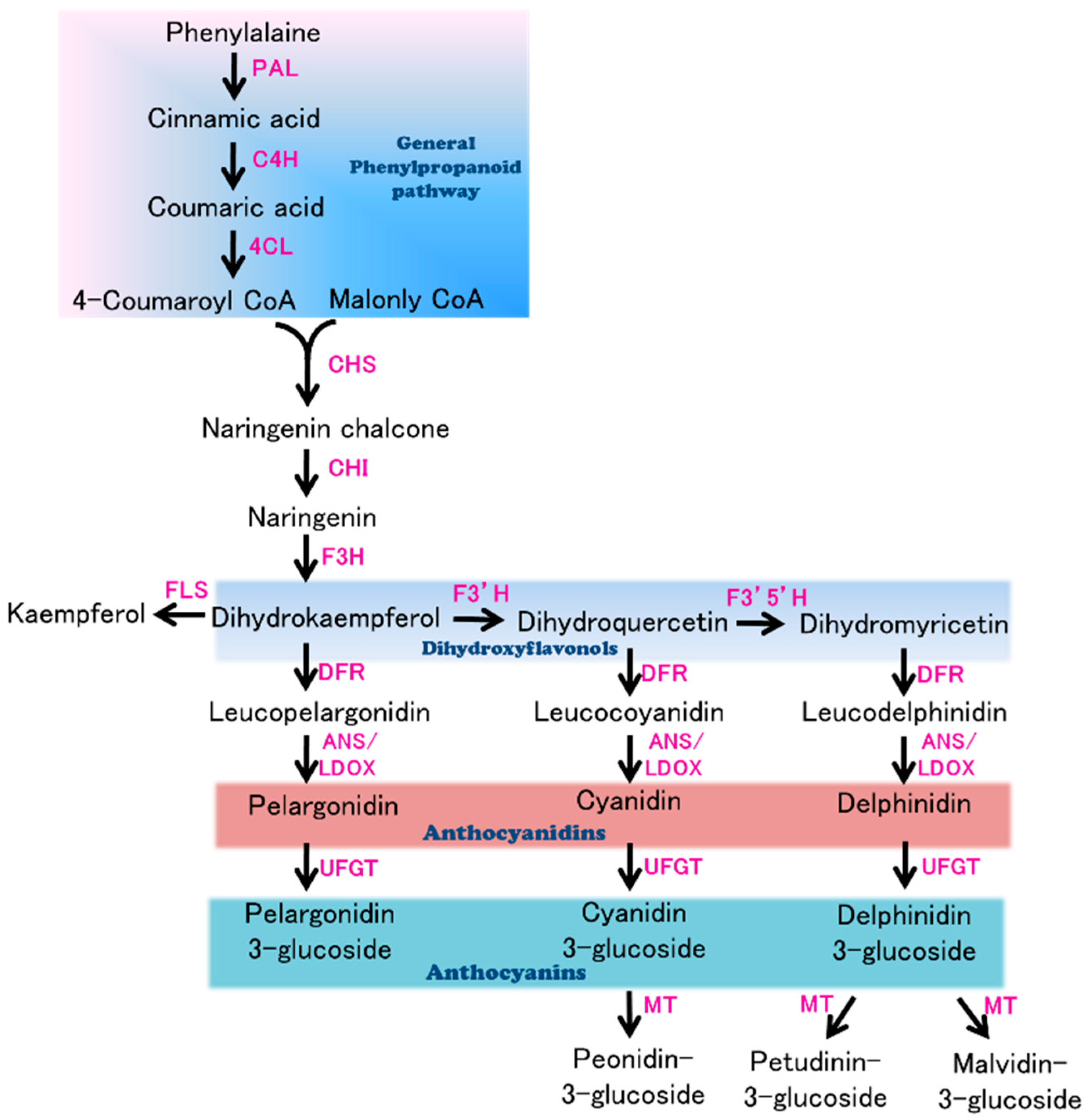
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Mapping Populations
2.2. DNA Extraction and SSR Identification
2.3. Mapping Strategy and Linkage Analysis
2.4. Candidate Gene Prediction and Identification
2.5. Quantitative Real-Time RT-PCR Analysis of Candidate Genes
3. Results
3.1. Inheritance of SmFAS
3.2. Linkage Mapping of SmFAS
3.3. Candidate Genes for Purple Flower Color in Eggplant
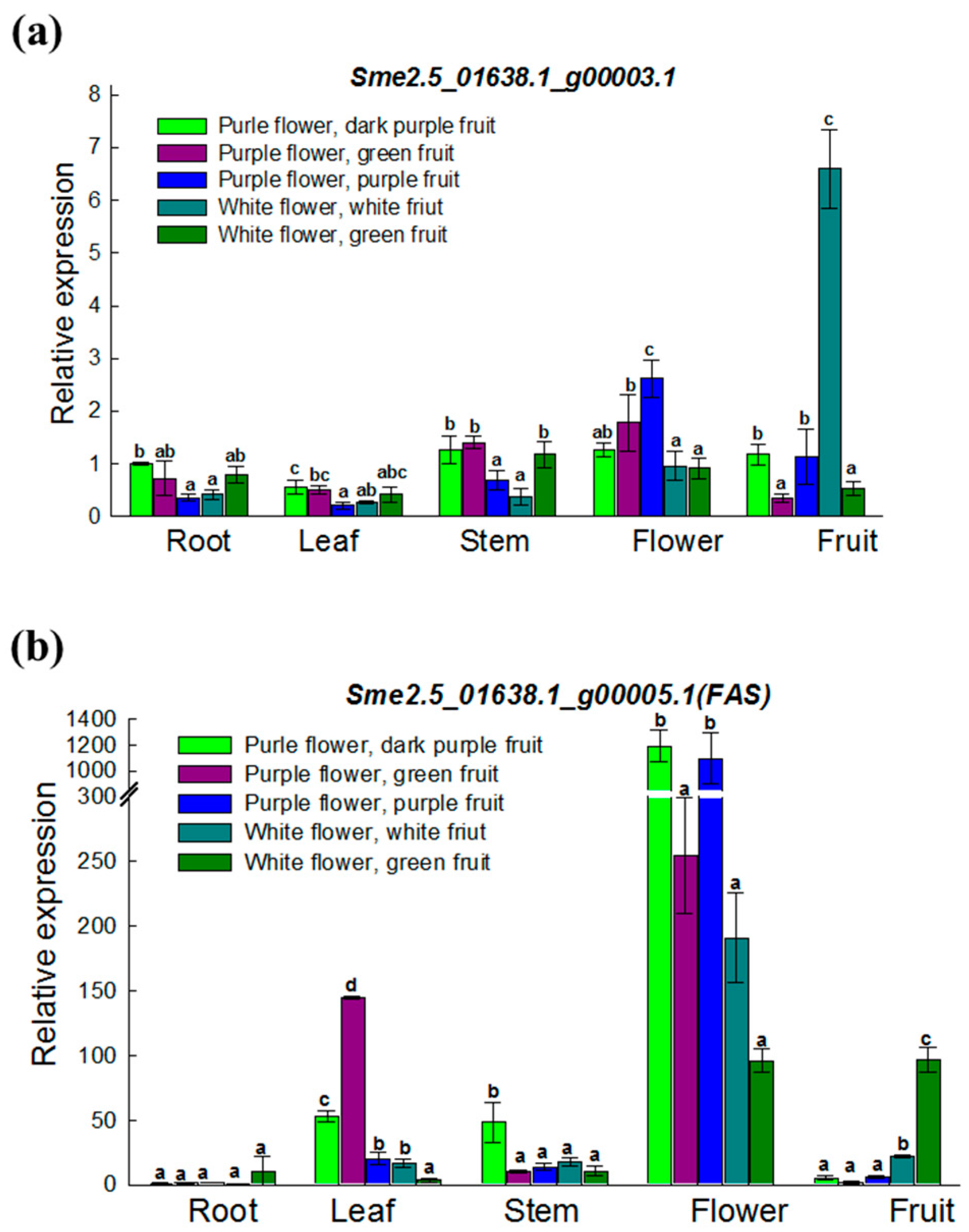
3.4. Expression Analysis of Two ANS Genes in Various Materials and Tissues
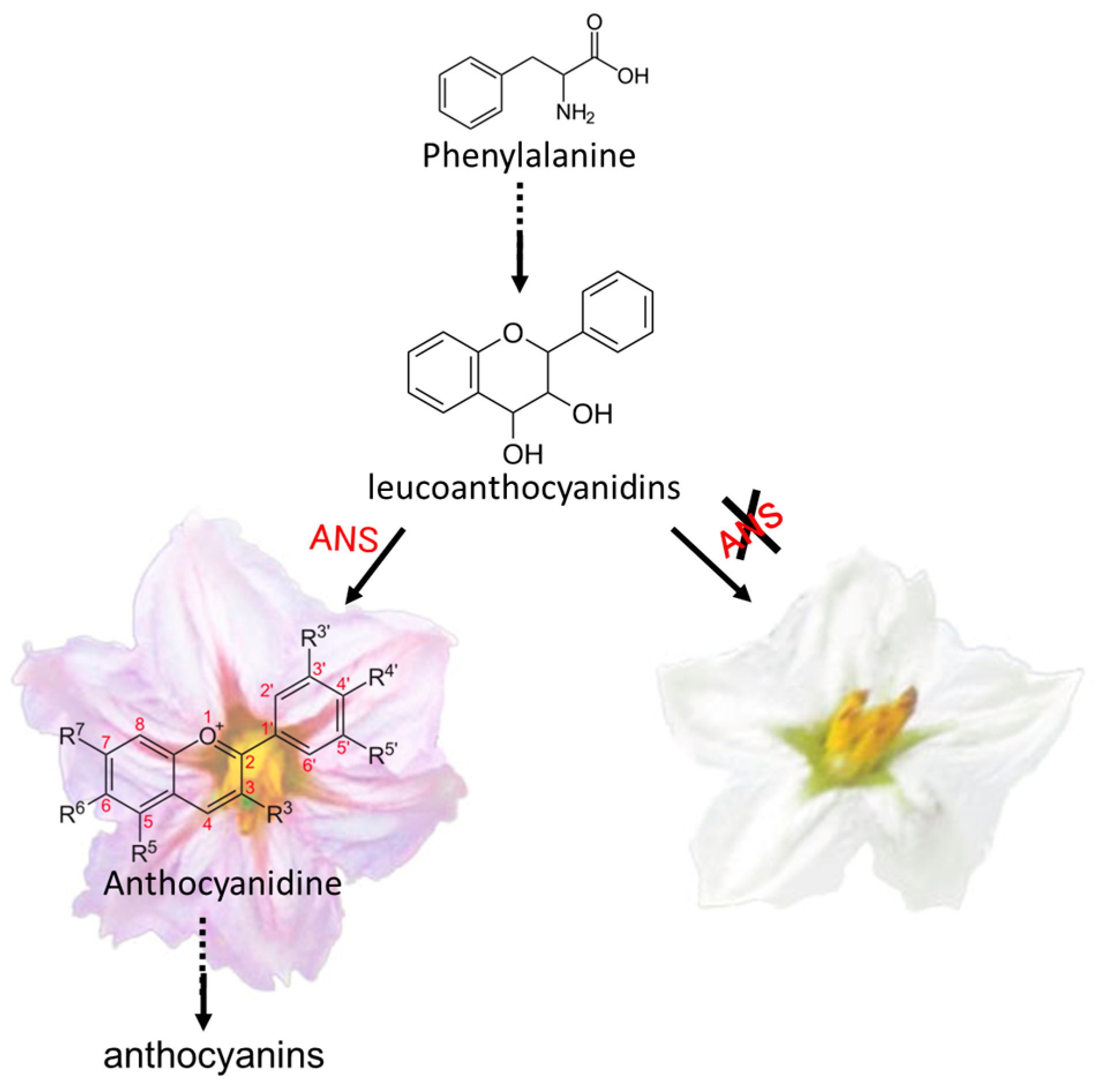
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef] [PubMed]
- Yoshikazu, T.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar]
- Pourcel, L.; Routaboul, J.M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Brenda, W.-S. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar]
- Katsumoto, Y.; Fukuchi-Mizutani, M.; Fukui, Y.; Brugliera, F.; Holton, T.A.; Karan, M.; Nakamura, N.; Yonekura-Sakakibara, K.; Togami, J.; Pigeaire, A.; et al. Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 2007, 48, 1589–1600. [Google Scholar] [CrossRef] [PubMed]
- Noda, Y.; Kneyuki, T.; Igarashi, K.; Mori, A.; Packer, L. Antioxidant activity of nasunin, an anthocyanin in eggplant peels. Toxicology 2000, 148, 119–123. [Google Scholar] [CrossRef]
- Doganlar, S.; Frary, A.; Daunay, M.C.; Lester, R.N.; Tanksley, S.D. A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the Solanaceae. Genetics 2002, 161, 1697–1711. [Google Scholar] [PubMed]
- Hirakawa, H.; Shirasawa, K.; Miyatake, K.; Nunome, T.; Negoro, S.; Ohyama, A.; Yamaguchi, H.; Sato, S.; Isobe, S.; Tabata, S.; et al. Draft genome sequence of eggplant (Solanum melongena L.): The representative solanum species indigenous to the old world. DNA Res. 2014, 21, 649–660. [Google Scholar] [CrossRef] [PubMed]
- Joseph, M.; Grotewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar]
- Francisco, D.; Jiménez, A.R.; Paredes-López, O. Natural pigments: Carotenoids, anthocyanins, and betalains—Characteristics, biosynthesis, processing, and stability. Crit. Rev. Food Sci. Nutr. 2000, 40, 173–289. [Google Scholar]
- Adami, M.; De Franceschi, P.; Brandi, F.; Liverani, A.; Giovannini, D.; Rosati, C.; Tartarini, S. Identifying a carotenoid cleavage dioxygenase (CCD4) gene controlling yellow/white fruit flesh color of peach. Plant Mol. Boil. Rep. 2013, 31, 1166–1175. [Google Scholar] [CrossRef]
- Liu, H.; Jiao, J.; Liang, X.; Liu, J.; Meng, H.; Chen, S.; Cheng, Z. Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2016, 129, 1247–1256. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Wan, H.; Wu, Z.; Wang, R.; Ruan, M.; Ye, Q.; Li, Z.; Zhou, G.; Yao, Z.; Yang, Y. A comprehensive analysis of carotenoid cleavage dioxygenases genes in Solanum lycopersicum. Plant Mol. Boil. Rep. 2016, 34, 512–523. [Google Scholar] [CrossRef]
- Gert, F.; Martens, S. Metabolic engineering and applications of flavonoids. Curr. Opin. Biotechnol. 2001, 12, 155–160. [Google Scholar]
- Springob, K.; Nakajima, J.; Yamazaki, M.; Saito, K. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat. Prod. Rep. 2003, 20, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.; Tian, J.; Zhang, J.; Song, T.; Yao, Y. A Malus crabapple chalcone synthase gene, McCHS, regulates red petal color and flavonoid biosynthesis. PLoS ONE 2014, 9, e110570. [Google Scholar] [CrossRef] [PubMed]
- Yoshikazu, T.; Tsuda, S.; Kusumi, T. Metabolic engineering to modify flower color. Plant Cell Physiol. 1998, 39, 1119–1126. [Google Scholar]
- Brenda, W.-S. Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 1999, 107, 142–149. [Google Scholar]
- Ahmadiani, N.; Robbins, R.J.; Collins, T.M.; Giusti, M.M. Anthocyanins contents, profiles, and color characteristics of red cabbage extracts from different cultivars and maturity stages. J. Agric. Food Chem. 2014, 62, 7524–7531. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.L.; Liu, X.J.; Guan, C.Y.; Chen, X.B.; Liu, Z.S. Cloning and expression analysis of an anthocyanidin synthase gene homolog from Brassica juncea. Mol. Breed. 2011, 28, 313–322. [Google Scholar] [CrossRef]
- Heppel, S.C.; Jaffé, F.W.; Takos, A.M.; Schellmann, S.; Rausch, T.; Walker, A.R.; Bogs, J. Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 2013, 82, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Jun, T. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015, 6, 261. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Ramachandra, R.S.; Ravishankar, G.A. Plant cell cultures: Chemical factories of secondary metabolites. Biotechnol. Adv. 2002, 20, 101–153. [Google Scholar] [CrossRef]
- Prinsi, B.; Negri, A.S.; Quattrocchio, F.M.; Koes, R.E.; Espen, L. Proteomics of red and white corolla limbs in petunia reveals a novel function of the anthocyanin regulator ANTHOCYANIN1 in determining flower longevity. J. Proteom. 2016, 131, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Díaz, J.R.; Pérez-Díaz, J.; Madrid-Espinoza, J.; González-Villanueva, E.; Moreno, Y.; Ruiz-Lara, S. New member of the R2R3-MYB transcription factors family in grapevine suppresses the anthocyanin accumulation in the flowers of transgenic tobacco. Plant Mol. Biol. 2016, 90, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Elomaa, P.; Uimari, A.; Mehto, M.; Albert, V.A.; Laitinen, R.A.; Teeri, T.H. Activation of anthocyanin biosynthesis in Gerbera hybrida (Asteraceae) suggests conserved protein-protein and protein-promoter interactions between the anciently diverged monocots and eudicots. Plant Physiol. 2003, 133, 1831–1842. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Peng, Q.; Zhao, J.; Owiti, A.; Ren, F.; Liao, L.; Wang, L.; Deng, X.; Jiang, Q.; Han, Y. Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 2016, 7, 1557. [Google Scholar] [CrossRef] [PubMed]
- Barchi, L.; Lanteri, S.; Portis, E.; Valè, G.; Volante, A.; Pulcini, L.; Ciriaci, T.; Acciarri, N.; Barbierato, V.; Toppino, L.; et al. A RAD tag derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PLoS ONE 2012, 7, e43740. [Google Scholar] [CrossRef] [PubMed]
- Cericola, F.; Portis, E.; Lanteri, S.; Toppino, L.; Barchi, L.; Acciarri, N.; Pulcini, L.; Sala, T.; Rotino, G.L. Linkage disequilibrium and genome-wide association analysis for anthocyanin pigmentation and fruit color in eggplant. BMC Genom. 2014, 15, 896. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L.; Määttä, K.; Pirttilä, A.M.; Törrönen, R.; Kärenlampi, S.; Hohtola, A. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 2002, 130, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Dooner, H.K.; Robbins, T.P.; Jorgensen, R.A. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 1991, 25, 173–199. [Google Scholar] [CrossRef] [PubMed]


| Population | Plant Tested | Purple:White | Mendelian Expectations | X2 | p |
|---|---|---|---|---|---|
| BN (purple) × SR (white) | |||||
| F1 | all | all purple | 1:0 | ||
| F2 | 174 | 137:37 | 3:1 | 1.295 | 0.255 |
| Scaffold | CDS | Gene | Arabodopsis Homologs | Annoations | Polymorphism |
|---|---|---|---|---|---|
| Sme2.5_01332.1 | Sme2.5_01332.1_g00003.1 | Solyc08g079820.2.1 | AT4G11980.1 | nudix hydrolase 14 | None |
| Sme2.5_01332.1_g00004.1 | Solyc08g079790.1.1 | AT1G12500.1 | sugar phosphate/phosphate translocator | None | |
| Sme2.5_01332.1_g00005.1 | Solyc08g079780.1.1 | AT3G17675.1 | mavicyanin-like protein | 1 SNP | |
| Sme2.5_01332.1_g00006.1 | Solyc08g079770.2.1 | AT1G12480.1 | guard cell S-type anion channel | None | |
| Sme2.5_01332.1_g00007.1 | Solyc08g079760.2.1 | AT4G22860.1 | protein TPX2-like | None | |
| Sme2.5_01332.1_g00008.1 | Solyc08g079750.2.1 | AT1G62960.1 | aminotransferase ACS10 | None | |
| Sme2.5_01332.1_g00009.1 | Solyc08g079740.2.1 | AT1G12460.1 | LRR receptor-like serine/threonine-protein kinase | None | |
| Sme2.5_01332.1_g00010.1 | Solyc12g044400.1.1 | AT3G16290.1 | inactive ATP-dependent zinc metalloprotease FTSH 12, chloroplastic | None | |
| Sme2.5_01332.1_g00011.1 | Solyc08g079730.1.1 | AT4G22790.1 | protein DETOXIFICATION 56-like | None | |
| Sme2.5_20506.1 | Sme2.5_20506.1_g00001.1 | Solyc08g079790.1.1 | AT1G12500.1 | sugar phosphate/phosphate translocator | None |
| Sme2.5_20506.1_g00002.1 | None | ||||
| Sme2.5_20506.1_g00003.1 | None | ||||
| Sme2.5_25093.1 | Sme2.5_25093.1_g00001.1 | Solyc08g079820.2.1 | AT4G11980.1 | nudix hydrolase 14 | None |
| Sme2.5_25093.1_g00002.1 | Solyc08g079830.2.1 | AT1G12520.1 | copper chaperone for superoxide dismutase | None | |
| Sme2.5_14718.1 | Sme2.5_14718.1_g00001.1 | Solyc08g079850.1.1 | AT1G04110.1 | subtilisin-like protease SBT1.2 | 2 SNPs |
| Sme2.5_05247.1 | Sme2.5_05247.1_g00002.1 | Solyc08g080010.1.1 | AT1G04110.1 | subtilisin-like protease SBT1.7 | None |
| Sme2.5_05247.1_g00003.1 | Solyc08g080000.2.1 | uncharacterized | None | ||
| Sme2.5_05247.1_g00004.1 | Solyc08g079980.1.1 | AT5G67360.1 | subtilisin-like protease SBT1.7 | None | |
| Sme2.5_05247.1_g00005.1 | Solyc08g079970.1.1 | AT5G67360.1 | subtilisin-like protease SBT1.7 | None | |
| Sme2.5_05247.1_g00006.1 | Solyc07g065420.1.1 | AT5G06990.1 | protein MIZU-KUSSEI 1 | None | |
| Sme2.5_05247.1_g00008.1 | Solyc08g079870.1.1 | AT5G67360.1 | subtilisin-like protease SBT1.7 | None | |
| Sme2.5_05247.1_g00009.1 | Solyc08g079870.1.1 | AT1G04110.1 | subtilisin-like protease SBT1.7 | 1 SNP | |
| Sme2.5_05247.1_g000010.1 | Solyc08g079850.1.1 | AT2G05920.1 | subtilisin-like protease SBT1.7 | 3 SNPs | |
| Sme2.5_05247.1_g000011.1 | subtilisin-like protease SBT1.7 | 2 SNPs | |||
| Sme2.5_01638.1 | Sme2.5_01638.1_g00002.1 | Solyc08g080030.2.1 | AT1G12550.1 | glyoxylate/hydroxypyruvate reductase HPR3-like | None |
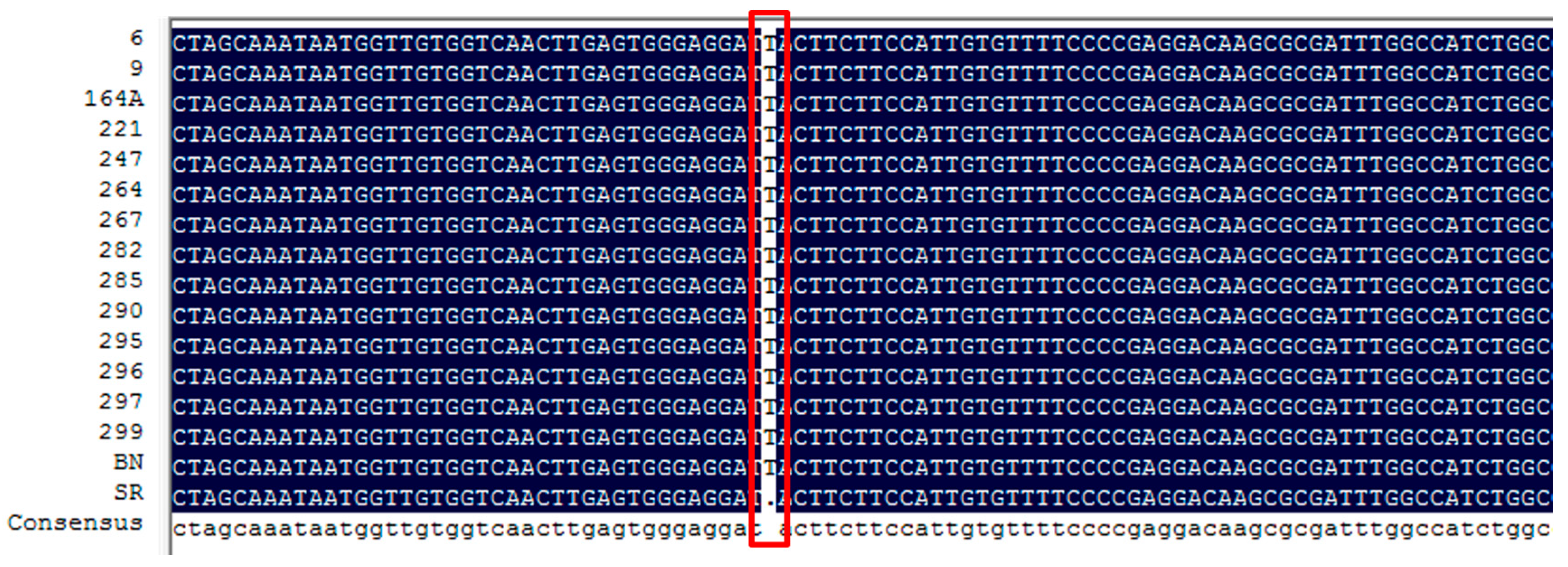
| Sme2.5_01638.1_g00003.1 | Solyc08g080040.2.1 | AT4G22880.2 | Anthocyanidin Synthase (ANS) gene | None | |
| Sme2.5_01638.1_g00004.1 | Solyc05g023820.2.1 | AT3G48800.1 | tuberosum myb-like protein | 2 SNPs | |
| Sme2.5_01638.1_g00005.1 | Solyc08g080040.2.1 | AT4G22880.2 | Anthocyanidin Synthase (ANS) gene | 4 SNPs and 1 InDel | |
| Sme2.5_01638.1_g00006.1 | Solyc08g080050.2.1 | AT4G22890.2 | PGR5-like protein | None |
| Eggplant Cultivar | Color of Flower | Color of Peel |
|---|---|---|
| 6 | purple | green |
| 9 | purple | white |
| 164A | purple | white |
| 247 | purple | white |
| 264 | purple | green |
| 267 | purple | green |
| 282 | purple | green |
| 285 | purple | white |
| 290 | purple | green |
| 295 | purple | white |
| 296 | purple | white |
| 297 | purple | white |
| BN | purple | purple |
| 221 | white | green |
| 299 | white | green |
| SR | white | green |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, M.; Xu, M.; Xiao, Y.; Cui, D.; Qin, Y.; Wu, J.; Wang, W.; Wang, G. Fine Mapping Identifies SmFAS Encoding an Anthocyanidin Synthase as a Putative Candidate Gene for Flower Purple Color in Solanum melongena L. Int. J. Mol. Sci. 2018, 19, 789. https://doi.org/10.3390/ijms19030789
Chen M, Xu M, Xiao Y, Cui D, Qin Y, Wu J, Wang W, Wang G. Fine Mapping Identifies SmFAS Encoding an Anthocyanidin Synthase as a Putative Candidate Gene for Flower Purple Color in Solanum melongena L. International Journal of Molecular Sciences. 2018; 19(3):789. https://doi.org/10.3390/ijms19030789
Chicago/Turabian StyleChen, Mengqiang, Mengyun Xu, Yao Xiao, Dandan Cui, Yongqiang Qin, Jiaqi Wu, Wenyi Wang, and Guoping Wang. 2018. "Fine Mapping Identifies SmFAS Encoding an Anthocyanidin Synthase as a Putative Candidate Gene for Flower Purple Color in Solanum melongena L." International Journal of Molecular Sciences 19, no. 3: 789. https://doi.org/10.3390/ijms19030789
APA StyleChen, M., Xu, M., Xiao, Y., Cui, D., Qin, Y., Wu, J., Wang, W., & Wang, G. (2018). Fine Mapping Identifies SmFAS Encoding an Anthocyanidin Synthase as a Putative Candidate Gene for Flower Purple Color in Solanum melongena L. International Journal of Molecular Sciences, 19(3), 789. https://doi.org/10.3390/ijms19030789





