Lignins: Biosynthesis and Biological Functions in Plants
Abstract
1. Introduction
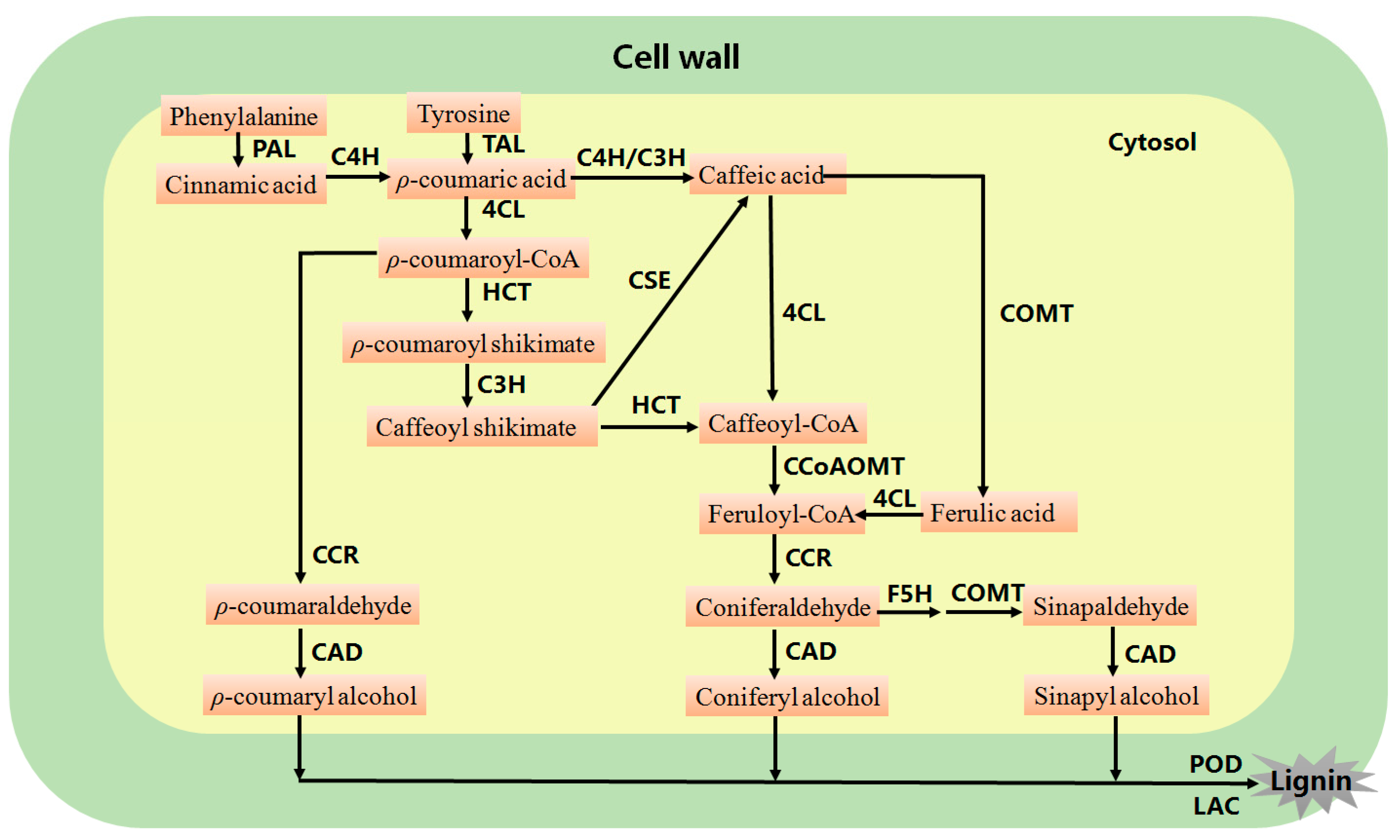
2. Genetic Modification of Lignin Biosynthesis
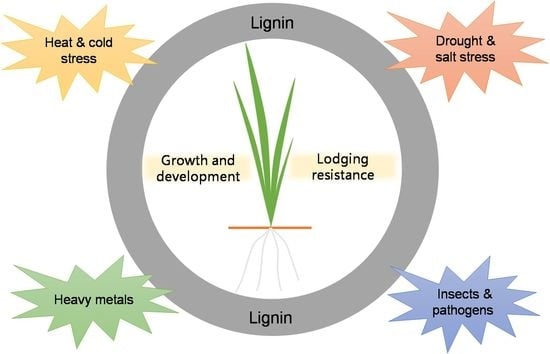
3. Role of Lignin in Plant Growth and Development
4. Role of Lignin in Plant Lodging Resistance
5. Relationship between Lignin Biosynthesis and Plant Stress Adaptation
5.1. Roles of Lignin Biosynthesis in Plant Insect Pests and Diseases Resistance
5.2. Role of Lignin Deposition in Plant Heavy Metal Tolerance
5.3. Role of Lignin in Plant Drought and Salt Stress Tolerance
5.4. Role of Lignin in Plant Temperature Stress Adaptability
6. Different Types of Lignin and Its Application
7. Conclusions and Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ralph, J.; Lundquist, K.; Brunow, G.; Lu, F.; Kim, H.; Schatz, P.F.; Marita, J.M.; Hatfield, R.D.; Ralph, S.A.; Christensen, J.H. Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenyl-propanoids. Phytochem. Rev. 2004, 3, 29–60. [Google Scholar] [CrossRef]
- Alejandro, S.; Lee, Y.; Tohge, T.; Sudre, D.; Osorio, S.; Park, J.; Bovet, L.; Geldner, N.; Fernie, A.R.; Martinoia, E. AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr. Biol. 2012, 22, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Liu, C. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 22728–22733. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Chapple, C. The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Miao, Y.C.; Zhang, K.W. Sequestration and transport of lignin monomeric precursors. Molecules 2011, 16, 710–727. [Google Scholar] [CrossRef] [PubMed]
- Ralph, J. Hydroxycinnamates in lignification. Phytochem. Rev. 2010, 9, 65–83. [Google Scholar] [CrossRef]
- Del Río, J.C.; Rencoret, J.; Prinsen, P.; Martínez, Á.T.; Ralph, J.; Gutiérrez, A. Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J. Agric. Food Chem. 2012, 60, 5922–5935. [Google Scholar] [CrossRef] [PubMed]
- Mottiar, Y.; Vanholme, R.; Boerjan, W.; Ralph, J.; Mansfield, S.D. Designer lignins: Harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 2016, 37, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Bashri, G.; Singh, A.; Prasad, S.M. Regulation of Xenobiotics in Higher Plants: Signalling and Detoxification; Springer: Singapore, 2016; pp. 39–56. [Google Scholar]
- Lan, W.; Lu, F.; Regner, M.; Zhu, Y.; Rencoret, J.; Ralph, S.A.; Zakai, U.I.; Morreel, K.; Boerjan, W.; Ralph, J. Tricin, a flavonoid monomer in monocot lignification. Plant Physiol. 2015, 167, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Eloy, N.; Voorend, W.; Lan, W.; Saleme, M.L.; Cesarino, I.; Vanholme, R.; Smith, R.A.; Goeminne, G.; Pallidis, A.; Morreel, K.; et al. Silencing chalcone synthase impedes the incorporation of tricin in lignin and increases lignin content. Plant Physiol. 2017, 173, 998–1016. [Google Scholar] [CrossRef] [PubMed]
- Del Río, J.C.; Rencoret, J.; Gutiérrez, A.; Kim, H.; Ralph, J. Hydroxystilbenes are monomers in palm fruit endocarp lignins. Plant Physiol. 2017, 174, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, M.; Benske, A.; Smith, R.A.; Watanabe, Y.; Tobimatsu, Y.; Ralph, J.; Demura, T.; Ellis, B.; Samuels, A.L. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 2014, 166, 798–807. [Google Scholar] [CrossRef] [PubMed]
- Ithal, N.; Recknor, J.; Nettleton, D.; Maier, T.; Baum, T.J.; Mitchum, M.G. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant Microbe Interact. 2007, 20, 510–525. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.C.; Sayre, K.D.; Kaul, J.N.; Narang, R.S. Growth and morphology of spring wheat (Triticum aestivum L.) culms and their association with lodging: Effects of genotypes, N levels and ethephon. Field Crop. Res. 2003, 84, 271–290. [Google Scholar] [CrossRef]
- Rest, B.V.D.; Rochange, S.F. Down-regulation of cinnamoyl-CoA reductase in tomato (Solanum lycopersicum L.) induces dramatic changes in soluble phenolic pools. J. Exp. Bot. 2006, 57, 1399–1411. [Google Scholar] [CrossRef] [PubMed]
- Shadle, G.; Chen, F.; Srinivasa Reddy, M.S.; Jackson, L.; Nakashima, J.; Dixon, R.A. Down-regulation of hydroxycinnamoyl CoA: Shikimate hydroxycinnamoyl transferase in transgenic alfalfa affects lignification, development and forage quality. Phytochemistry 2007, 68, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Derikvand, M.M.; Sierra, J.B.; Ruel, K.; Pollet, B.; Do, C.T.; Thévenin, J.; Buffard, D.; Jouanin, L.; Lapierre, C. Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 2008, 227, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Moura, J.C.M.S.; Bonine, C.A.V.; Viana, J.D.O.F.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Storme, V.; Vanholme, B.; Sundin, L.; Christensen, J.H.; Goeminne, G.; Halpin, C.; Rohde, A.; Morreel, K.; Boerjan, W. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 2012, 24, 3506–3529. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.; Yu, J.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.; Shen, J.; Li, L. Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol. 2011, 157, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Donaldson, L.; Kim, H.; Phillips, L.; Flint, H.; Steward, D.; Torr, K.; Koch, G.; Schmitt, U.; Ralph, J. Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiol. 2009, 149, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Thevenin, J.; Pollet, B.; Letarnec, B.; Saulnier, L.; Gissot, L.; Maiagrondard, A.; Lapierre, C.; Jouanin, L. The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol. Plant 2011, 4, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, R.; Déjardin, A.; Desmet, S.; Hoengenaert, L.; Vanholme, R.; Morreel, K.; Laurans, F.; Kim, H.; Santoro, N.; Foster, C.; et al. Different metabolic routes for coniferaldehyde and sinapaldehyde with CINNAMYL ALCOHOL DEHYDROGENASE1 deficiency. Plant Physiol. 2017, 175, 1018–1039. [Google Scholar] [CrossRef] [PubMed]
- Giordano, A.; Liu, Z.; Panter, S.N.; Dimech, A.M.; Shang, Y.; Wijesinghe, H.; Fulgueras, K.; Ran, Y.; Mouradov, A.; Rochfort, S. Reduced lignin content and altered lignin composition in the warm season forage grass Paspalum dilatatum by down-regulation of a Cinnamoyl CoA Reductase Gene. Transgenic Res. 2014, 23, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.; Rochfort, S.; Liu, Z.; Ran, Y.; Griffith, M.; Badenhorst, P.; Louie, G.V.; Bowman, M.E.; Smith, K.F.; Noel, J.P.; et al. Functional analyses of caffeic acid O-methyltransferase and Cinnamoyl-CoA-Reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 2010, 22, 3357–3373. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, B.; Cesarino, I.; Goeminne, G.; Kim, H.; Marroni, F.; Van Acker, R.; Vanholme, R.; Morreel, K.; Ivens, B.; Pinosio, S.; et al. Breeding with rare defective alleles (BRDA): A natural Populus nigra HCT mutant with modified lignin as a case study. New Phytol. 2013, 198, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Coleman, H.D.; Park, J.; Nair, R.; Chapple, C.; Mansfield, S.D. RNAi-mediated suppression of p-coumaroyl-CoA 3′-hydroxylase in hybrid poplar impacts lignin deposition and soluble secondary metabolism. Proc. Natl. Acad. Sci. USA 2008, 105, 4501–4506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Bhuiya, M.; Pazo, J.R.; Miao, Y.; Kim, H.; Ralph, J.; Liu, C. An engineered Monolignol 4-O-Methyltransferase depresses lignin biosynthesis and confers novel metabolic capability in Arabidopsis. Plant Cell 2012, 24, 3135–3152. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Tobimatsu, Y.; Phillips, L.; Flint, H.; Geddes, B.; Lu, F.; Ralph, J. Syringyl lignin production in conifers: Proof of concept in a Pine tracheary element system. Proc. Natl. Acad. Sci. USA 2015, 112, 6218–6223. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Koshiba, T.; Tobimatsu, Y.; Suzuki, S.; Murakami, S.; Yamamura, M.; Rahman, M.M.; Takano, T.; Hattori, T.; Sakamoto, M.; et al. Regulation of CONIFERALDEHYDE 5-HYDROXYLASE expression to modulate cell wall lignin structure in rice. Planta 2017, 246, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Cesarino, I.; Rataj, K.; Xiao, Y.; Sundin, L.; Goeminne, G.; Kim, H.; Cross, J.; Morreel, K.; Araujo, P.; et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 2013, 341, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.M.; Escamilla-Trevino, L.; Yarce, J.C.S.; Kim, H.; Ralph, J.; Chen, F.; Dixon, R.A. An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J. 2016, 86, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Saleme, M.; Cesarino, I.; Vargas, L.; Kim, H.; Vanholme, R.; Goeminne, G.; Van, R.A.; Fonseca, F.; Pallidis, A.; Voorend, W.; et al. Silencing CAFFEOYL SHIKIMATE ESTERASE affects lignification and improves saccharification. Plant Physiol. 2017, 175, 1040–1057. [Google Scholar] [CrossRef] [PubMed]
- Eudes, A.; Dutta, T.; Deng, K.; Jacquet, N.; Sinha, A.; Benites, V.T.; Eek, B.; Richel, A.; Sattler, S.E.; Northen, T.R.; et al. SbCOMT (Bmr12) is involved in the biosynthesis of tricin-lignin in sorghum. PLoS ONE 2017, 12, e0178160. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Guo, Z.; Gu, F.; Ke, S.; Sun, D.; Dong, S.; Liu, W.; Huang, M.; Xiao, W.; Yang, G. 4-Coumarate-CoA ligase-like gene OsAAE3 negatively mediates the rice blast resistance, floret development and lignin biosynthesis. Front. Plant Sci. 2016, 7, 2041. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; He, Y.; Kabahuma, M.; Chaya, T.; Kelly, A.; Borrego, E.; Bian, Y.; El, K.F.; Yang, L.; Teixeira, P. A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat. Genet. 2017, 49, 1364–1372. [Google Scholar] [CrossRef] [PubMed]
- Herrero, J.; Fernándezpérez, F.; Yebra, T.; Novouzal, E.; Pomar, F.; Pedreño, M.Á.; Cuello, J.; Guéra, A.; Estebancarrasco, A.; Zapata, J.M. Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 2013, 237, 1599–1612. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.; Dixon, R.A. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Wang, Y.; Wang, L.; Hu, P.; Wang, Y.; Jia, Y.; Zhang, C.; Zhang, Y.; Zhang, Y.; Wang, C. Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 2017, 15, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, Y.; Wang, Z.; Guo, X.; Liu, F.; Jiang, J.; Liu, G. BpMADS12 gene role in lignin biosynthesis of Betula platyphylla Suk by transcriptome analysis. J. For. Res. 2016, 27, 1111–1120. [Google Scholar] [CrossRef]
- Zeng, J.K.; Li, X.; Zhang, J.; Ge, H.; Yin, X.R.; Chen, K.S. Regulation of loquat fruit low temperature response and lignification involves interaction of heat shock factors and genes associated with lignin biosynthesis. Plant Cell Environ. 2016, 39, 1780–1789. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.; Chauhan, R.; Gill, T.; Swarnkar, M.K.; Sreenivasulu, Y.; Kumar, S.; Kumar, N.; Shankar, R.; Ahuja, P.S.; Singh, A.K. Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol. Biol. 2015, 87, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Tronchet, M.; Balagué, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Berthet, S.; Demontcaulet, N.; Pollet, B.; Bidzinski, P.; Cezard, L.; Bris, P.L.; Borrega, N.; Herve, J.; Blondet, E.; Balzergue, S.; et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 2011, 23, 1124–1137. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, C.Y.; Song, W.; Park, D.; Kwon, S.; Lee, H.; Bang, J.; Kwak, S. Overexpression of sweetpotato swpa4 peroxidase results in increased hydrogen peroxide production and enhances stress tolerance in tobacco. Planta 2008, 227, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Shigeto, J.; Itoh, Y.; Hirao, S.; Ohira, K.; Fujita, K.; Tsutsumi, Y. Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem. J. Integr. Plant Biol. 2015, 57, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Cosio, C.; Ranocha, P.; Francoz, E.; Burlat, V.; Zheng, Y.; Perry, S.E.; Ripoll, J.J.; Yanofsky, M.; Dunand, C. The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol. 2016, 213, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, C.; Zhu, M.; Yu, Y.; Zhang, Y.; Wei, Z. Generation and characterization of transgenic poplar plants overexpressing a cotton laccase gene. Plant Cell Tissue Organ Cult. 2008, 93, 303–310. [Google Scholar] [CrossRef]
- Wang, Y.; Bouchabke-Coussa, O.; Lebris, P.; Antelme, S.; Soulhat, C.; Gineau, E.; Dalmais, M.; Bendahmane, A.; Morin, H.; Mouille, G.; et al. LACCASE5 is required for lignification of the Brachypodium distachyon Culm. Plant Physiol. 2015, 168, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Legay, S.; Lacombe, E.; Goicoechea, M.; Briere, C.; Seguin, A.; Mackay, J.; Grimapettenati, J. Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci. 2007, 173, 542–549. [Google Scholar] [CrossRef]
- Legay, S.; Sivadon, P.; Blervacq, A.; Pavy, N.; Baghdady, A.; Tremblay, L.; Levasseur, C.; Ladouce, N.; Lapierre, C.; Seguin, A.; et al. EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol. 2010, 188, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Richardson, E.A.; Ye, Z.H. Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 2007, 225, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Giraldo, L.; Shadle, G.; Shen, H.; Barrosrios, J.; Fresquet, C.S.; Wang, H.; Dixon, R.A. Combining enhanced biomass density with reduced lignin level for improved forage quality. Plant Biotechnol. J. 2016, 14, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.D.; Smith, J.A.; Mazarei, M.; Zhang, J.; Turner, G.B.; Decker, S.R.; Sykes, R.W.; Poovaiah, C.R.; Baxter, H.L.; Mann, D.G.J.; et al. Downregulation of a UDP-arabinomutase gene in switchgrass (Panicum virgatum L.) results in increased cell wall lignin while reducing arabinose-glycans. Front. Plant Sci. 2016, 7, 1580. [Google Scholar] [CrossRef] [PubMed]
- Fornalé, S.; Rencoret, J.; García-Calvo, L.; Encina, A.; Rigau, J.; Gutiérrez, A.; Del Río, J.C.; Caparros-Ruiz, D. Changes in cell wall polymers and degradability in maize mutants lacking 3′- and 5′-O-Methyltransferases involved in lignin biosynthesis. Plant Cell Physiol. 2017, 58, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Kim, J.I.; Tobimatsu, Y.; Ciesielski, P.N.; Anderson, N.A.; Ximenes, E.; Maeda, J.; Ralph, J.; Donohoe, B.S.; Ladisch, M. Disruption of mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 2014, 509, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Liljegren, S.J.; Ditta, G.S.; Eshed, Y.; Savidge, B.; Bowman, J.L.; Yanofsky, M.F. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 2000, 404, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Davis, E.; Gardner, D.; Cai, X.; Wu, Y. Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 2006, 224, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Schilmiller, A.L.; Stout, J.; Weng, J.K.; Humphreys, J.; Ruegger, M.O.; Chapple, C. Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 2009, 60, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Berry, P.M.; Sterling, M.; Spink, J.H.; Baker, C.J.; Sylvesterbradley, R.; Mooney, S.J.; Tams, A.R.; Ennos, A.R. Understanding and reducing lodging in cereals. Adv. Agron. 2004, 84, 217–271. [Google Scholar]
- Tanaka, K.; Murata, K.; Yamazaki, M.; Onosato, K.; Miyao, A.; Hirochika, H. Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall. Plant Physiol. 2003, 133, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Peng, S.; Visperas, R.M.; Ereful, N.; Bhuiya, M.S.U.; Julfiquar, A.W. Lodging-related morphological traits of hybrid rice in a tropical irrigated ecosystem. Field Crop. Res. 2007, 101, 240–248. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, Y. Rice brittleness mutants: A way to open the ‘black box’ of monocot cell wall biosynthesis. J. Integr. Plant Biol. 2011, 53, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Chen, X.; Yin, Y.; Lu, K.; Yang, W.; Tang, Y.; Wang, Z. Lodging resistance of winter wheat (Triticum aestivum L.): Lignin accumulation and its related enzymes activities due to the application of paclobutrazol or gibberellin acid. Field Crop. Res. 2014, 157, 1–7. [Google Scholar] [CrossRef]
- Zheng, M.; Jin, C.; Shi, Y.; Li, Y.; Yin, Y.; Yang, D.; Luo, Y.; Pang, D.; Xu, X.; Li, W. Manipulation of lignin metabolism by plant densities and its relationship with lodging resistance in wheat. Sci. Rep. 2017, 7, 41805. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Liu, X.B.; She, H.Z.; Gao, Z.; Ruan, R.W.; Wu, D.Q.; Yi, Z.L. The lignin synthesis related genes and lodging resistance of Fagopyrum esculentum. Biol. Plant. 2017, 61, 1–9. [Google Scholar] [CrossRef]
- Dorairaj, D.; Ismail, M.R.; Sinniah, U.R.; Tan, K.B. Influence of silicon on growth, yield, and lodging resistance of MR219, a lowland rice of Malaysia. J. Plant Nutr. 2017, 40. [Google Scholar] [CrossRef]
- Jie, K.; Sun, Y.; Zhou, M.; Zhang, P.; Zuo, Q.; Wu, J.; Zhou, G. The effect of nitrogen application and planting density on the radiation use efficiency and the stem lignin metabolism in rapeseed (Brassica napus L.). Field Crop. Res. 2016, 199, 89–98. [Google Scholar]
- Zhang, W.; Wu, L.; Ding, Y.; Xiong, Y.; Wu, X.; Fei, W.; Li, G.; Liu, Z.; She, T.; Ding, C. Nitrogen fertilizer application affects lodging resistance by altering secondary cell wall synthesis in japonica rice (Oryza sativa). J. Plant Res. 2017, 130, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Hu, D.; Liu, X.; She, H.; Ruan, R.; Yang, H.; Yi, Z.; Wu, D. Effects of uniconazole on the lignin metabolism and lodging resistance of culm in common buckwheat (Fagopyrum esculentum M.). Field Crop. Res. 2015, 180, 46–53. [Google Scholar] [CrossRef]
- Kong, L.; Sun, M.; Wang, F.; Liu, J.; Feng, B.; Si, J.; Zhang, B.; Li, S.; Li, H. Effects of high NH4+ on K+ uptake, culm mechanical strength and grain filling in wheat. Front. Plant Sci. 2014, 5, 703. [Google Scholar] [CrossRef] [PubMed]
- Schützendübel, A.; Polle, A. Plant responses to abiotic stresses: Heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 2002, 53, 1351–1365. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zheng, L.; He, F.; Zhao, F.J.; Shen, Z.; Zheng, L. Transcriptional and physiological analyses identify a regulatory role for hydrogen peroxide in the lignin biosynthesis of copper-stressed rice roots. Plant Soil 2015, 387, 323–336. [Google Scholar] [CrossRef]
- Jannoey, P.; Pongprasert, W.; Lumyong, S.; Roytrakul, S.; Nomura, M. Comparative proteomic analysis of two rice cultivars (Oryza sativa L.) contrasting in brown planthopper (BPH) stress resistance. Plant Omics 2015, 8, 96–105. [Google Scholar]
- Santiago, R.; Barrosrios, J.; Malvar, R.A. Impact of cell wall composition on maize resistance to pests and diseases. Int. J. Mol. Sci. 2013, 14, 6960–6980. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Yu, J.; Bai, J.; Zhu, Z.; Wang, X. Induced defense responses in rice plants against small brown planthopper infestation. Crop J. 2014, 2, 55–62. [Google Scholar] [CrossRef]
- Jannoey, P.; Channei, D.; Kotcharerk, J.; Pongprasert, W.; Nomura, M. Expression analysis of genes related to rice resistance against brown planthopper, Nilaparvata lugens. Rice Sci. 2017, 24, 163–172. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, L.; Zhang, H.; Du, X.; An, C.; Xia, X.; Chen, F.; Jiang, J.; Chen, S. CmMYB19 over-expression improves aphid tolerance in Chrysanthemum by promoting lignin synthesis. Int. J. Mol. Sci. 2017, 18, 619. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, T.; Mizukubo, T.; Abe, H.; Seo, S. Sclareol induces plant resistance to root-knot nematode partially through ethylene-dependent enhancement of lignin accumulation. Mol. Plant Microbe Interact. 2015, 28, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Tianpei, X.; Li, D.; Qiu, P.; Luo, J.; Zhu, Y.; Li, S. Scorpion peptide LqhIT2 activates phenylpropanoid pathways via jasmonate to increase rice resistance to rice leafrollers. Plant Sci. 2015, 230, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yang, Q.; Ma, R. Erwinia carotovora ssp. carotovora infection induced “defense lignin” accumulation and lignin biosynthetic gene expression in chinese cabbage (Brassica rapa L. ssp. pekinensis). J. Integr. Plant Biol. 2007, 49, 993–1002. [Google Scholar] [CrossRef]
- Karkonen, A.; Koutaniemi, S. Lignin biosynthesis studies in plant tissue cultures. J. Integr. Plant Biol. 2010, 52, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.H.; Zhu, H.H.; Han, J.Q. Wheat ROP proteins modulate defense response through lignin metabolism. Plant Sci. 2017, 262, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Kar, I.; Mukherjee, A.K.; Acharya, P. Elicitor-induced defense responses in Solanum lycopersicum against Ralstonia solanacearum. Sci. World J. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Liu, Z.; Zhu, L.; Zhang, C.; Chen, Y.; Zhou, Y.; Li, F.; Li, X. Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. 2012, 44, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.F.; He, Y.; Strauch, R.; Olukolu, B.A.; Nielsen, D.; Li, X.; Balint-Kurti, P.J. Maize homologs of Hydroxycinnamoyltransferase, a key enzyme in lignin biosynthesis, bind the nucleotide binding leucine-rich repeat Rp1 proteins to modulate the defense response. Plant Physiol. 2015, 169, 2230. [Google Scholar] [PubMed]
- Chen, G.; Liu, Y.; Wang, R.; Zhang, J.; Owens, G. Cadmium adsorption by willow root: The role of cell walls and their subfractions. Environ. Sci. Pollut. Res. Int. 2013, 20, 5665–5672. [Google Scholar] [CrossRef] [PubMed]
- Dalcorso, G.; Farinati, S.; Furini, A. Regulatory networks of cadmium stress in plants. Plant Signal. Behav. 2010, 5, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Demirbas, A. Adsorption of lead and cadmium ions in aqueous solutions onto modified lignin from alkali glycerol delignication. J. Hazard. Mater. 2004, 109, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhang, S.; Shan, X.Q. Adsorption of metal ions on lignin. J. Hazard. Mater. 2008, 151, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, N.H.; Liu, W.; Liu, G.; Selvaraj, G.; Wei, Y.; King, J. Transcriptional regulation of genes involved in the pathways of biosynthesis and supply of methyl units in response to powdery mildew attack and abiotic stresses in wheat. Plant Mol. Biol. 2007, 64, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Kováčik, J.; Bačkor, M. Phenylalanine ammonia-lyase and phenolic compounds in chamomile tolerance to cadmium and copper excess. Water Air Soil Pollut. 2007, 185, 185–193. [Google Scholar] [CrossRef]
- Tahara, K.; Norisada, M.; Hogetsu, T.; Kojima, K. Aluminum tolerance and aluminum-induced deposition of callose and lignin in the root tips of Melaleuca and Eucalyptus species. J. For. Res 2005, 10, 325–333. [Google Scholar] [CrossRef]
- Lima, J.D.; Mazzafera, P.; Moraes, W.D.S.; Silva, R.B.D. Chá: Aspectos relacionados à qualidade e perspectivas tea: Aspects related to the quality and prospects. Ciênc. Rural 2009, 39, 1258–1266. [Google Scholar] [CrossRef]
- Ghanati, F.; Morita, A.; Yokota, H. Deposition of suberin in roots of soybean induced by excess boron. Plant Sci. 2005, 168, 397–405. [Google Scholar] [CrossRef]
- Mao, C.; Yi, K.; Yang, L.; Zheng, B.; Wu, Y.; Liu, F.; Wu, P. Identification of aluminium-regulated genes by cDNA-AFLP in rice (Oryza sativa L.): Aluminium-regulated genes for the metabolism of cell wall components. J. Exp. Bot. 2004, 55, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Peng, K.; Chen, Y.; Wang, G.; Shen, Z. Roles of apoplastic peroxidases, laccases, and lignification in the manganese tolerance of hyperaccumulator Phytolacca americana. Acta Physiol. Plant 2012, 34, 151–159. [Google Scholar] [CrossRef]
- Lin, C.C.; Chen, L.M.; Liu, Z.H. Rapid effect of copper on lignin biosynthesis in soybean roots. Plant Sci. 2005, 168, 855–861. [Google Scholar] [CrossRef]
- Van de Mortel, J.E.; Villanueva, L.A.; Schat, H.; Kwekkeboom, J.; Coughlan, S.; Moerland, P.D.; Van Themaat, E.V.L.; Koornneef, M.; Aarts, M.G.M. Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsisthaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 2006, 142, 1127–1147. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.J.; Cheng, L.M.; Liu, Z.H. Rapid effect of cadmium on lignin biosynthesis in soybean roots. Plant Sci. 2007, 172, 632–639. [Google Scholar] [CrossRef]
- Akgül, M.; Çöpür, Y.; Temiz, S. A comparison of kraft and kraft-sodium borohydrate brutia pine pulps. Build. Environ. 2007, 42, 2586–2590. [Google Scholar] [CrossRef]
- Ederli, L.; Reale, L.; Ferranti, F.; Pasqualini, S. Responses induced by high concentration of cadmium in Phragmites australis roots. Physiol. Plant. 2004, 121, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Van de Mortel, J.E.; Schat, H.; Moerland, P.D.; Ver Loren van Themaat, E.; Ent, S.V.D.; Blankestijn, H.; Ghandilyan, A.; Tsiatsiani, S.; Aarts, M.G.M. Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 2008, 31, 301–324. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Le, L.; Wang, X.; Shen, Z.; Zheng, L. Comprehensive analysis of rice laccase gene (OsLAC) family and ectopic expression of OsLAC10 enhances tolerance to copper stress in Arabidopsis. Int. J. Mol. Sci. 2017, 18, 209. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Jia, W.; Lv, S.; Bao, H.; Miao, F.; Zhang, X.; Wang, J.; Li, J.; Li, D.; Zhu, C. Comparative transcriptome combined with morpho-physiological analyses revealed key factors for differential cadmium accumulation in two contrasting sweet sorghum genotypes. Plant Biotechnol. J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.; Nakamura, T.; Komatsu, S. Differential responses of microsomal proteins and metabolites in two contrasting cadmium (Cd)-accumulating soybean cultivars under Cd stress. Amino Acids 2012, 42, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.K.; Shukla, P.S.; Gupta, K.; Jha, B. Bioengineering for salinity tolerance in plants: State of the art. Mol. Biotechnol. 2012, 54, 102–123. [Google Scholar] [CrossRef] [PubMed]
- Monties, B.; Fukushima, K. Occurrence, Function and Biosynthesis of Lignins. In Biopolymers. Lignin, Humic Substances and Coal; Hofrichter, M., Steinbuchel, A., Eds.; Wiley: Weinheim, Germany, 2001; Volume 1, pp. 1–64. [Google Scholar]
- Mourasobczak, J.; Souza, U.; Mazzafera, P. Drought stress and changes in the lignin content and composition in Eucalyptus. BMC Proc. 2011, 5. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.C.; Xu, Y.Q.; Li, G.J.; Liao, Y.; Fu, F.L. Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J. Appl. Genet. 2009, 50, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Linker, R.; Gepstein, S.; Tanimoto, E.; Yamamoto, R.; Neumann, P.M. Progressive inhibition by water deficit of cell wall extensibility and growth along the elongation zone of maize roots is related to increased lignin metabolism and progressive stelar accumulation of wall phenolics. Plant Physiol. 2006, 140, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Vishwakarma, R.K.; Arafat, Y.A.; Gupta, S.K.; Khan, B.M. Abiotic stress induces change in Cinnamoyl CoA Reductase (CCR) protein abundance and lignin deposition in developing seedlings of Leucaena leucocephala. Physiol. Mol. Biol. Plants 2015, 21, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Valliyodan, B.; Zhang, J.; Lenoble, M.E.; Yu, O.; Rogers, E.E.; Nguyen, H.T.; Sharp, R.E. Regulation of growth response to water stress in the soybean primary root. I. Proteomic analysis reveals region-specific regulation of phenylpropanoid metabolism and control of free iron in the elongation zone. Plant Cell Environ. 2010, 33, 223–243. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Kim, Y.H.; Jeong, J.C.; Kim, C.Y.; Lee, H.S.; Bang, J.W.; Kwak, S.S. Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta 2011, 233, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Kováčik, J.; Grúz, J.; Klejdus, B.; Štork, F.; Marchiosi, R.; Ferrarese Filho, O. Lignification and related parameters in copper-exposed Matricaria chamomilla roots: Role of H2O2 and NO in this process. Plant Sci. 2010, 179, 383–389. [Google Scholar] [CrossRef]
- Christensen, J.H.; Christensen, O.B. A summary of the PRUDENCE model projections of changes in European climate by the end of this century. Clim. Chang. 2007, 81, 7–30. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed]
- Guy, C.; Kaplan, F.; Kopka, J.; Selbig, J.; Hincha, D.K. Metabolomics of temperature stress. Physiol. Plant. 2008, 132, 220–235. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Dhanaraj, A.L.; Arora, R.; Rowland, L.J.; Fu, Y.; Sun, L. Identification of cold acclimation-responsive Rhododendron genes for lipid metabolism, membrane transport and lignin biosynthesis: Importance of moderately abundant ESTs in genomic studies. Plant Cell Environ. 2006, 29, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Gindl, W.; Grabner, M.; Wimmer, R. The influence of temperature on latewood lignin content in treeline Norway spruce compared with maximum density and ring width. Trees 2000, 14, 409–414. [Google Scholar] [CrossRef]
- Yun, Z.; Gao, H.; Ping, L.; Liu, S.; Tao, L.; Shuai, J.; Qiang, X.; Xu, J.; Cheng, Y.; Deng, X. Comparative proteomic and metabolomic profiling of citrus fruit with enhancement of disease resistance by postharvest heat treatment. BMC Plant Biol. 2013, 13, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Tobimatsu, Y.; Zhou, R.; Pattathil, S.; Gallego-Giraldo, L.; Fu, C.; Jackson, L.A.; Hahn, M.G.; Kim, H.; Chen, F. Loss of function of cinnamyl alcohol dehydrogenase 1 leads to unconventional lignin and a temperature-sensitive growth defect in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2013, 110, 13660–13665. [Google Scholar] [CrossRef] [PubMed]
- Gellerstedt, G.; Henriksson, G. Lignins: Major Sources, Structure and Properties. In Monomers, Polymers and Composites from Renewable Resources; Elsevier: Amsterdam, The Netherlands, 2008; pp. 201–224. [Google Scholar]
- Chen, F.; Tobimatsu, Y.; Havkinfrenkel, D.; Dixon, R.A.; Ralph, J. A polymer of caffeyl alcohol in plant seeds. Proc. Natl. Acad. Sci. USA 2012, 109, 1772–1777. [Google Scholar] [CrossRef] [PubMed]
- Constant, S.; Wienk, H.L.J.; Frissen, A.E.; Peinder, P.D.; Boelens, R.; Es, D.S.V.; Grisel, R.J.H.; Weckhuysen, B.M.; Huijgen, W.J.J.; Gosselink, R.J.A. New insights into the structure and composition of technical lignins: A comparative characterisation study. Green Chem. 2016, 18, 2651–2665. [Google Scholar] [CrossRef]
- Lupoi, J.S.; Singh, S.; Parthasarathi, R.; Simmons, B.A.; Henry, R.J. Recent innovations in analytical methods for the qualitative and quantitative assessment of lignin. Renew. Sustain. Energy Rev. 2015, 49, 871–906. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; Britovsek, G.; Cairney, J.; Eckert, C.A.; Frederick, W.J., Jr.; Hallett, J.P.; Leak, D.J.; Liotta, C.L. The path forward for biofuels and biomaterials. Science 2006, 311, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Monteil-Rivera, F.; Phuong, M.; Ye, M.; Halasz, A.; Hawari, J. Isolation and characterization of herbaceous lignins for applications in biomaterials. Ind. Crop. Prod. 2013, 41, 356–364. [Google Scholar] [CrossRef]
- Azadfar, M.; Gao, A.H.; Bule, M.V.; Chen, S. Structural characterization of lignin: A potential source of antioxidants guaiacol and 4-vinylguaiacol. Int. J. Biol. Macromol. 2015, 75, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Vinardell, M.P.; Mitjans, M. Lignins and their derivatives with beneficial effects on human health. Int. J. Mol. Sci. 2017, 18, 1219. [Google Scholar] [CrossRef] [PubMed]
| Genes | Plant Species | Functions | References |
|---|---|---|---|
| 4CL | Oryza sativa (OsAAE3) | Decreased lignin accumulation and increased sensitivity to rice blast | [37] |
| CCR | Paspalum dilatatum (PdCCR1) | Increased lignin content and altered lignin deposition | [26] |
| CAD | Populus trichocarpa (PtrCAD1) | Modified lignin content and structure | [25] |
| MOMT | Arabidopsis thaliana (MOMT4) | Depressed lignin biosynthesis and increased saccharification yields | [30] |
| COMT | Sorghum bicolor (SbCOMT) | Methylate the tricin precursors and participated in S lignin biosynthesis | [36] |
| CSE | Populus tremula × P. alba (PtxaCSE1/2) | Participated in lignin biosynthesis and affected saccharification | [35] |
| CCoAOMT | Zea mays (ZmCCoAOMT2) | Participated in lignin biosynthesis and resistance to multiple pathogens | [38] |
| HCT | Populus nigra (HCT1) | Modified H lignin disposition | [28] |
| F5H | Oryza sativa (OsCAld5H1) | Increased the content of S units, decreased G lignin | [31] |
| POD | Arabidopsis thaliana (AtPrx72) | Participated in lignin biosynthesis, growth and development of plant | [39] |
| LAC | Arabidopsis thaliana (LAC11) | Participated in lignin biosynthesis and the development of root, anthers and vascular | [40] |
| ABC | Arabidopsis thaliana (AtABCG29) | Participated in p-coumaryl alcohol transport | [2] |
| MYB | Betula platyphylla (BplMYB46) | Increased salt/osmotic stress tolerance and enhanced lignin deposition | [41] |
| MADS | Betula platyphylla (BpMADS12) | Participated in lignin biosynthesis and regulation of brassinosteroid signaling pathway | [42] |
| HSF | Eriobotrya japonica (EjHSF3) | Participated in fruit lignification | [43] |
| SOD | Potentilla atrosanguinea (SOD) | Enhanced lignin accumulation and stress tolerance of Arabidopsis | [44] |
| APX | Rheum australe (APX) | Enhanced lignin accumulation and stress tolerance of Arabidopsis | [44] |
| DIR | Gossypium hirsutum (GhDIR1) | Promoted the degree of lignification and enhanced Verticillium dahliae resistance | [45] |
| CHS | Zea mays (CHS C2) | Increased lignin content of maize | [11] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. https://doi.org/10.3390/ijms19020335
Liu Q, Luo L, Zheng L. Lignins: Biosynthesis and Biological Functions in Plants. International Journal of Molecular Sciences. 2018; 19(2):335. https://doi.org/10.3390/ijms19020335
Chicago/Turabian StyleLiu, Qingquan, Le Luo, and Luqing Zheng. 2018. "Lignins: Biosynthesis and Biological Functions in Plants" International Journal of Molecular Sciences 19, no. 2: 335. https://doi.org/10.3390/ijms19020335
APA StyleLiu, Q., Luo, L., & Zheng, L. (2018). Lignins: Biosynthesis and Biological Functions in Plants. International Journal of Molecular Sciences, 19(2), 335. https://doi.org/10.3390/ijms19020335