Functional Characterization of the Steroid Reductase Genes GmDET2a and GmDET2b from Glycine max
Abstract
:1. Introduction
2. Results
2.1. Identification of Steroid Reductases in Soybean
2.2. Phylogenetic Analysis of Steroid Reductases
2.3. Transcript Levels of GmDET2a and GmDET2b in the Seedling Stage
2.4. GmDET2a and GmDET2b Were Negatively Regulated by Epibrassinosteroid
2.5. Responses of GmDET2a and GmDET2b to Nutrient Deficiencies
2.6. Ectopic Expression of GmDET2a and GmDET2b in Atdet2-1 Rescued the Growth and Development Defects
2.7. Over-Expression of GmDET2a or GmDET2b Rescued the Responses of BR Biosynthesis-Related Genes to Exogenous BR
3. Discussion
4. Materials and Methods
4.1. Soybean and Arabidopsis Growth
4.2. Extraction of Genomic DNA, RNA and Reserve Transcription of mRNA
4.3. Determination of Soybean GmDET2 Family Structure
4.4. Alignment of DET2s
4.5. Phylogenetic Analysis
4.6. Analysis of Expression Patterns of Soybean DET2 Family at the Seedling Stage
4.7. Overexpression of GmDET2a and GmDET2b in Arabidopsis det2
4.8. Phenotypic Analysis of Transgenic Arabidopsis
4.9. Determination of the Expression Levels of BR Biosynthesis-Related Genes in Col-0, det2-1 and Transgenic Lines
4.10. Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AA | amino acid |
| BAK1 | BRI1-associated receptor kinase 1 |
| BIN2 | brassinosteroid insensitive 2 |
| BKI1 | BRI1 kinase inhibitor 1 |
| BR | brassinosteroid |
| BRI1 | brassinosteroid insensitive 1 |
| Brz | brassinazole |
| BZR1 | brassinazole-resistant 1 |
| d | day |
| h | hour |
| ID | island domain |
| K | potassium |
| N | nitrogen |
| ORF | open reading frame |
| P | phosphorus |
| PR | primary root |
| qRT-PCR | quantitative real-time PCR |
| S | sulfate |
References
- Müssig, C.; Shin, G.H.; Altmann, T. Brassinosteroids promote root growth in Arabidopsis. Plant Physiol. 2003, 133, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Hacham, Y.; Holland, N.; Butterfield, C.; Ubeda-Tomas, S.; Bennett, M.J.; Chory, J.; Savaldi-Goldstein, S. Brassinosteroid perception in the epidermis controls root meristem size. Development 2011, 138, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Kim, S.Y.; Nam, K.H. Genes encoding plant-specific class III peroxidases are responsible for increased cold tolerance of the brassinosteroid-insensitive 1 mutant. Mol. Cells 2012, 34, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Leubner-Metzger, G. Brassinosteroids and gibberellins promote tobacco seed germination by distinct pathways. Planta 2001, 213, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Chory, J.; Nagpal, P.; Peto, C.A. Phenotypic and Genetic Analysis of det2, a New Mutant That Affects Light-Regulated Seedling Development in Arabidopsis. Plant cell 1991, 3, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Szekeres, M.; Németh, K.; Koncz-Kálmán, Z.; Mathur, J.; Kauschmann, A.; Altmann, T.; Rédei, G.P.; Nagy, F.; Schell, J.; Koncz, C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 1996, 85, 171–182. [Google Scholar] [CrossRef]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Ross, J.J.; Reid, J.B. Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 2005, 138, 2396–2405. [Google Scholar] [CrossRef] [PubMed]
- Nakashita, H.; Yasuda, M.; Nitta, T.; Asami, T.; Fujioka, S.; Arai, Y.; Sekimata, K.; Takatsuto, S.; Yamaguchi, I.; Yoshida, S. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003, 33, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Dhaubhadel, S.; Browning, K.S.; Gallie, D.R.; Krishna, P. Brassinosteroid functions to protect the translational machinery and heat-shock protein synthesis following thermal stress. Plant J. 2002, 29, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Bajguz, A.; Hayat, S. Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem. 2009, 47, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef]
- Wang, X.; Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 2006, 313, 1118–1122. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- Li, J.; Nam, K.H.; Vafeados, D.; Chory, J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001, 127, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [PubMed]
- He, J.X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Peng, P.; Schmitz, R.J.; Decker, A.D.; Tax, F.E.; Li, J. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002, 130, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroidregulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Yan, Z.; Zhu, Y.; Li, J. Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 2008, 1, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Cano-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-Garca, S.; Cheng, J.C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.; Wang, H.; Walker, J.C.; Li, J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004, 40, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Ihara, Y.; Wu, X.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000, 12, 1591–1606. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Fujioka, S.; Sunohara, H.; Kamiya, N.; Hong, Z.; Inukai, Y.; Miura, K.; Takatsuto, S.; Yoshida, S.; Ueguchi-Tanaka, M.; et al. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 2006, 140, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Montoya, T.; Nomura, T.; Farrar, K.; Kaneta, T.; Yokota, T.; Bishop, G.J. Cloning the tomato curl3 gene highlights the putative dual role of the leucine-rich repeat receptor kinase tBRI1/SR160 in plant steroid hormone and peptide hormone signaling. Plant cell 2002, 14, 3163–3176. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Bishop, G.J.; Kaneta, T.; Reid, J.B.; Chory, J.; Yokota, T. The LKA gene is a brassinosteroid insensitive 1 homolog of pea. Plant J. 2003, 36, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Chono, M.; Honda, I.; Zeniya, H.; Yoneyama, K.; Saisho, D.; Takeda, K.; Takatsuto, S.; Hoshino, T.; Watanabe, Y. A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol. 2003, 133, 1209–1219. [Google Scholar]
- Sun, Y.; Fokar, M.; Asami, T.; Yoshida, S.; Allen, R.D. Characterization of the brassinosteroid insensitive 1 genes of cotton. Plant Mol. Biol. 2004, 54, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, S.; Wu, C.; Han, T.; Wang, Q. Isolation and characterization of the brassinosteroid receptor gene (GmBRI1) from Glycine max. Int. J. Mol. Sci. 2014, 15, 3871–3888. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Tao, P.; Xu, F.; Wu, A.; Huo, W.; Wang, J. Functional Characterization of Soybean Glyma04g39610 as a Brassinosteroid Receptor Gene and Evolutionary Analysis of Soybean Brassinosteroid Receptors. Int. J. Mol. Sci. 2016, 17, 897. [Google Scholar] [CrossRef] [PubMed]
- Bajguz A, T.A. The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 2003, 62, 1027–1046. [Google Scholar] [CrossRef]
- Fujioka, S.; Yokota, T. Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 2003, 54, 137–164. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Biswas, M.; Chao, A.; Russell, D.; Chory, J. Conservation of function between mammalian and plant steroid 5α-reductases. Proc. Natl. Acad. Sci. USA 1997, 94, 3554–3559. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, T.; Fujioka, S.; Choe, S.; Takatsuto, S.; Yoshida, S.; Yuan, H.; Feldmann, K.; Tax, F. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999, 121, 743–752. [Google Scholar]
- Sathiyamoorthy, P.; Nakamura, S. In vitro root induction by 24-epibrassinolide on hypocotyl segments of soybean [Glycine max (L.) Merr.]. Plant Growth Regul. 1990, 9, 73–76. [Google Scholar] [CrossRef]
- Zurek, D.M.; Rayle, D.L.; McMorris, T.C.; Clouse, S.D. Investigation of Gene Expression, Growth Kinetics, and Wall Extensibility during Brassinosteroid-Regulated Stem Elongation. Plant Physiol. 1994, 104, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhai, Z.; Tian, X.; Duan, L.; Li, Z. Brassinolide alleviated the adverse effect of water deficits on photosynthesis and the antioxidant of soybean (Glycine max L.). Plant Growth Regul. 2008, 56, 257–264. [Google Scholar] [CrossRef]
- Li, J.; Nagpal, P.; Vitart, V.; McMorris, T.; Chory, J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 1996, 272, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Severin, A.J.; Woody, J.L.; Bolon, Y.T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.; Taylor, W.; Thornton, J. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Bancos, S.; Nomura, T.; Sato, T.; Moln’ar, G.; Bishop, G.J.; Koncz, C.; Yokota, T.; Nagy, F.; Szekeres, M. Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 2002, 130, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Asami, T.; Yoshida, S.; Nakamura, Y.; Matsuo, T.; Okamoto, S. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 2005, 138, 1117–1125. [Google Scholar] [CrossRef] [PubMed]
- Terakado, J.; Fujihara, S.; Goto, S.; Kuratani, R.; Suzuki, Y.; Yoshida, S.; Yoneyama, T. Systemic Effect of a Brassinosteroid on Root Nodule Formation in Soybean as Revealed by the Application of Brassinolide and Brassinazole. Soil Sci. Plant Nutr. 2005, 51, 389–395. [Google Scholar] [CrossRef]
- Kutschera, U.; Wang, Z.Y. Brassinosteroid action in flowering plants: A Darwinian perspective. J. Exp. Bot. 2012, 63, 3511–3522. [Google Scholar] [CrossRef] [PubMed]
- Hayat, S.; Ahmad, A. Brassinosteroids: A Class of Plant Hormone; Springer: New York, NY, USA, 2011. [Google Scholar]
- Rosati, F.; Bardazzi, I.; De Blasi, P.; Simi, L.; Scarpi, D.; Guarna, A.; Serio, M.; Racchi, M.L.; Danza, G. 5alpha-Reductase activity in Lycopersicon esculentum: cloning and functional characterization of LeDET2 and evidence of the prealsence of two isoenzymes. J. Steroid Biochem. Mol. Biol. 2005, 96, 287–299. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Xiao, Y.; Li, X.; Lu, X.; Deng, W.; Li, D.; Hou, L.; Hu, M.; Li, Y.; Pei, Y. GhDET2, a steroid 5α-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J. 2007, 51, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Basse, C.W.; Kerschbamer, C.; Brustmann, M.; Altmann, T.; Kahmann, R. Evidence for a Ustilago maydis steroid 5alpha-reductase by functional expression in Arabidopsis det2-1 mutants. Plant Physiol. 2002, 129, 717–732. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.P.; Pandey, B.K.; Deveshwar, P.; Narnoliya, L.; Parida, S.K.; Giri, J. JAZ Repressors: Potential Involvement in Nutrients Deficiency Response in Rice and Chickpea. Front. Plant Sci. 2015, 6, 975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.D.; Ling, L.Z.; Yi, T.S. Evolution and divergence of SBP-box genes in land plants. BMC Genom. 2015, 16, 787. [Google Scholar] [CrossRef] [PubMed]
- Victoria, F.d.C.; Bervald, C.M.P.; da Maia, L.C.; de Sousa, R.O.; Panaud, O.; de Oliveira, A.C. Phylogenetic relationships and selective pressure on gene families related to iron homeostasis in land plants. Genome 2012, 55, 883–900. [Google Scholar] [CrossRef] [PubMed]
- Bao, F.; Shen, J.; Brady, S.R.; Muday, G.K.; Asami, T.; Yang, Z. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol. 2004, 134, 1624–1631. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhu, W.; Chen, Y.; Ito, S.; Asami, T.; Wang, X. Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. eLife 2014, 3, e02525. [Google Scholar] [CrossRef] [PubMed]
- He, J.X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.L.; Gendron, N.; Sun, C.Q.; Wang, Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Liu, Q.; Chen, L.; Kuang, J.; Walk, T.; Wang, J.; Liao, H. Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genom. 2013, 14, 66. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method forAgrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Warnes, G.R.; Ben, B.; Lodewijk, B.; Robert, G.; Wolfgang, H.; Andy, L.; Thomas, L. GR Gplots: Various R programming tools for plotting data. R Packag. Version 2009, 3, 1. [Google Scholar]
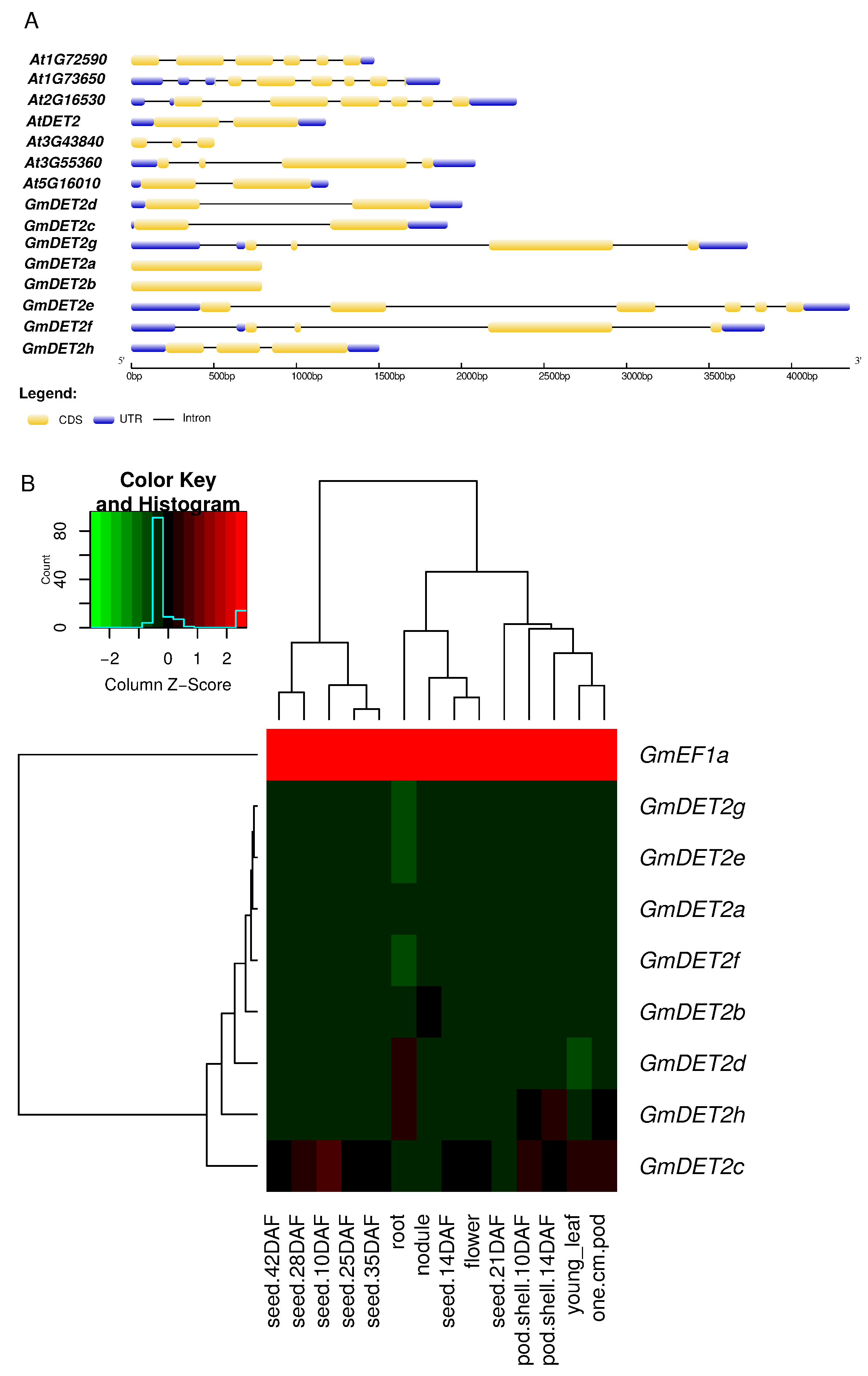
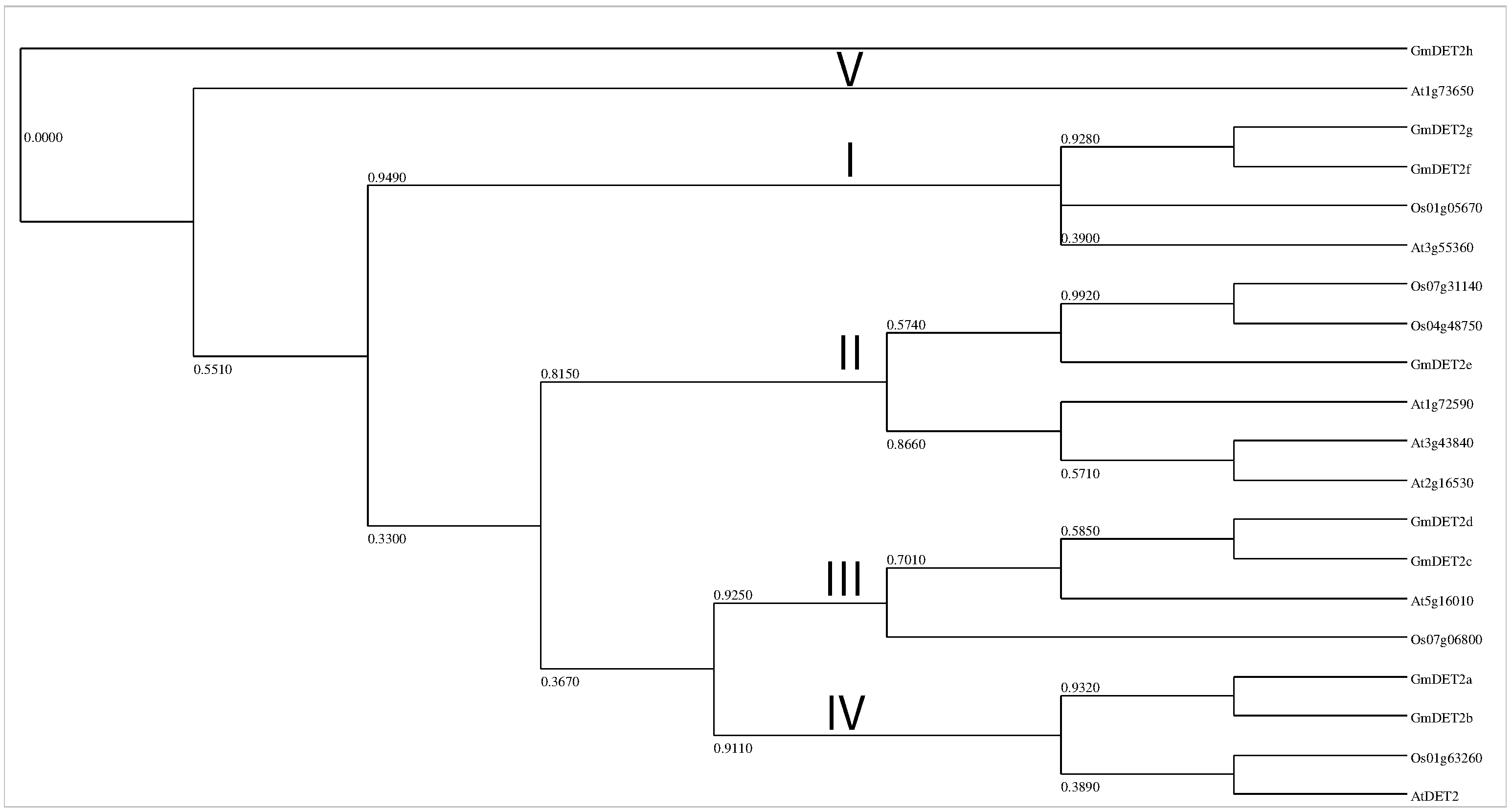
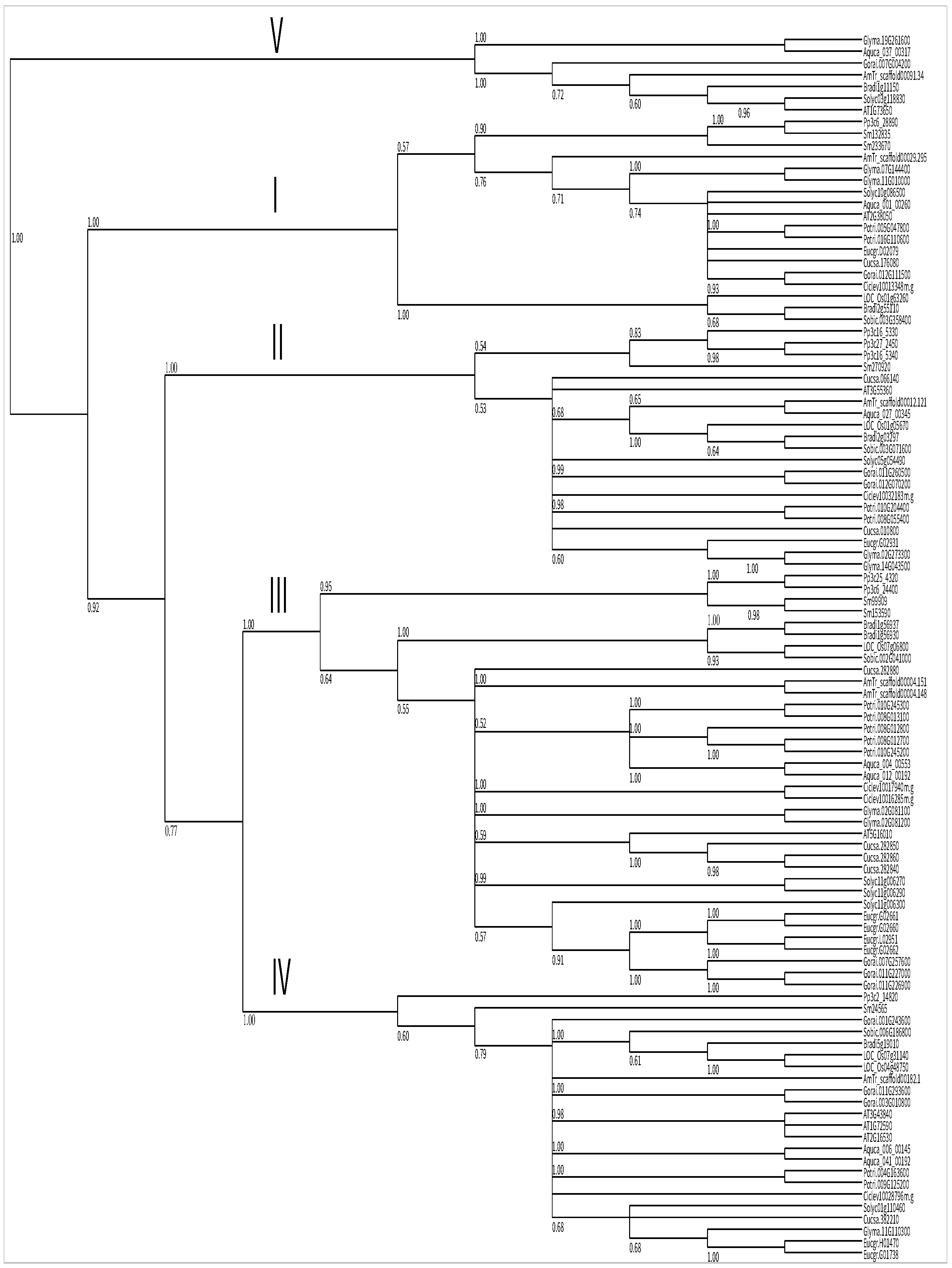

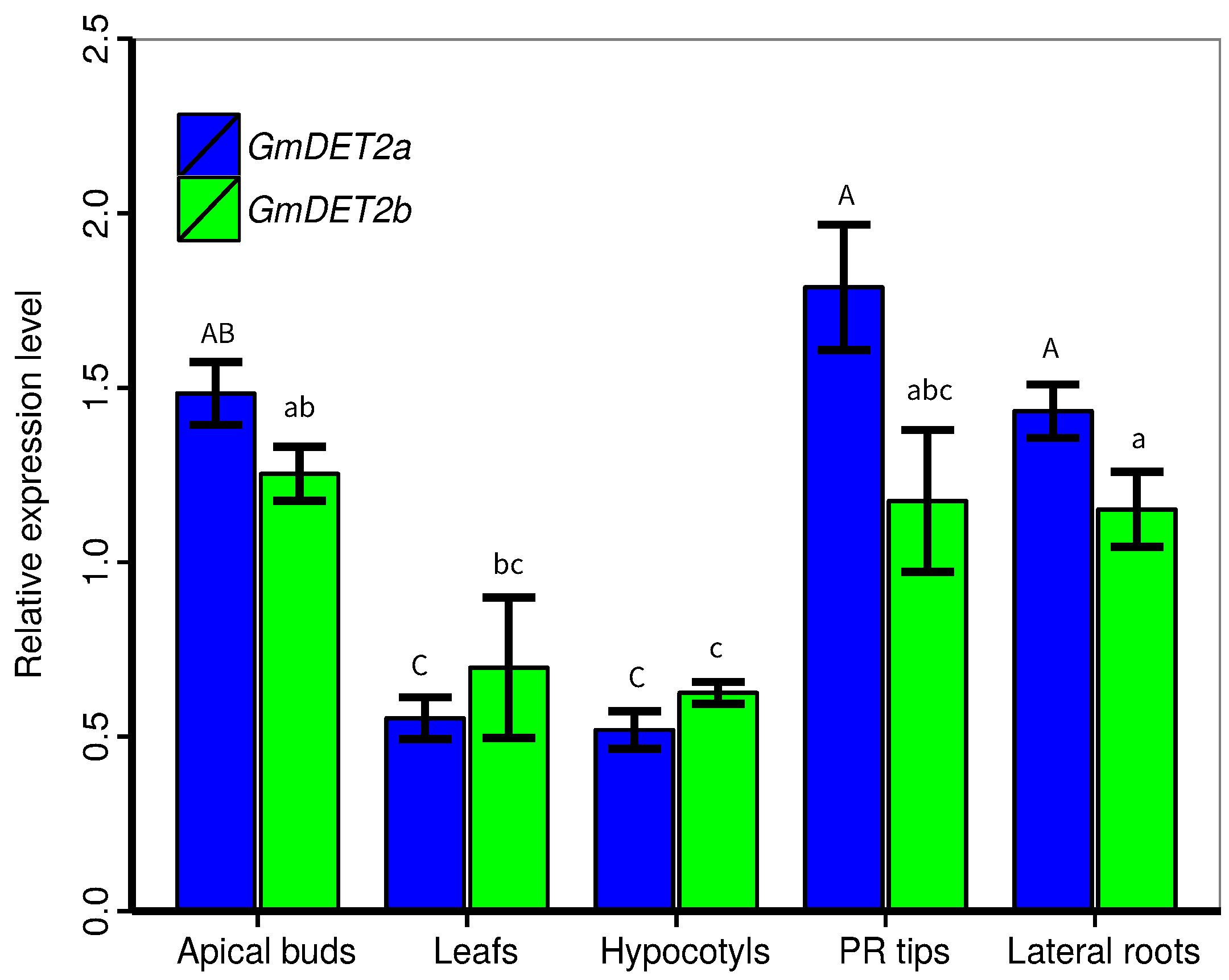



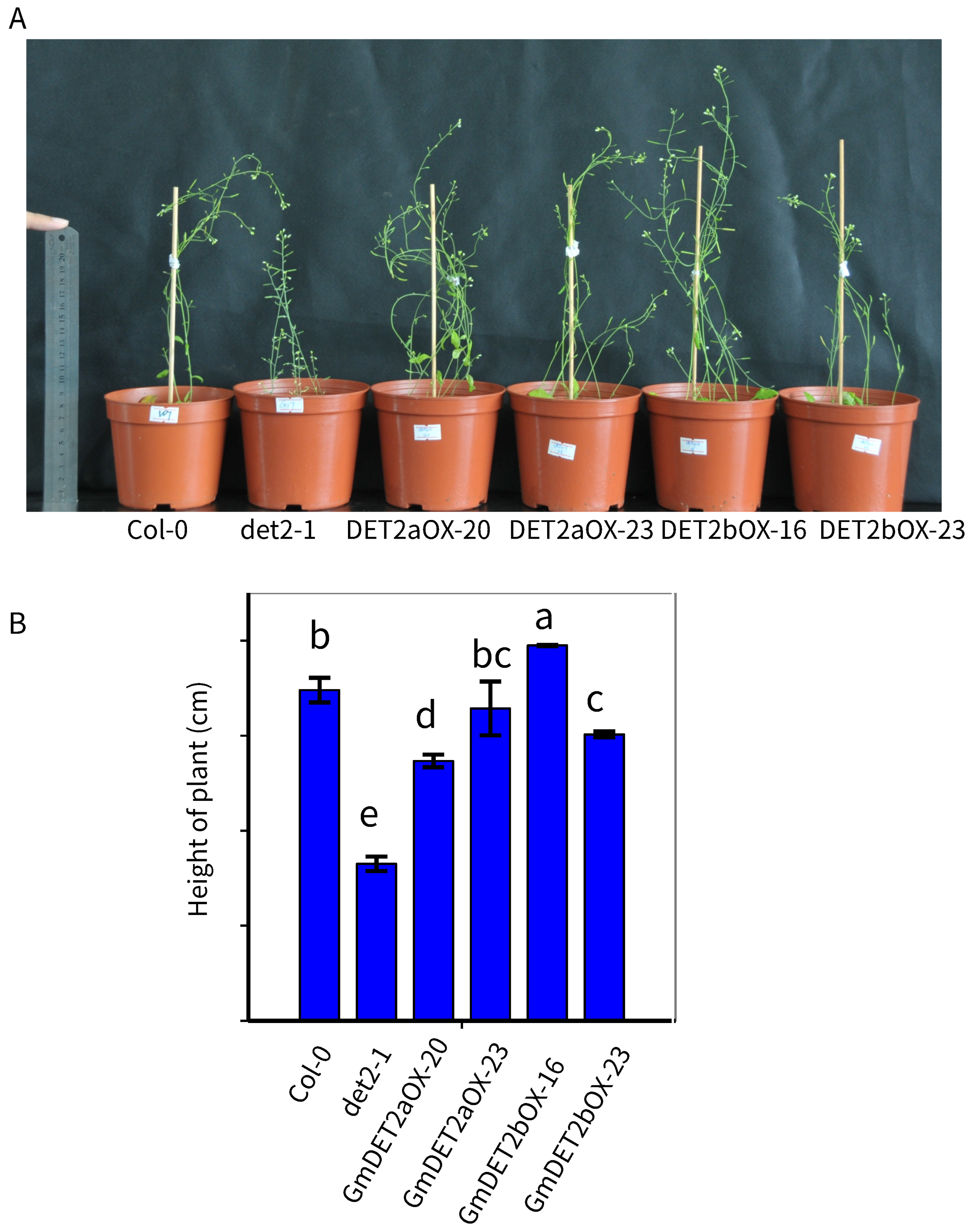

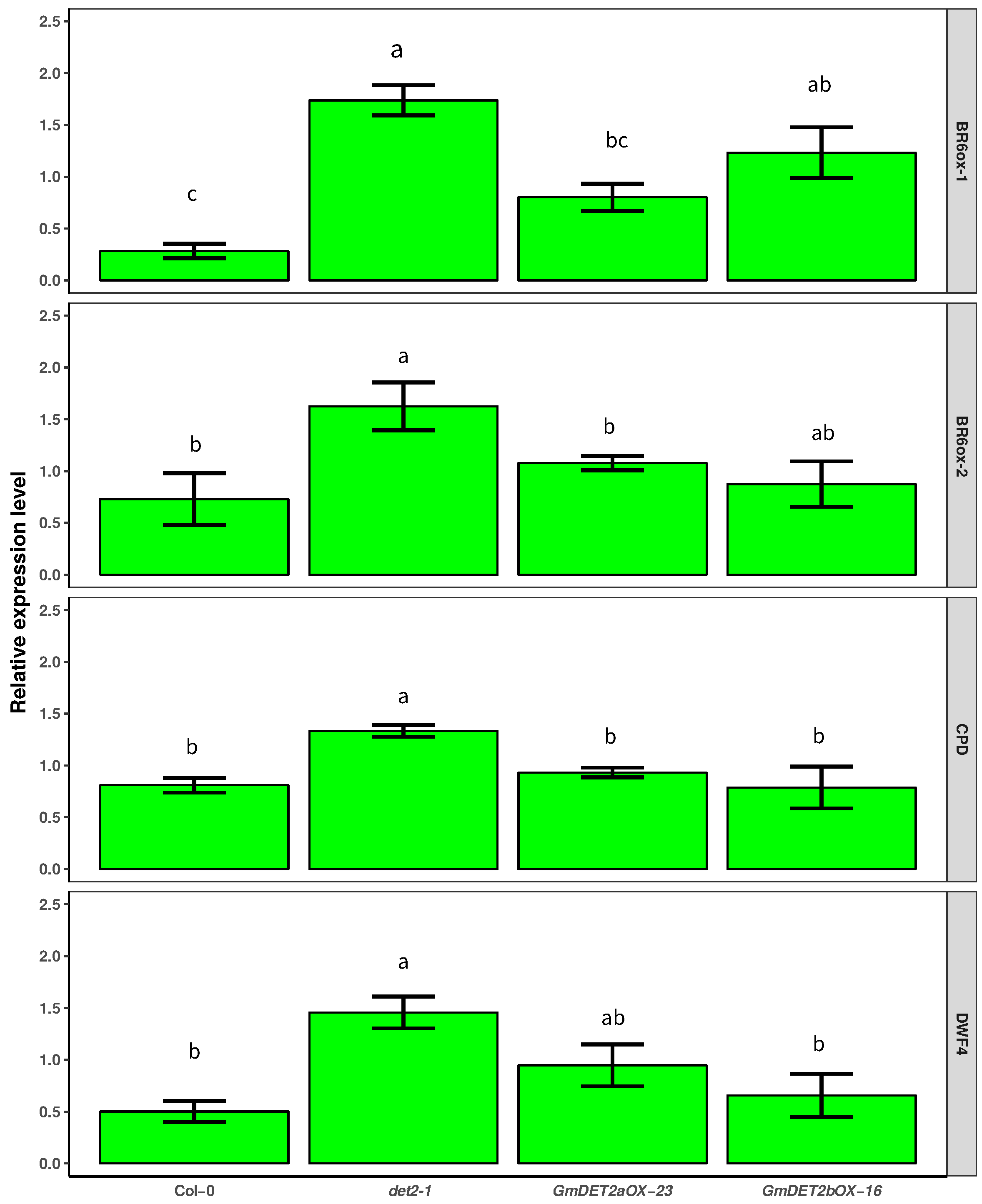
| Name | Locus | Alias | EST | Length (AA) | Putative Localization | Int/Ext |
|---|---|---|---|---|---|---|
| GmDET2a | Glyma.07G144400 | Glyma07g17410 | yes | 263 | Plas | 0/1 |
| GmDET2b | Glyma.11G010000 | Glyma11g01210 | yes | 263 | Plas | 0/1 |
| GmDET2c | Glyma.02G081200 | Glyma02g08970 | yes | 266 | Cyto | 1/2 |
| GmDET2d | Glyma.02G081100 | Glyma02g08950 | yes | 266 | Plas | 1/2 |
| GmDET2e | Glyma.11G110300 | Glyma11g11780 | yes | 342 | Nucl | 5/6 |
| GmDET2f | Glyma.14G043500 | Glyma14g04780 | yes | 309 | Plas | 3/4 |
| GmDET2g | Glyma.02g273300 | Glyma02g44090 | yes | 309 | ? | 4/5 |
| GmDET2h | Glyma.19g261600 | Glyma19g45120 | yes | 317 | ? | 2/3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huo, W.; Li, B.; Kuang, J.; He, P.; Xu, Z.; Wang, J. Functional Characterization of the Steroid Reductase Genes GmDET2a and GmDET2b from Glycine max. Int. J. Mol. Sci. 2018, 19, 726. https://doi.org/10.3390/ijms19030726
Huo W, Li B, Kuang J, He P, Xu Z, Wang J. Functional Characterization of the Steroid Reductase Genes GmDET2a and GmDET2b from Glycine max. International Journal of Molecular Sciences. 2018; 19(3):726. https://doi.org/10.3390/ijms19030726
Chicago/Turabian StyleHuo, Weige, Bodi Li, Jiebing Kuang, Pingan He, Zhihao Xu, and Jinxiang Wang. 2018. "Functional Characterization of the Steroid Reductase Genes GmDET2a and GmDET2b from Glycine max" International Journal of Molecular Sciences 19, no. 3: 726. https://doi.org/10.3390/ijms19030726






