Insights into the Mechanisms of Chloroplast Division
Abstract
:1. Introduction
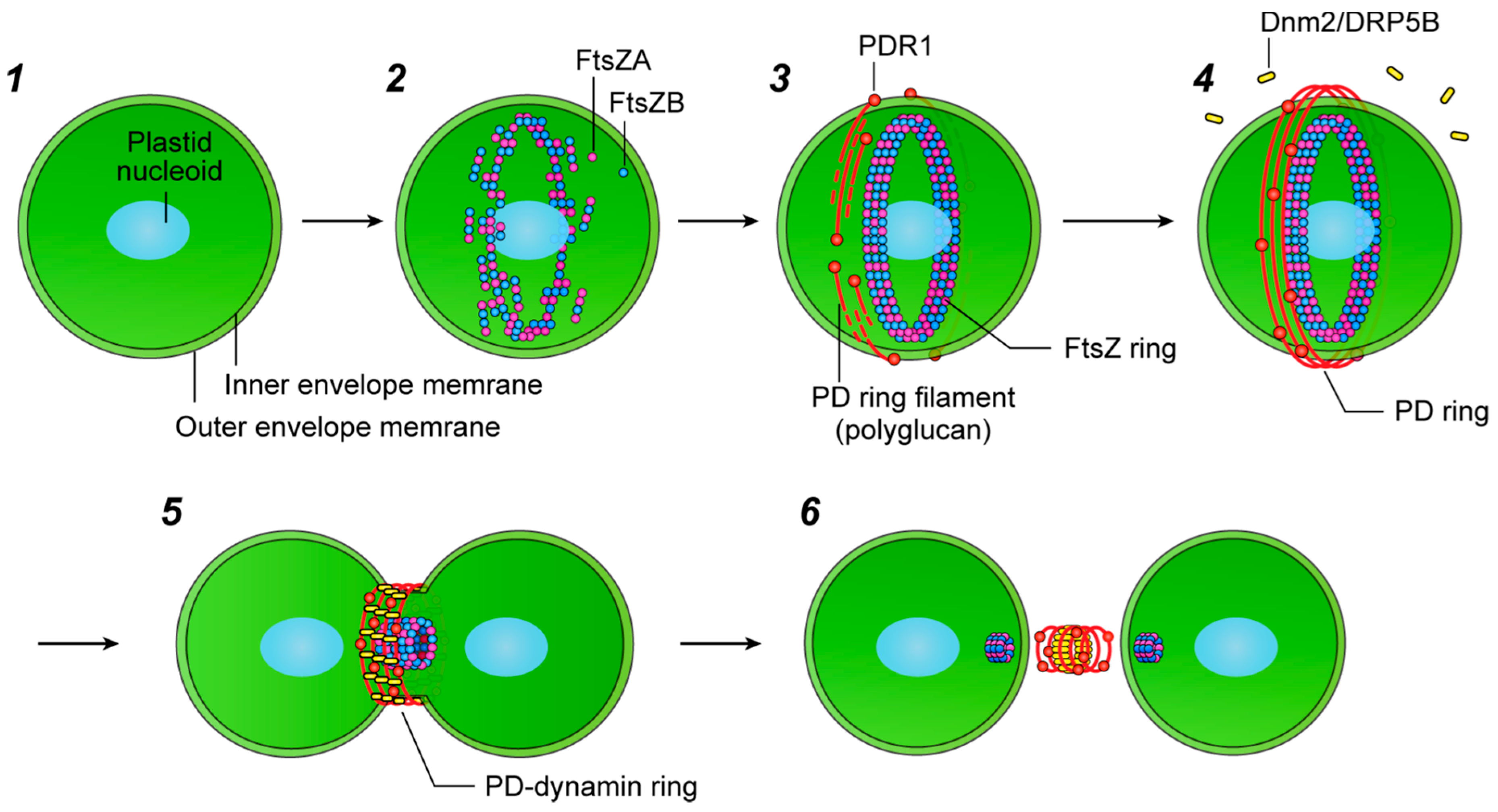
2. Structure and Assembly of the Plastid-Division Machinery
3. The FtsZ Ring
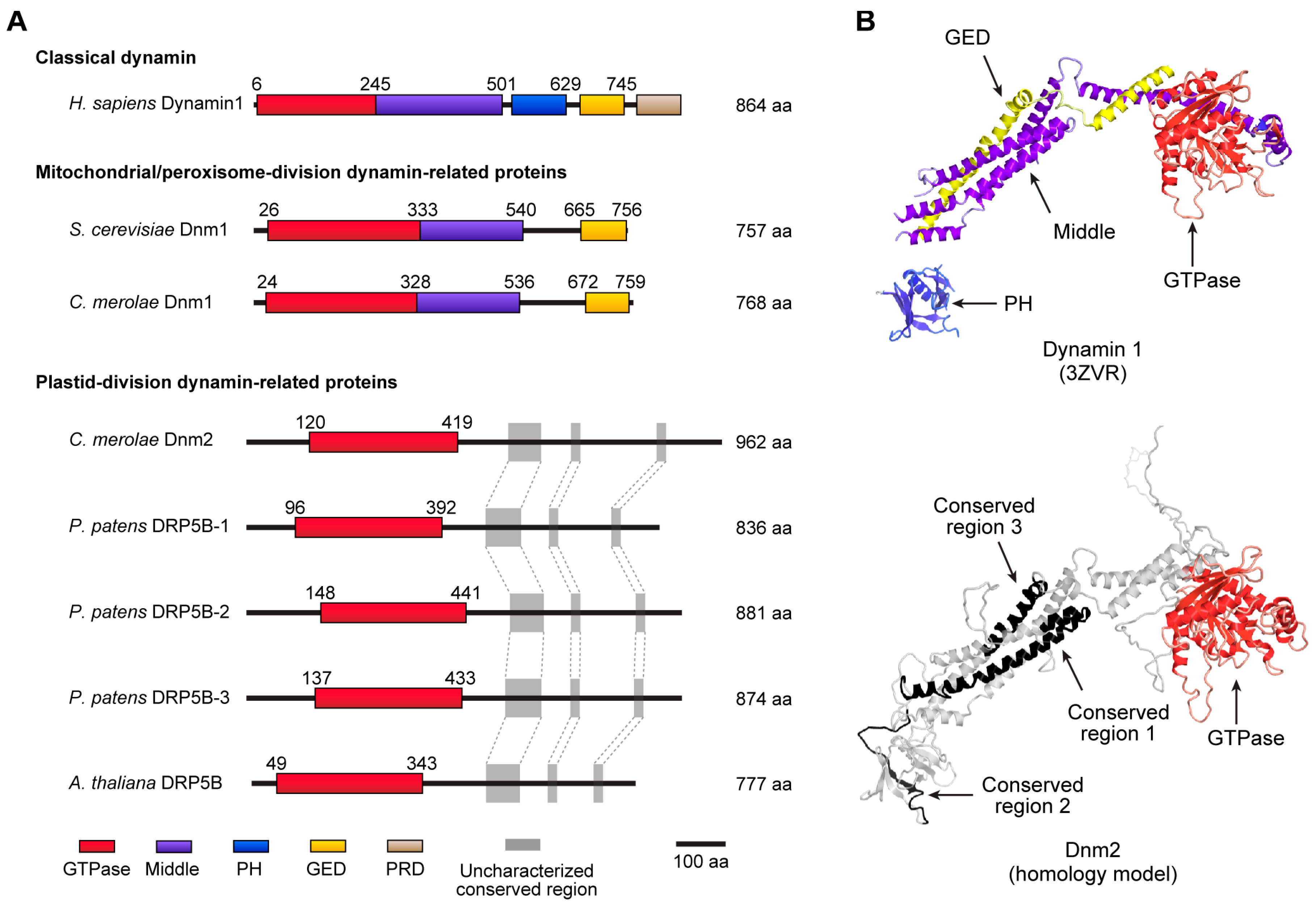
4. The Dynamin Ring
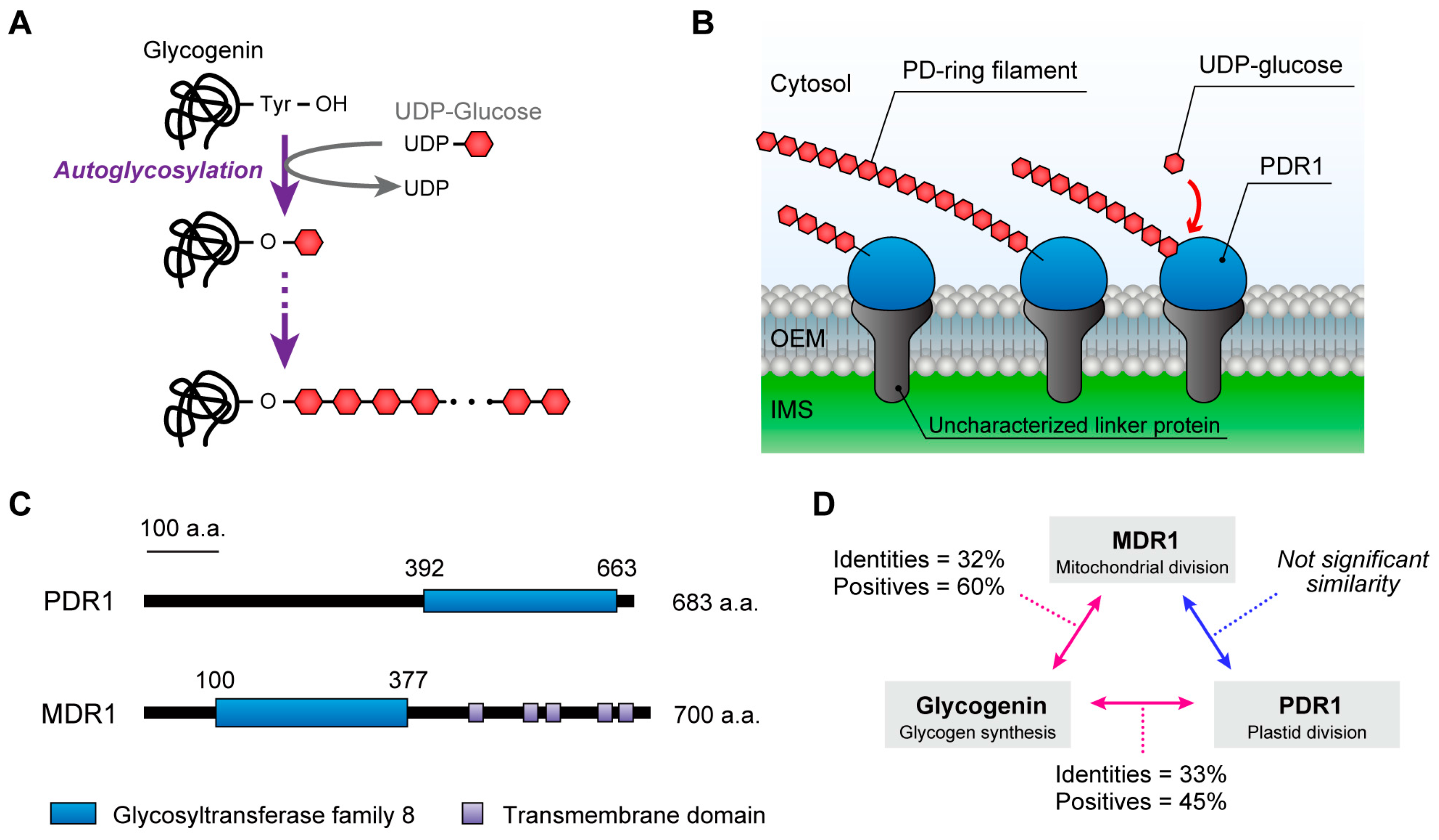
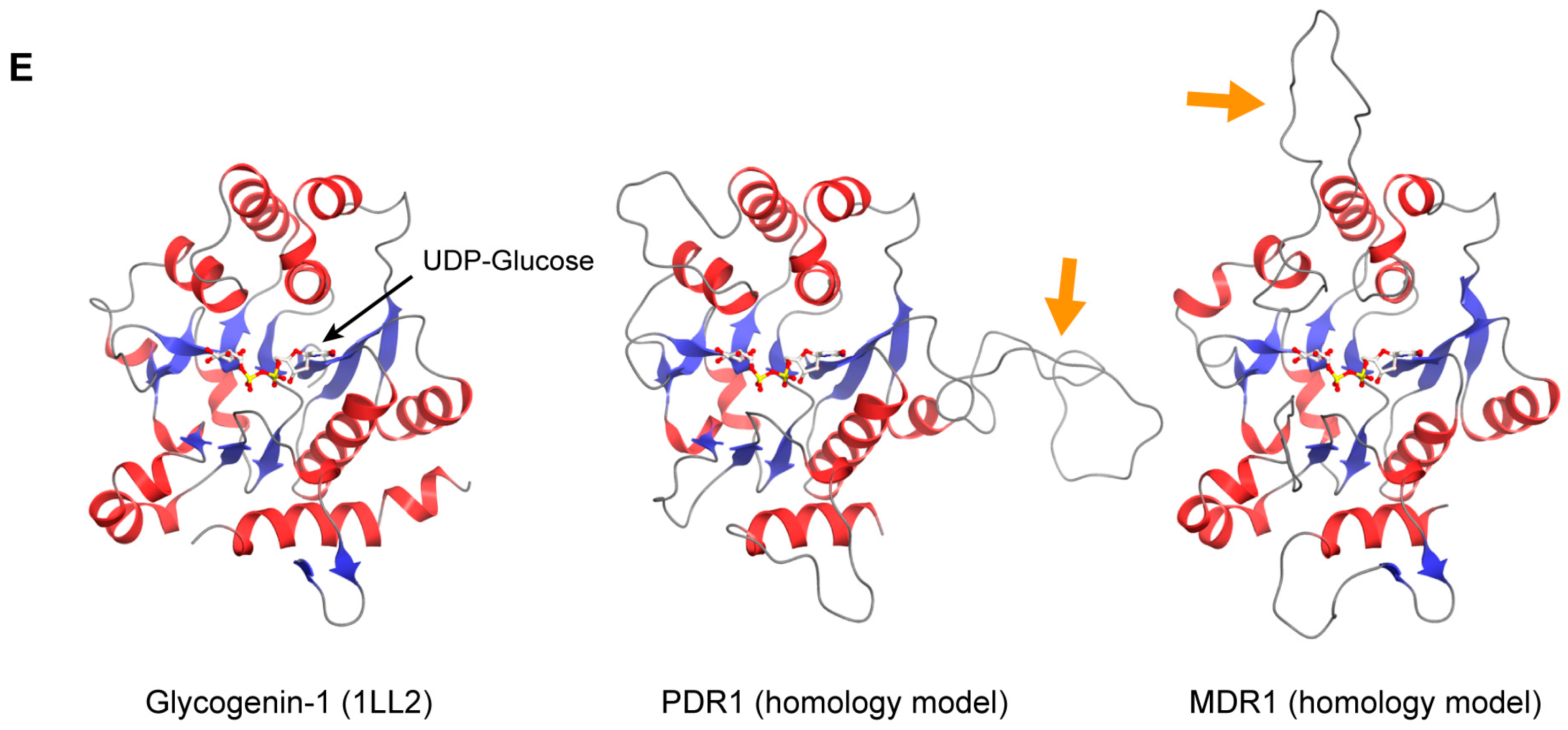
5. The PD Ring
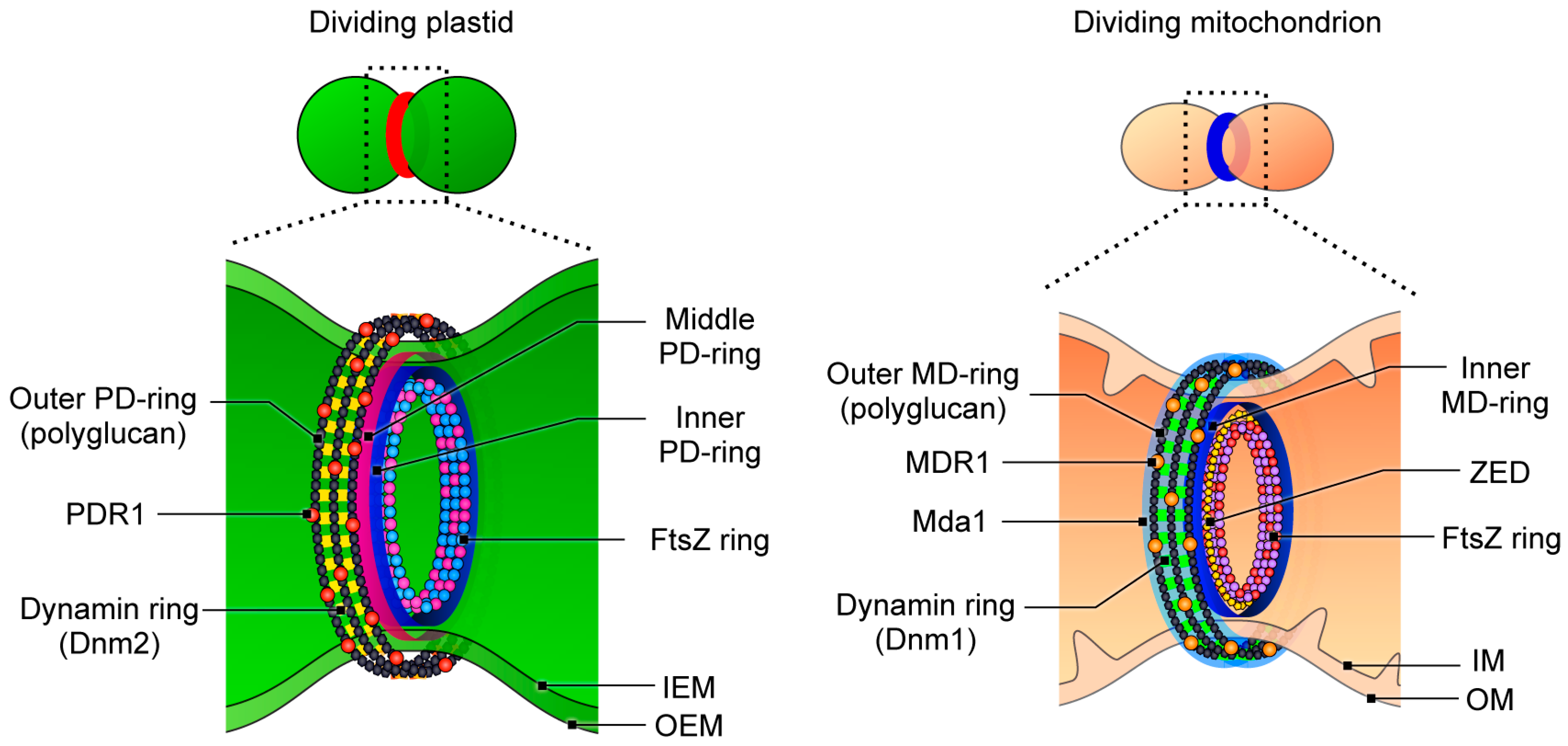
6. The Homology between Plastid- and Mitochondrial-Division Machinery
7. Conclusions and Perspectives
Acknowledgments
Conflicts of Interest
References
- Osteryoung, K.W.; Nunnari, J. The division of endosymbiotic organelles. Science 2003, 302, 1698–1704. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, T.; Misumi, O.; Nishida, K.; Yagisawa, F.; Yoshida, Y.; Fujiwara, T.; Kuroiwa, H. Vesicle, mitochondrial, and plastid division machineries with emphasis on dynamin and electron-dense rings. Int. Rev. Cell Mol. Biol. 2008, 271, 97–152. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Nakanishi, H.; Kabeya, Y. Structure, regulation, and evolution of the plastid division machinery. Int. Rev. Cell Mol. Biol. 2011, 291, 115–153. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Miyagishima, S.Y.; Kuroiwa, H.; Kuroiwa, T. The plastid-dividing machinery: Formation, constriction and fission. Curr. Opin. Plant Biol. 2012, 15, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, T.; Kuroiwa, H.; Sakai, A.; Takahashi, H.; Toda, K.; Itoh, R. The division apparatus of plastids and mitochondria. Int. Rev. Cytol. 1998, 181, 1–41. [Google Scholar] [PubMed]
- Chen, C.; MacCready, J.S.; Ducat, D.C.; Osteryoung, K.W. The molecular machinery of chloroplast division. Plant Physiol. 2018, 176, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Nishida, K.; Kuroiwa, T. An evolutionary puzzle: Chloroplast and mitochondrial division rings. Trends Plant Sci. 2003, 8, 432–438. [Google Scholar] [CrossRef]
- TerBush, A.D.; Yoshida, Y.; Osteryoung, K.W. FtsZ in chloroplast division: Structure, function and evolution. Curr. Opin. Cell Biol. 2013, 25, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Nishida, K.; Mori, T.; Matsuzaki, M.; Higashiyama, T.; Kuroiwa, H.; Kuroiwa, T. A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell Online 2003, 15, 655–665. [Google Scholar] [CrossRef]
- Gao, H.; Kadirjan-Kalbach, D.; Froehlich, J.E.; Osteryoung, K.W. ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc. Natl. Acad. Sci. USA 2003, 100, 4328–4333. [Google Scholar] [CrossRef] [PubMed]
- Osteryoung, K.W.; Pyke, K.A. Division and dynamic morphology of plastids. Annu. Rev. Plant Biol. 2014, 65, 443–472. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Kuroiwa, H.; Misumi, O.; Yoshida, M.; Ohnuma, M.; Fujiwara, T.; Yagisawa, F.; Hirooka, S.; Imoto, Y.; Matsushita, K.; et al. Chloroplasts divide by contraction of a bundle of nanofilaments consisting of polyglucan. Science 2010, 329, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Kuroiwa, H.; Misumi, O.; Nishida, K.; Yagisawa, F.; Fujiwara, T.; Nanamiya, H.; Kawamura, F.; Kuroiwa, T. Isolated chloroplast division machinery can actively constrict after stretching. Science 2006, 313, 1435–1438. [Google Scholar] [CrossRef] [PubMed]
- Glynn, J.M.; Yang, Y.; Vitha, S.; Schmitz, A.J.; Hemmes, M.; Miyagishima, S.-Y.; Osteryoung, K.W. PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J. 2009, 59, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, C.; Froehlich, J.E.; Terbush, A.D.; Osteryoung, K.W. Roles of Arabidopsis PARC6 in coordination of the chloroplast division complex and negative regulation of FtsZ assembly. Plant Physiol. 2016, 170, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Glynn, J.M.; Froehlich, J.E.; Osteryoung, K.W. Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 2008, 20, 2460–2470. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, J.; Sun, Q.; Yu, X.; Zhang, W.; Jia, N.; An, C.; Li, Y.; Dong, Y.; Han, F.; et al. Structural insights into the coordination of plastid division by the ARC6-PDV2 complex. Nat. Plants 2017, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sumiya, N.; Fujiwara, T.; Kobayashi, Y.; Misumi, O.; Miyagishima, S.Y. Development of a heat-shock inducible gene expression system in the red alga Cyanidioschyzon merolae. PLoS ONE 2014, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sumiya, N.; Fujiwara, T.; Era, A.; Miyagishima, S.Y. Chloroplast division checkpoint in eukaryotic algae. Proc. Natl. Acad. Sci. USA 2016, 113, E7629–E7638. [Google Scholar] [CrossRef] [PubMed]
- Sumiya, N.; Miyagishim, S.Y. Hierarchal order in the formation of chloroplast division machinery in the red alga Cyanidioschyzon merolae. Commun. Integr. Biol. 2017, 10, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Mogi, Y.; TerBush, A.D.; Osteryoung, K.W. Chloroplast FtsZ assembles into a contractible ring via tubulin-like heteropolymerization. Nat. Plants 2016, 2, 16095. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Takahara, M.; Mori, T.; Kuroiwa, H.; Higashiyama, T.; Kuroiwa, T. Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 2001, 13, 2257–2268. [Google Scholar] [CrossRef]
- Miyagishima, S.Y.; Takahara, M.; Kuroiwa, T. Novel filaments 5 nm in diameter constitute the cytosolic ring of the plastid division apparatus. Plant Cell 2001, 13, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Sumiya, N.; Hirata, A.; Kawano, S. Multiple FtsZ ring formation and reduplicated chloroplast DNA in Nannochloris bacillaris (Chlorophyta, Trebouxiophyceae) under phosphate-enriched culture. J. Phycol. 2008, 44, 1476–1489. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, H.; Mori, T.; Takahara, M.; Miyagishima, S.Y.; Kuroiwa, T. Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 2002, 215, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Osteryoung, K.W.; Vierling, E. Conserved cell and organelle division. Nature 1995, 376, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Takahara, M.; Takahashi, H.; Matsunaga, S.Y.; Miyagishima, S.; Takano, H.; Sakai, A.; Kawano, S.; Kuroiwa, T. A putative mitochondrial FtsZ gene is present in the unicellular primitive red alga Cyanidioschyzon merolae. Mol. Gen. Genet. 2000, 264, 452–460. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Kuroiwa, H.; Takahara, M.; Miyagishima, S.Y.; Kuroiwa, T. Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol. 2001, 42, 555–559. [Google Scholar] [CrossRef] [PubMed]
- Vitha, S.; McAndrew, R.S.; Osteryoung, K.W. FtsZ ring formation at the chloroplast division site in plants. J. Cell Biol. 2001, 153, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Löwe, J.; Amos, L.A. Crystal structure of the bacterial cell-division protein FtsZ. Nature 1998, 391, 203–206. [Google Scholar] [CrossRef]
- Oliva, M.A.; Cordell, S.C.; Löwe, J. Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol. 2004, 11, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Nozaki, H.; Nishida, K.; Nishida, K.; Matsuzaki, M.; Kuroiwa, T. Two types of FtsZ proteins in mitochondria and red-lineage chloroplasts: The duplication of FtsZ is implicated in endosymbiosis. J. Mol. Evol. 2004, 58, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.J.; Glynn, J.M.; Olson, B.J.S.C.; Stokes, K.D.; Osteryoung, K.W. Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol. Plant 2009, 2, 1211–1222. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, M.; Misumi, O.; Shin-I, T.; Maruyama, S.; Takahara, M.; Miyagishima, S.Y.; Mori, T.; Nishida, K.; Yagisawa, F.; Nishida, K.; et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 2004, 428, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N.; John, B.; Mirkovic, N.; Fiser, A.; Ilyin, V.A.; Pieper, U.; Stuart, A.C.; Marti-Renom, M.A.; Madhusudhan, M.S.; Yerkovich, B.; et al. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003, 31, 3375–3380. [Google Scholar] [CrossRef] [PubMed]
- TerBush, A.D.; Osteryoung, K.W. Distinct functions of chloroplast FtsZ1 and FtsZ2 in Z-ring structure and remodeling. J. Cell Biol. 2012, 199, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Shimada, H.; Koizumi, M.; Kuroki, K.; Mochizuki, M.; Fujimoto, H.; Ohta, H.; Masuda, T.; Takamiya, K. ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 2004, 45, 960–967. [Google Scholar] [CrossRef] [PubMed]
- Maple, J.; Fujiwara, M.T.; Kitahata, N.; Lawson, T.; Baker, N.R.; Yoshida, S.; Møller, S.G. GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabidopsis. Curr. Biol. 2004, 14, 776–781. [Google Scholar] [CrossRef] [PubMed]
- Shpetner, H.S.; Vallee, R.B. Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell 1989, 59, 421–432. [Google Scholar] [CrossRef]
- Praefcke, G.J.K.; McMahon, H.T. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004, 5, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Bleazard, W.; McCaffery, J.M.; King, E.J.; Bale, S.; Mozdy, A.; Tieu, Q.; Nunnari, J.; Shaw, J.M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999, 1, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Faelber, K.; Posor, Y.; Gao, S.; Held, M.; Roske, Y.; Schulze, D.; Haucke, V.; Noé, F.; Daumke, O. Crystal structure of nucleotide-free dynamin. Nature 2011, 477, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Ford, M.G.J.; Jenni, S.; Nunnari, J. The crystal structure of dynamin. Nature 2011, 477, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, E.; Takechi, K.; Sato, H.; Yamada, T.; Takio, S.; Takano, H. Three dynamin-related protein 5B genes are related to plastid division in Physcomitrella patens. Plant Sci. 2011, 180, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Kuwayama, H.; Urushihara, H.; Nakanishi, H. Evolutionary linkage between eukaryotic cytokinesis and chloroplast division by dynamin proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 15202–15207. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.; Takahara, M.; Miyagishima, S.Y.; Kuroiwa, H.; Matsuzaki, M.; Kuroiwa, T. Dynamic recruitment of dynamin for final mitochondrial severance in a primitive red alga. Proc. Natl. Acad. Sci. USA 2003, 100, 2146–2151. [Google Scholar] [CrossRef] [PubMed]
- Imoto, Y.; Kuroiwa, H.; Yoshida, Y.; Ohnuma, M.; Fujiwara, T.; Yoshida, M.; Nishida, K.; Yagisawa, F.; Hirooka, S.; Miyagishima, S.; Misumi, O.; Kawano, S.; Kuroiwa, T. Single-membrane-bounded peroxisome division revealed by isolation of dynamin-based machinery. Proc. Natl. Acad. Sci. USA 2013, 110, 9583–9588. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, T. The primitive red algae Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. BioEssays 1998, 20, 344–354. [Google Scholar] [CrossRef]
- Hashimoto, H. Double ring structure around the constricting neck of dividing plastids of Avena sativa. Protoplasma 1986, 135, 166–172. [Google Scholar] [CrossRef]
- Miyagishima, S.Y.; Itoh, R.; Toda, K.; Takahashi, H.; Kuroiwa, H.; Kuroiwa, T. Identification of a triple ring structure involved in plastid division in the primitive red alga Cyanidioschyzon merolae. J. Electron Microsc. 1998, 47, 269–272. [Google Scholar] [CrossRef]
- Kuroiwa, H.; Mori, T.; Takahara, M.; Miyagishima, S.Y.; Kuroiwa, T. Multiple FtsZ rings in a pleomorphic chloroplast in embryonic cap cells of Pelargonium zonale. Cytologia 2001, 66, 227–233. [Google Scholar] [CrossRef]
- Lomako, J.; Lomako, W.M.; Whelan, W.J. Glycogenin: The primer for mammalian and yeast glycogen synthesis. Biochim. Biophys. Acta 2004, 1673, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Kuroiwa, H.; Shimada, T.; Yoshida, M.; Ohnuma, M.; Fujiwara, T.; Imoto, Y.; Yagisawa, F.; Nishida, K.; Hirooka, S.; et al. Glycosyltransferase MDR1 assembles a dividing ring for mitochondrial proliferation comprising polyglucan nanofilaments. Proc. Natl. Acad. Sci. USA 2017, 114, 13284–13289. [Google Scholar] [CrossRef] [PubMed]
- Kuroiwa, T.; Nishida, K.; Yoshida, Y.; Fujiwara, T.; Mori, T.; Kuroiwa, H.; Misumi, O. Structure, function and evolution of the mitochondrial division apparatus. Biochim. Biophys. Acta 2006, 1763, 510–521. [Google Scholar] [CrossRef] [PubMed]
- Van der Bliek, A.M.; Shen, Q.; Kawajiri, S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb. Perspect. Biol. 2013, 5, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.R.; Nunnari, J. Mitochondrial form and function. Nature 2014, 505, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Reddy, P.H.; Iijima, M.; Sesaki, H. Mitochondrial division and fusion in metabolism. Curr. Opin. Cell Biol. 2015, 33, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kanesaki, Y.; Tanaka, A.; Kuroiwa, H.; Kuroiwa, T.; Tanaka, K. Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc. Natl. Acad. Sci. USA 2009, 106, 803–807. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Imamura, S.; Hanaoka, M.; Tanaka, K. A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat. Cell Biol. 2011, 13, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Fujiwara, T.; Sumiya, N.; Hirooka, S.; Nakano, A.; Kabeya, Y.; Nakamura, M. Translation-independent circadian control of the cell cycle in a unicellular photosynthetic eukaryote. Nat. Commun. 2014, 5, 3807. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Ehara, T.; Osafune, T.; Kuroiwa, H.; Kawano, S.; Kuroiwa, T. Behavior of mitochondria, chloroplasts and their nuclei during the mitotic cycle in the ultramicroalga Cyanidioschyzon merolae. Eur. J. Cell Biol. 1994, 63, 280–288. [Google Scholar] [PubMed]
- Ohnuma, M.; Yokoyama, T.; Inouye, T.; Sekine, Y.; Tanaka, K. Polyethylene glycol (PEG)-mediated transient gene expression in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2008, 49, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Ohnuma, M.; Misumi, O.; Fujiwara, T.; Watanabe, S.; Tanaka, K.; Kuroiwa, T. Transient gene suppression in a red alga, Cyanidioschyzon merolae 10D. Protoplasma 2009, 236, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Kanesaki, Y.; Hirooka, S.; Era, A.; Sumiya, N.; Yoshikawa, H.; Tanaka, K.; Miyagishima, S.Y. A nitrogen source-dependent inducible and repressible gene expression system in the red alga Cyanidioschyzon merolae. Front. Plant Sci. 2015, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Ohnuma, M.; Kuroiwa, T.; Ohbayashi, R.; Hirooka, S.; Miyagishima, S.Y. Development of a double nuclear gene-targeting method by two-step transformation based on a newly established chloramphenicol-selection system in the red alga Cyanidioschyzon merolae. Front. Plant Sci. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, Y. Insights into the Mechanisms of Chloroplast Division. Int. J. Mol. Sci. 2018, 19, 733. https://doi.org/10.3390/ijms19030733
Yoshida Y. Insights into the Mechanisms of Chloroplast Division. International Journal of Molecular Sciences. 2018; 19(3):733. https://doi.org/10.3390/ijms19030733
Chicago/Turabian StyleYoshida, Yamato. 2018. "Insights into the Mechanisms of Chloroplast Division" International Journal of Molecular Sciences 19, no. 3: 733. https://doi.org/10.3390/ijms19030733




