De Novo Assembly and Characterization of the Xenocatantops brachycerus Transcriptome
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sequencing Analysis and Assembly
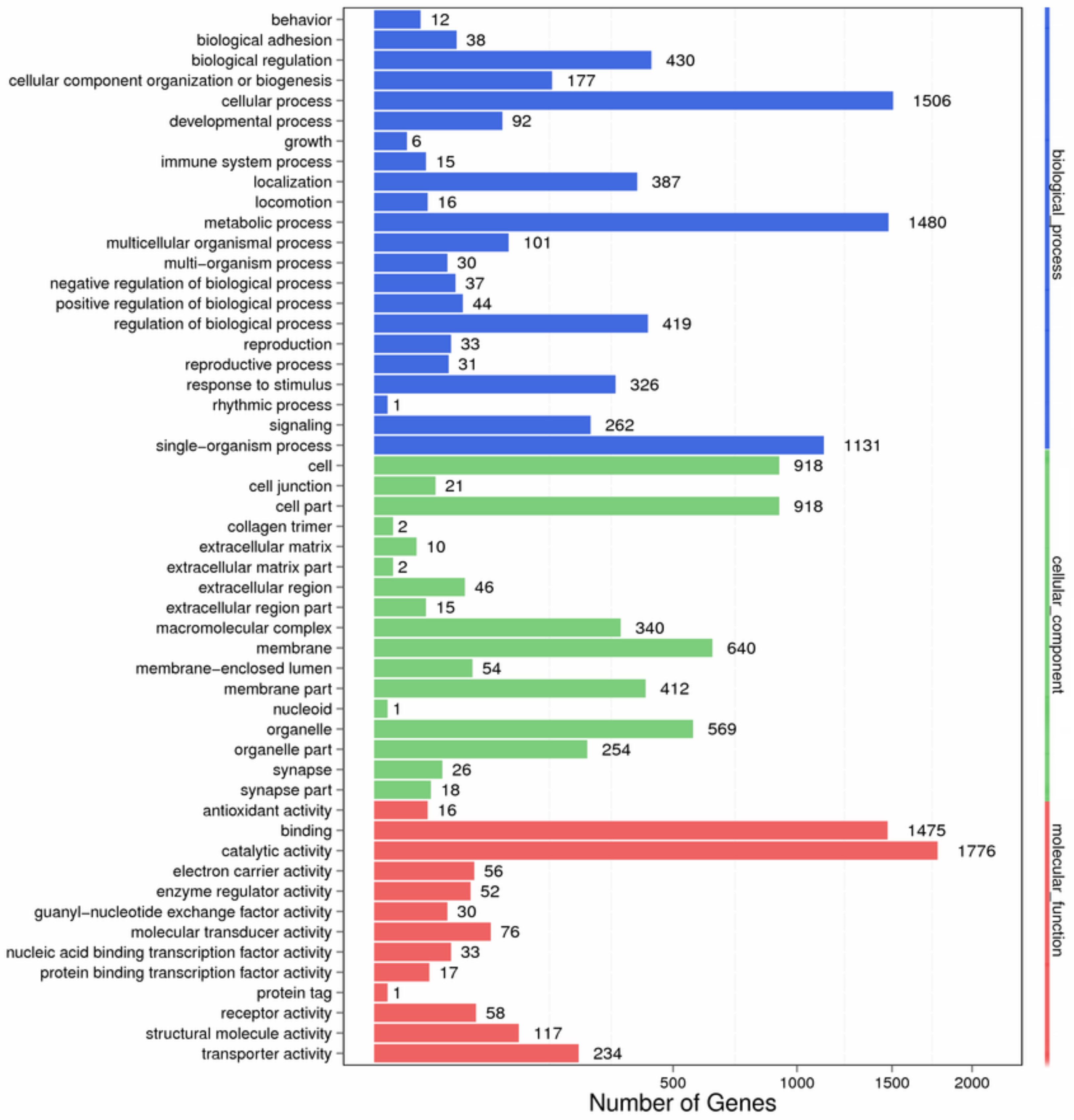
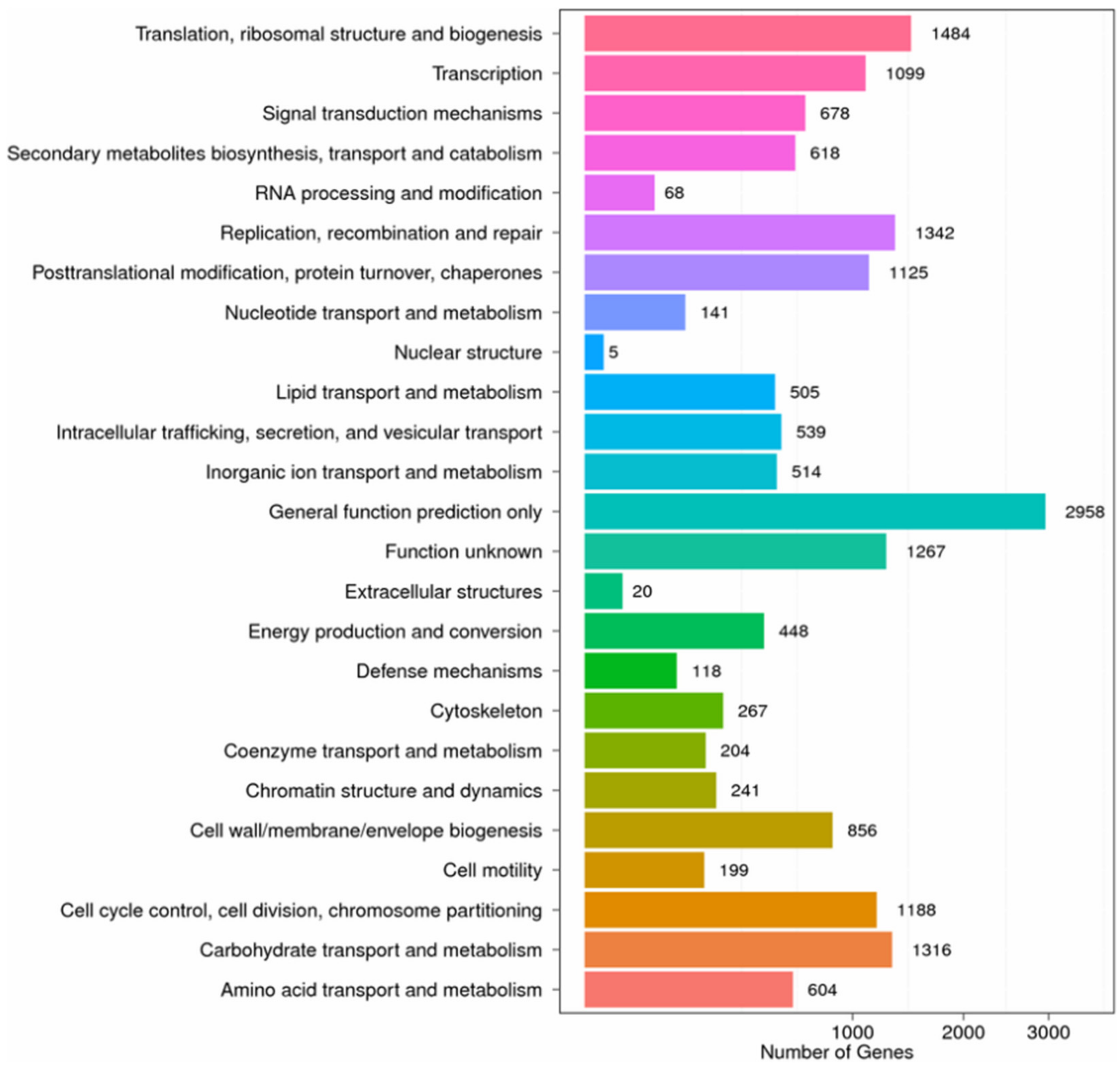
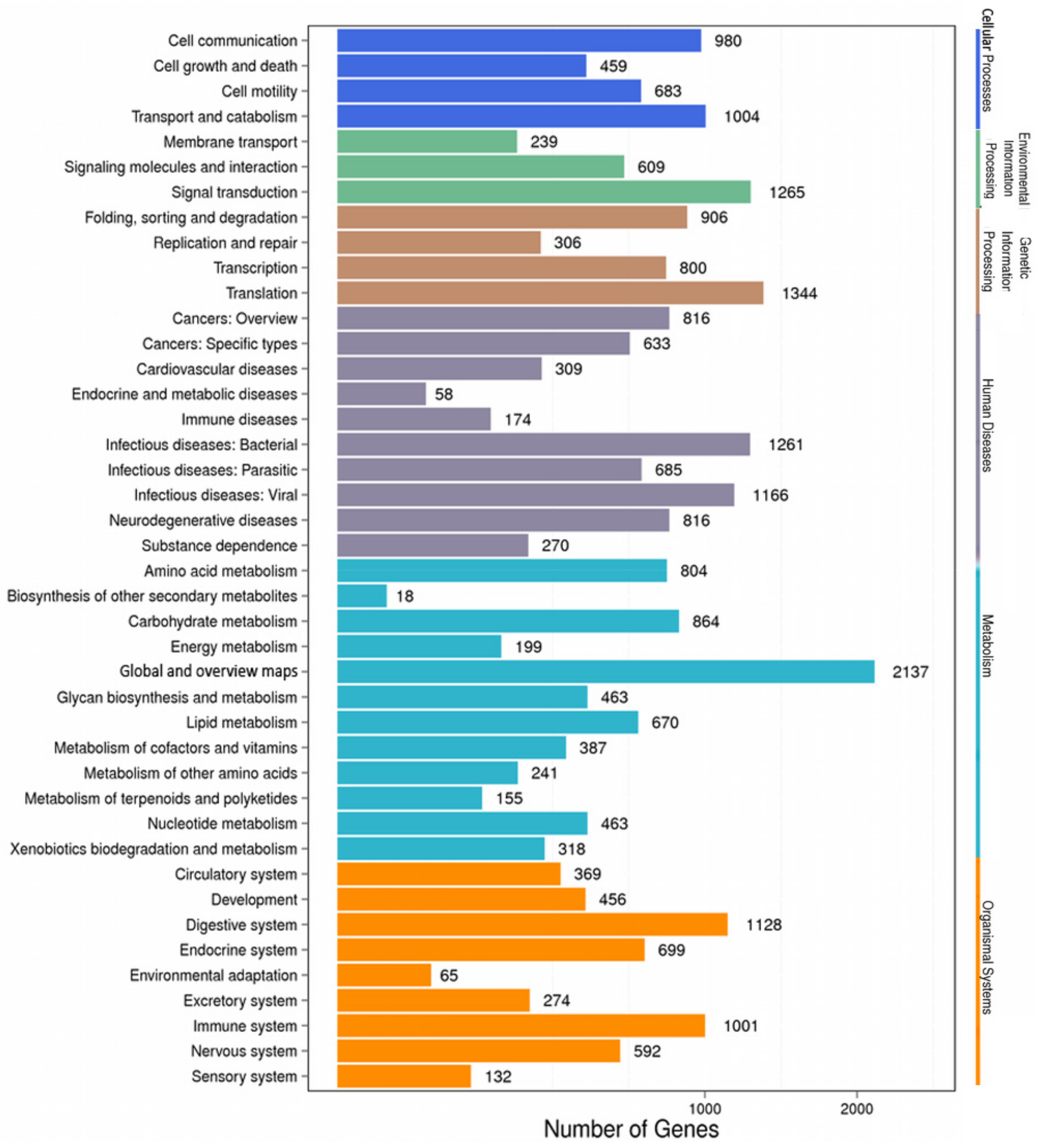
2.2. Functional Annotation
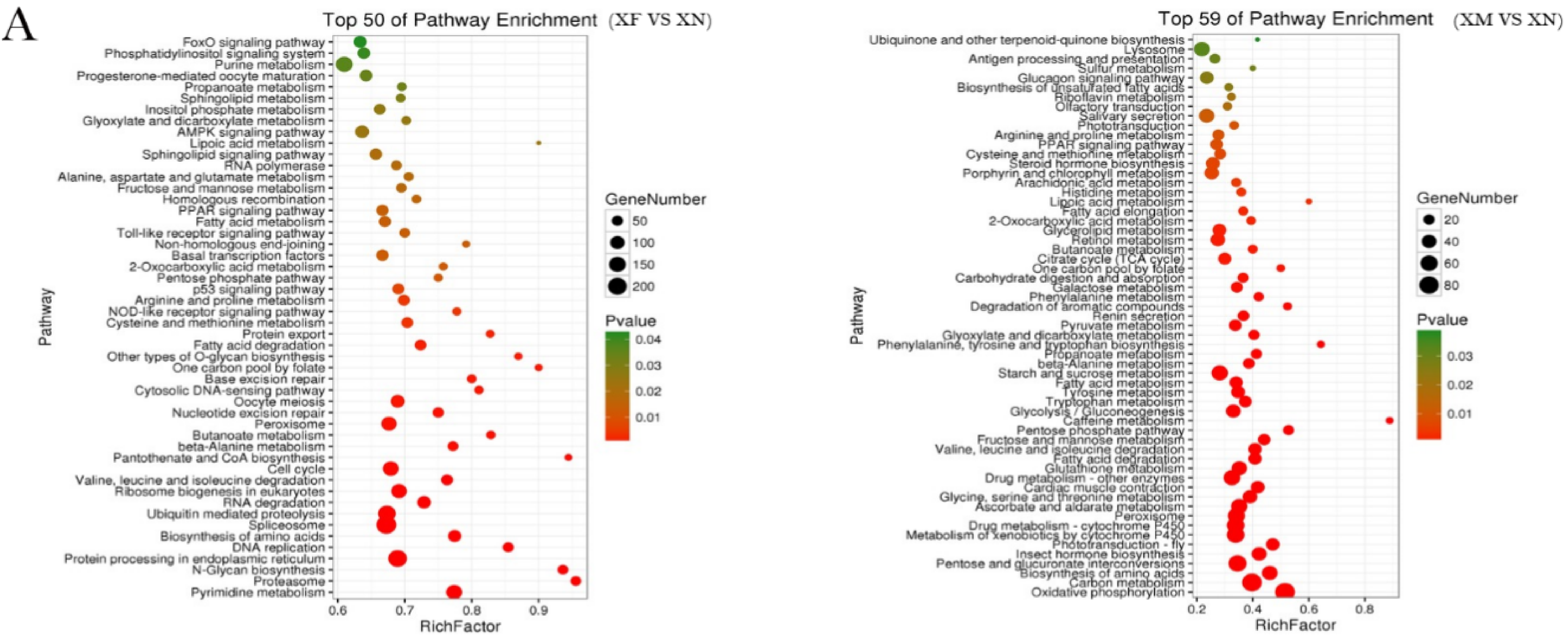
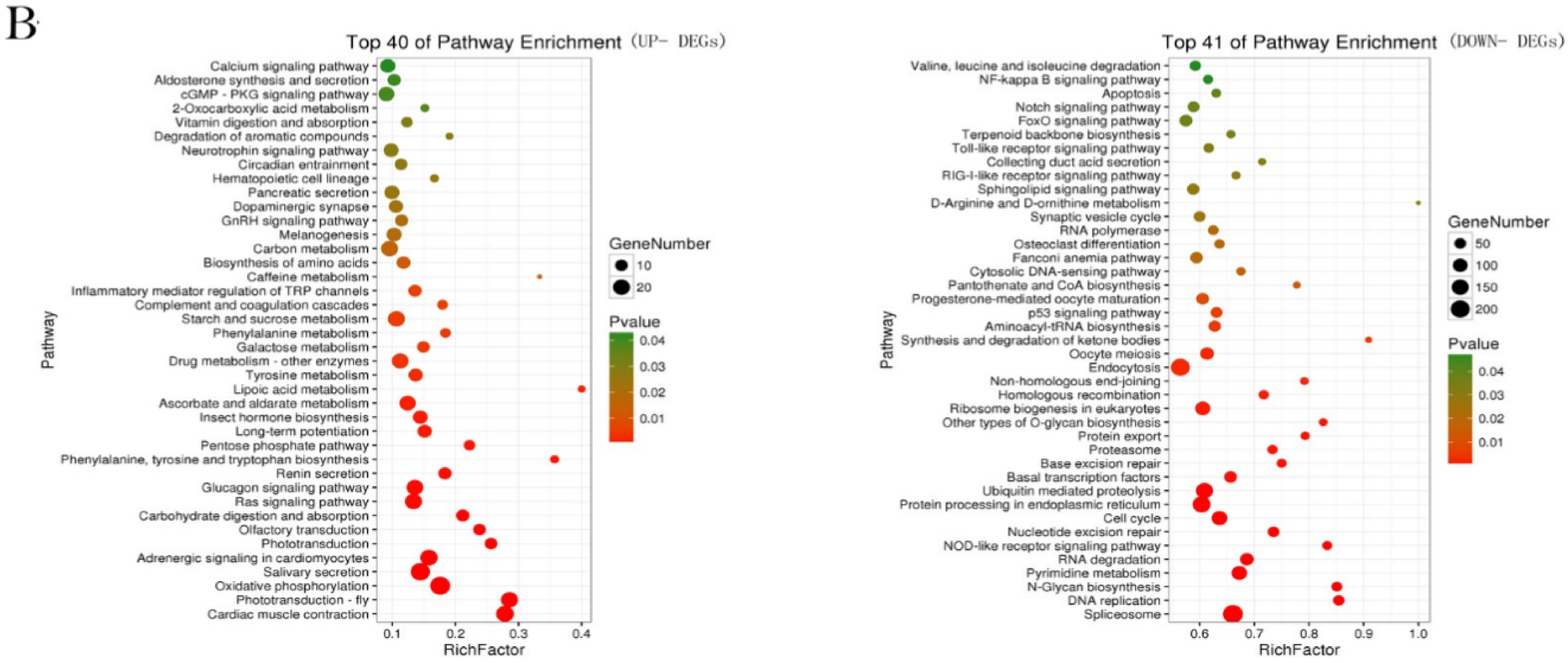
2.3. Functional Enrichment Analysis of DEGs
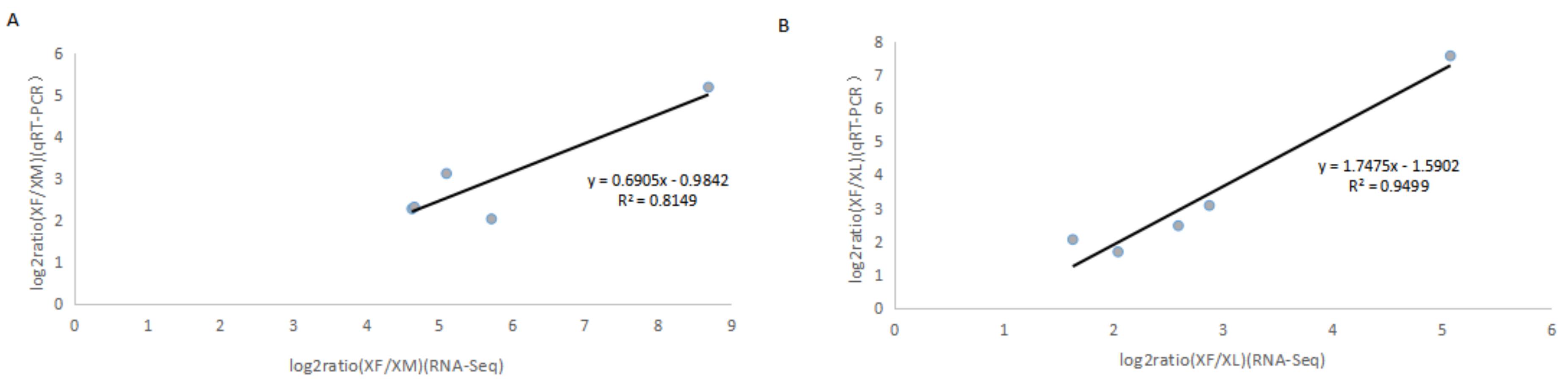
2.4. Validation of RNA-Seq Gene Expression Data
2.5. Candidate Genes Involved in Growth, Development, Immunity, and Nutritional and Bioactive Compounds Metabolism
3. Materials and Methods
3.1. Ethics Statement
3.2. Species Collection, RNA Extraction, and Illumina Sequencing
3.3. De Novo Assembly and Annotation
3.4. Differentially Expressed Unigenes (DEGs)
3.5. Quantitative Real-Time RT-PCR (qRT-PCR)
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Yang, Y.; Tian, W.; Wang, C.; Luo, Y. Current occurrence of locust plague and utilization and industrialization of grasshopper resource in Guangdong Province, China. J. Environ. Entomol. 2013, 35, 372–380. [Google Scholar]
- Deveson, E.; Martinez, A. Locusts in southern settler societies: Argentine and Australian experience and responses, 1880–1940. In Environmental History in the Making; Springer: Cham, Switzerland, 2017; pp. 259–286. [Google Scholar]
- Duan, Y.F. The Study on Nutritional Value and Biological Function of Locust. Ph.D. Thesis, Shaanxi Normal University, Xi’an, China, 2005. [Google Scholar]
- Sun, T.; Shang, Z.; Liu, Z.; Long, R. Nutrient Composition of Four Species of Grasshoppers from Alpine Grasslands in the Qilian Mountain of the Tibetan Plateau, China. Philipp. Agric. Sci. 2010, 93, 97–103. [Google Scholar]
- Blásquez, J.R.-E.; Moreno, J.M.P.; Camacho, V.H.M. Could Grasshoppers Be a Nutritive Meal? Food Nutr. Sci. 2012, 3, 164–175. [Google Scholar] [CrossRef]
- Xiong, Z.Y.; Xi, B.X.; Zhang, K.R.; He, D.P. Analysis and Evaluation of Nutritional Components of Four Species of Grasshoppers. J. Nutr. 1999, 21, 474–477. [Google Scholar]
- Finke, M.D. Complete nutrient content of four species of commercially available feeder insects fed enhanced diets during growth. Zoo Biol. 2015, 34, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Ssepuuya, G.; Mukisa, I.M.; Nakimbugwe, D. Nutritional composition, quality, and shelf stability of processed Ruspolianitidula (edible grasshoppers). Food Sci. Nutr. 2017, 5, 103–112. [Google Scholar] [CrossRef] [PubMed]
- WHO/FAO/UNU Expert Consultation. Protein and Amino Acid Requirements in Human Nutrition; World Health Organization Technical Report Series; World Health Organization: Geneva, Switzerland, 2007; pp. 1–265. ISBN 9241209356. [Google Scholar]
- Qin, L.P.; Liu, Z.Y.; Sun, T. Advance in nutritive value of grassland grasshopper. Pratacult. Sci. 2013, 30, 141–147. [Google Scholar]
- Food and Agriculture Organization of the United Nations (FAO). Edible Insects: Future Prospects for Food and Feed Security; Food and Agriculture Organization of the United Nations: Rome, Italy, 2013. [Google Scholar]
- Feng, Y.; Chen, X.M.; Zhao, M.; He, Z.; Sun, L.; Wang, C.Y.; Ding, W.F. Edible insects in China: Utilization and prospects. Insect Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, J. Edible insects in Japan. In Ecological Implications of Minilivestock: Potential of Insects, Rodents, Frogs and Snails; Science Publishers, Inc.: Boca Raton, FL, USA, 2005; pp. 251–262. [Google Scholar]
- Yang, L.F.; Siriamornpun, S.; Li, D. Polyunsaturated fatty acid content of edible insects in Thailand. J. Food Lipids 2006, 13, 277–285. [Google Scholar] [CrossRef]
- Yang, Y.P.; Ke, Y.L.; Wu, W.J.; Tian, W.J.; Luo, Y.L.; Li, W. Mass-rearing of two Catantopinae live feed for entomophagous birds. Guangdong Agric. Sci. 2015, 7, 99–104. [Google Scholar]
- Jizng, Z.; Shihong, Y.; Jingyou, W. Reasearch of karyotypes and c-banding of Xenocatantops humilis brachychrus. Dep. Biol. Sci. Biotechnol. 1997, 15, 33–36. [Google Scholar]
- Yang, J.; Liu, Y.; Liu, N. The complete mitochondrial genome of the Xenocatantops brachycerus (Orthoptera: Catantopidae). Mitochondrial DNA Part A 2016, 27, 2844–2845. [Google Scholar] [CrossRef]
- Morozova, O.; Hirst, M.; Marra, M.A. Applications of New Sequencing Technologies for Transcriptome Analysis. Ann. Rev. Genom. Hum. Genet. 2009, 10, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Hittinger, C.T.; Johnston, M.; Tossberg, J.T.; Rokas, A. Leveraging skewed transcript abundance by RNA-Seq to increase the genomic depth of the tree of life. Proc. Natl. Acad. Sci. USA 2010, 107, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wei, W.; Chu, Y.; Zhang, L.; Shen, J.; An, C. De novo transcriptome analysis of wing development-related signaling pathways in Locusta migratoria manilensis and Ostrinia furnacalis (Guenee). PLoS ONE 2014, 9, e106770. [Google Scholar] [CrossRef] [PubMed]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Bando, T.; Ishimaru, Y.; Kida, T.; Hamada, Y.; Matsuoka, Y.; Nakamura, T.; Ohuchi, H.; Noji, S.; Mito, T. Analysis of RNA-Seq data reveals involvement of JAK/STAT signaling during leg regeneration in the cricket Gryllus bimaculatus. Development 2013, 140, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Liu, F.; Lu, H.; Huang, Y. Characterization and analysis of a de novo transcriptome from the pygmy grasshopper Tetrix japonica. Mol. Ecol. Resour. 2017, 17, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Liu, F.; Lu, H.; Yuan, H.; Zhang, Q.; Huang, Y. De novo assembly and characterization of the transcriptome of grasshopper Shirakiacris shirakii. Int. J. Mol. Sci. 2016, 17, 1110. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Cong, B.; Wang, L.; Gao, Y.; Zhang, H.; Dong, H.; Lin, Z. Differential Gene Expression Analysis of the Epacromiuscoerulipes (Orthoptera: Acrididae) Transcriptome. J. Insect Sci. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Chai, Y.; Zhang, L.; Zhao, Z.; Gao, L.L.; Ma, R. Transcriptome Analysis and Identification of Major Detoxification Gene Families and Insecticide Targets in Grapholitamolesta (Busck) (Lepidoptera: Tortricidae). J. Insect Sci. 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Deli, X.; Dehua, W. Advances in ecological immunology. Acta Ecol. Sin. 2012, 32, 6251–6258. [Google Scholar] [CrossRef]
- Sheldon, B.C.; Verhulst, S. Ecological immunology: Costly parasite defenses and trade-offs in evolutionary ecology. Trends Ecol. Evol. 1996, 11, 317–321. [Google Scholar] [CrossRef]
- Freitak, D.; Wheat, C.W.; Heckel, D.G.; Vogel, H. Immune system responses and fitness costs associated with consumption of bacteria in larvae of Trichoplusiani. BMC Biol. 2007, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Bell, L.R.; Maine, E.M.; Schedl, P.; Cline, T.W. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell 1988, 55, 1037–1046. [Google Scholar] [CrossRef]
- Salz, H.K.; Erickson, J.W. Sex determination in Drosophila: The view from the top. Fly 2010, 4, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, A.F.; Hoy, M.A. Expression analysis of Drosophila doublesex, transformer-2, intersex, fruitless-like, and vitellogenin homologs in the parahaploid predator Metaseiulus occidentalis (Chelicerata: Acari: Phytoseiidae). Exp. Appl. Acarol. 2015, 65, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Min, K.J.; Jones, N.; Borst, D.W.; Rankin, M.A. Increased juvenile hormone levels after long-duration flight in the grasshopper, Melanoplus sanguinipes. J. Insect Physiol. 2004, 50, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Kang, L. Molecular Mechanisms of Phase Change in Locusts. Ann. Rev. Entomol. 2014, 59, 225–244. [Google Scholar] [CrossRef] [PubMed]
- Perrimon, N.; Pitsouli, C.; Shilo, B.-Z. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef] [PubMed]
- Beermann, A.; Pruhs, R.; Lutz, R.; Schroder, R. A context-dependent combination of Wnt receptors controls axis elongation and leg development in a short germ insect. Development 2011, 138, 2793–2805. [Google Scholar] [CrossRef] [PubMed]
- Futahashi, R.; Shirataki, H.; Narita, T.; Mita, K.; Fujiwara, H. Comprehensive microarray-based analysis for stage-specific larval camouflage pattern-associated genes in the swallowtail butterfly, Papilioxuthus. BMC Biol. 2012, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Pires-daSilva, A.; Sommer, R.J. The evolution of signaling pathways in animal development. Nat. Rev. Genet. 2003, 4, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Yang, P.; Zhang, Z.; Wu, G.X.; Yang, B. Transcriptomic Immune Response of Tenebrio molitor Pupae to Parasitization by Scleroderma guani. PLoS ONE 2013, 8, e54411. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Q.; Zhu, Y.F.; Ma, C.; Fabrick, J.A.; Kanost, M.R. Pattern recognition proteins in Manducasexta plasma. Insect Biochem. Mol. Biol. 2002, 32, 1287–1293. [Google Scholar] [CrossRef]
- Royet, J.; Dziarski, R. Peptidoglycan recognition proteins: Pleiotropic sensors and effectors of antimicrobial defenses. Nat. Rev. Microbiol. 2007, 5, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.-L.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defense mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Ligoxygakis, P.; Bulet, P.; Reichhart, J.-M. Critical evaluation of the role of the Toll-like receptor 18-Wheeler in the host defense of Drosophila. EMBO Rep. 2002, 3, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gong, Z.J.; Rao, X.J.; Li, M.Y.; Li, S.G.; Jurenka, R.A. Identification of putative carboxylesterase and glutathione S-transferase genes from the antennae of the Chilo suppressalis (Lepidoptera: Pyralidae). J. Insect Sci. 2015, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Lee, S.M.; Jung, C.; Meyer-Rochow, V.B. Nutritional composition of five commercial edible insects in South Korea. J. Asia Pac. Entomol. 2017, 20, 686–694. [Google Scholar] [CrossRef]
- Belluco, S.; Losasso, C.; Maggioletti, M.; Alonzi, C.C.; Paoletti, M.G.; Ricci, A. Edible insects in a food safety and nutritional perspective: A critical review. Compr. Rev. Food Sci. Food Saf. 2013, 12, 296–313. [Google Scholar] [CrossRef]
- Yao, J.; Yao, S. The development and utilization of grasshopper resource in Guizhou. J. Guizhou Norm Univ. Nat. Sci. 2006, 24, 19–24. [Google Scholar]
- Paul, A.; Frederich, M.; Uyttenbroeck, R.; Hatt, S.; Malik, P.; Lebecque, S.; Hamaidia, M.; Miazek, K.; Goffin, D.; Willems, L.; et al. Grasshoppers as a food source? A review. Biotechnol. Agron. Soc. Environ. 2016, 20, 337–352. [Google Scholar]
- Borrelli, G.M.; Trono, D. Recombinant Lipases and Phospholipases and Their Use as Biocatalysts for Industrial Applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M. Phospholipids for functional food. Eur. J. Lipid Sci. Technol. 2001, 103, 98–101. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Tsai, B.P.J.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Iseli, C.; Jongeneel, C.V.; Bucher, P. ESTScan: A program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. In Proceedings of the Seventh International Conference on Intelligent Systems for Molecular Biology, Heidelberg, Germany, 6–10 August 1999; pp. 138–148. [Google Scholar]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, C.; Gao, Z.; Min, J.; Gu, Y.; Jian, J.; Jiang, X.; Cai, H.; Ebersberger, I.; Xu, M. The draft genome of blunt snout bream (Megalobrama amblycephala) reveals the development of intermuscular bone and adaptation to herbivorous diet. Gigascience 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.-M.; Yan, Z.-H.; Zhang, J.-E.; Li, H.-Y. Immune related genes expression in juveniles of an invasive snail after challenged with Lipopolysaccharide. Invertebr. Surviv. J. 2017, 14, 295–302. [Google Scholar]
- Li, S.; Liu, H.; Bai, J.; Zhu, X. Transcriptome assembly and identification of genes and SNPs associated with growth traits in largemouth bass (Micropterus salmoides). Genetica 2017, 145, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Item | Number | Percentage |
|---|---|---|
| Total | 43,187 | 100% |
| nr-annotated | 21,978 | 50.89% |
| nt-annotated | 12,071 | 27.95% |
| Swiss-Prot-annotated | 17,692 | 40.97% |
| KEGG-annotated | 15,762 | 36.50% |
| COG-annotated | 9127 | 21.13% |
| InterPro-annotated | 15,099 | 34.96% |
| GO-annotated | 3272 | 7.58% |
| All annotated | 24,717 | 57.23% |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Zhang, X.; Qiu, Z.; Huang, Y. De Novo Assembly and Characterization of the Xenocatantops brachycerus Transcriptome. Int. J. Mol. Sci. 2018, 19, 520. https://doi.org/10.3390/ijms19020520
Zhao L, Zhang X, Qiu Z, Huang Y. De Novo Assembly and Characterization of the Xenocatantops brachycerus Transcriptome. International Journal of Molecular Sciences. 2018; 19(2):520. https://doi.org/10.3390/ijms19020520
Chicago/Turabian StyleZhao, Le, Xinmei Zhang, Zhongying Qiu, and Yuan Huang. 2018. "De Novo Assembly and Characterization of the Xenocatantops brachycerus Transcriptome" International Journal of Molecular Sciences 19, no. 2: 520. https://doi.org/10.3390/ijms19020520





