Whole Genome Resequencing from Bulked Populations as a Rapid QTL and Gene Identification Method in Rice
Abstract
:1. Introduction
2. Conventional Bulk Segregant Analysis (BSA)
3. MutMap
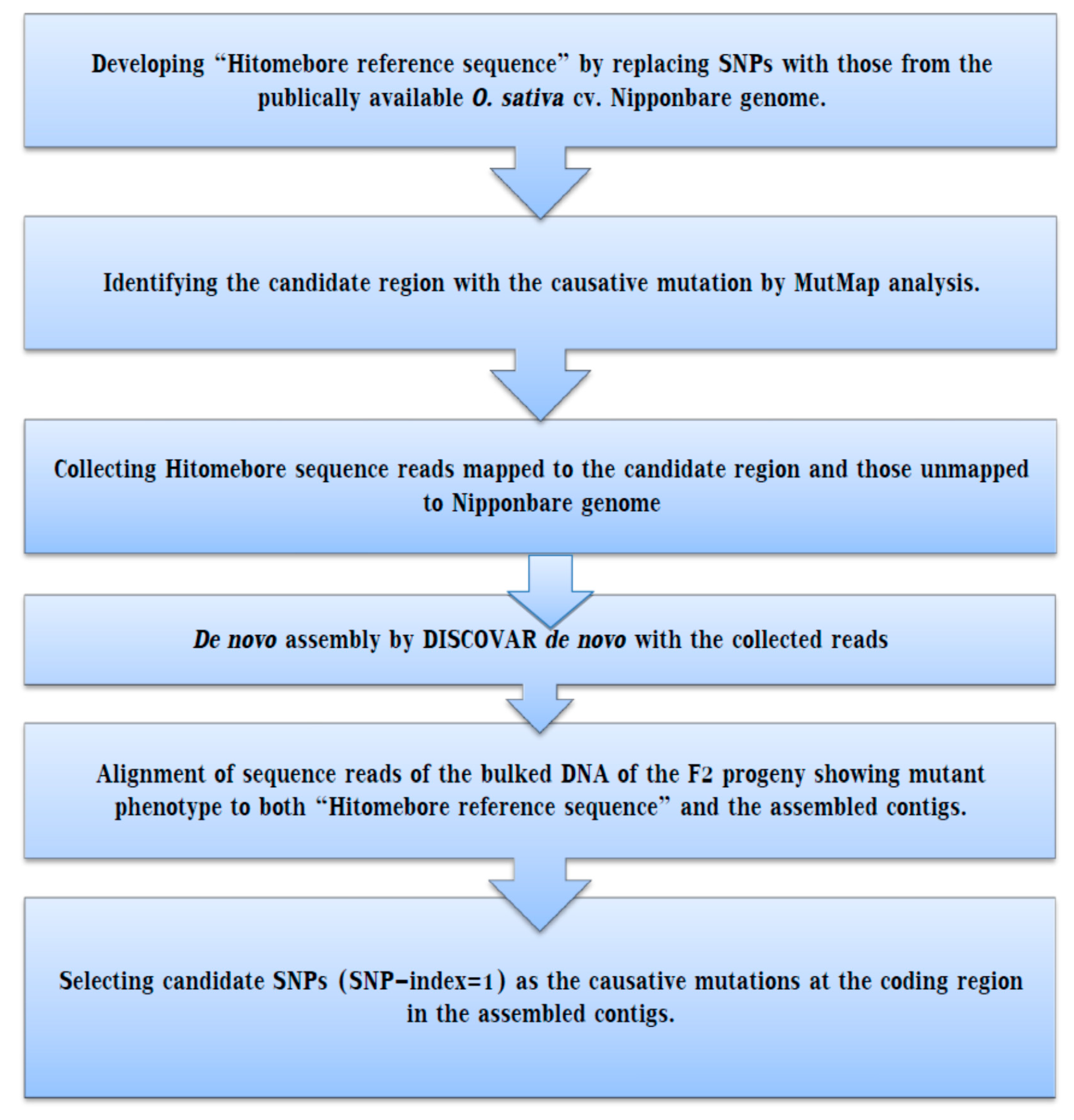
4. MutMap Gap
5. MutMap+
6. Modified MutMap
7. QTL Seq
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Takeda, S.; Matsuoka, M. Genetic approaches to crop improvement: Responding to environmental and population changes. Nat. Rev. Genet. 2008, 9, 444–457. [Google Scholar] [CrossRef] [PubMed]
- Burke, J.M.; Burger, J.C.; Chapman, M.A. Crop evolution: From genetics to genomics. Curr. Opin. Genet. Dev. 2007, 17, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Watanabe, S.; Yamada, T.; Tsubokura, Y.; Nakashima, H.; Zhai, H.; Anai, T.; Sato, S.; Yamazaki, T.; Lü, S.; et al. Positional cloning and characterization reveal the molecular basis for soybean maturity locus E1 that regulates photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2012, 109, E2155–E2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funatsuki, H.; Suzuki, M.; Hirose, A.; Inaba, H.; Yamada, T.; Hajika, M.; Komatsu, K.; Katayama, T.; Sayama, T.; Ishimoto, M.; et al. Molecular basis of a shattering resistance boosting global dissemination of soybean. Proc. Natl. Acad. Sci. USA 2014, 111, 17797–17802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peters, J.L.; Cnudde, F.; Gerats, T. Forward genetics and map-based cloning approaches. Trends Plant Sci. 2003, 8, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Gallavotti, A.; Whipple, C.J. Positional cloning in maize (Zea mays subsp. mays, Poaceae). Appl. Plant Sci. 2015, 188, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Terauchi, R.; McCouch, S.R. Harvesting the promising fruits of genomics: Applying genome sequencing technologies to crop breeding. PLoS Biol. 2014, 12, e1001883. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Wang, P.; Xu, Y. Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol. J. 2016, 14, 1941–1955. [Google Scholar] [CrossRef] [Green Version]
- Kale, S.M.; Jaganathan, D.; Ruperao, P.; Chen, C.; Punna, R.; Kudapa, H.; Thudi, M.; Roorkiwal, M.; Katta, M.A.; Doddamani, D.; et al. Prioritization of candidate genes in ‘QTL-hotspot’ region for drought tolerance in chickpea (Cicer arietinum L.). Sci. Rep. 2015, 5, 15296. [Google Scholar] [CrossRef]
- Qi, X.; Li, M.-W.; Xie, M.; Liu, X.; Ni, M.; Shao, G.; Song, C.; Yim, A.K.-Y.; Tao, Y.; Wong, F.-L.; et al. Identification of a novel salt tolerance gene in wild soybean by whole genome sequencing. Nat. Commun. 2014, 5, 4340. [Google Scholar] [CrossRef]
- Xu, X.; Zeng, L.; Tao, Y.; Vuong, T.; Wan, J.; Boerma, R.; Noe, J.; Li, Z.; Finnerty, S.; Pathan, S.M.; et al. Pinpointing genes underlying the quantitative trait loci for root-knot nematode resistance in palaeopolyploid soybean by whole genome resequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 13469–13474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michelmore, R.W.; Paran, I.; Kesseli, R.V. Identification of markers linked to disease-resistance genes by bulked segregant analysis–a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 1991, 88, 9828–9832. [Google Scholar] [CrossRef] [PubMed]
- Quarrie, S.A.; Lazic-Jancic, V.; Kovacevic, D.; Steed, A.; Pekic, S. Bulk segregant analysis with molecular markers and its use for improving drought resistance in maize. J. Exp. Bot. 1999, 50, 1299–1306. [Google Scholar] [CrossRef] [Green Version]
- Mansur, L.M.; Orf, J.; Lark, K.G. Determining the linkage of quantitative trait loci to RFLP markers using extreme phenotypes of recombinant inbreds of soybean (Glycine max L Merr). Theor. Appl. Genet. 1993, 86, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Yi, B.; Chen, Y.N.; Lei, S.L.; Tu, J.X.; Fu, T.D. Fine mapping of the recessive genic male-sterile gene (Bnms1) in Brassica napus L. Theor. Appl. Genet. 2006, 113, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Whipple, C.J.; Kebrom, T.H.; Weber, A.L.; Yang, F.; Hall, D.; Meeley, R.; Schmidt, R.; Doebley, J.; Brutnell, T.P.; Jackson, D.P. Grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 2011, 108, E506–E512. [Google Scholar] [CrossRef] [PubMed]
- Schneeberger, K.; Weigel, D. Fast-forward genetics enabled by new sequencing technologies. Trends Plant Sci. 2011, 16, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Schneeberger, K.; Ossowski, S.; Lanz, C.; Juul, T.; Petersen, A.H.; Nielsen, K.L.; Jorgensen, J.E.; Weigel, D.; Andersen, S.U. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat. Methods. 2009, 6, 550–551. [Google Scholar] [CrossRef] [PubMed]
- Austin, R.S.; Vidaurre, D.; Stamatiou, G.; Breit, R.; Provart, N.J.; Bonetta, D.; Zhang, J.; Fung, P.; Gong, Y.; Wang, P.W.; et al. Next-generation mapping of Arabidopsis genes. Plant J. 2011, 67, 715–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, N.; Sakamoto, T.; Kurata, T.; Tasaka, M. Identification of EMSinduced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 2011, 52, 716–722. [Google Scholar] [CrossRef]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M.; et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, B.; James, G.V.; Konrad, K.; Schneeberger, K.; Turck, F. Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol. 2012, 160, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Lindner, H.; Raissig, M.T.; Sailer, C.; Shimosato-Asano, H.; Bruggmann, R.; Grossniklaus, U. SNP-ratio mapping (SRM): Identifying lethal alleles and mutations in complex genetic backgrounds by next-generation sequencing. Genetics 2012, 191, 1381–1386. [Google Scholar] [CrossRef] [PubMed]
- Fekih, R.; Takagi, H.; Tamiru, M.; Abe, A.; Natsume, S.; Yaegashi, H.; Sharma, S.; Sharma, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap+: Genetic Mapping and Mutant Identification without Crossing in Rice. PLoS ONE 2013, 8, e68529. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Uemura, A.; Yaegashi, H.; Tamiru, M.; Abe, A.; Mitsuoka, C.; Utsushi, H.; Natsume, S.; Kanzaki, H.; Matsumura, H.; et al. MutMap-Gap: whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol. 2013, 200, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Tamiru, M.; Abe, A.; Yoshida, K.; Uemura, A.; Yaegashi, H.; Obara, T.; Oikawa, K.; Utsushi, H.; Kanzaki, E.; et al. MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat. Biotechnol. 2015, 33, 445–449. [Google Scholar] [CrossRef]
- Daware, A.; Das, S.; Srivastava, R.; Badoni, S.; Singh, A.K.; Agarwal, P.; Parida, S.K.; Tyagi, A.K. An efficient strategy combining SSR markers- and advanced QTL-seq-driven QTL mapping unravels candidate genes regulating grain weight in rice. Front. Plant Sci. 2016, 7, 1535. [Google Scholar] [CrossRef]
- Zheng, W.; Wang, Y.; Wang, L.; Ma, Z.; Zhao, J.; Wang, P.; Zhang, L.; Liu, Z.; Lu, X. Genetic mapping and molecular marker development for Pi65 (t), a novel broad spectrum resistance gene to rice blast using next-generation sequencing. Theor. Appl. Genet. 2016, 129, 1035–1044. [Google Scholar] [CrossRef]
- Ogiso-Tanaka, E.; Tanaka, T.; Tanaka, K.; Nonoue, Y.; Sasaki, T.; Fushimi, E.; Koide, Y.; Okumoto, Y.; Yano, M.; Saito, H. Detection of novel QTLs qDTH4.5 and qDTH6.3, which confer late heading under short-day conditions, by SSR marker-based and QTL seq analysis. Breeding Sci. 2017, 67, 101–109. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Sun, L.; Xu, P.; Tu, R.; Meng, S.; Wu, W.; Anis, G.B.; Hussain, K.; Riaz, A.; et al. WB1, a Regulator of Endosperm Development in Rice, Is Identified by a Modified MutMap Method. Int. J. Mol. Sci. 2018, 19, 2159. [Google Scholar] [CrossRef]
- Liu, S.Z.; Yeh, C.T.; Tang, H.M.; Nettleton, D.; Schnable, P.S. Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE 2012, 7, e36406. [Google Scholar] [CrossRef] [PubMed]
- Mascher, M.; Jost, M.; Kuon, J.E.; Himmelbach, A.; Assfalg, A.; Beier, S.; Scholz, U.; Graner, A.; Stein, N. Mapping-by-sequencing accelerates forward genetics in barley. Genome Biol. 2014, 5, R78. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.W.; Hofstad, A.N.; Sreekanta, S.; Fu, F.; Kono, T.J.Y.; O’Rourke, J.A.; Vance, C.P.; Muehlbauer, G.J.; Stupar, R.M. Fast neutron-induced structural rearrangements at a soybean NAP1 locus result in gnarled trichomes. Theor. Appl. Genet. 2016, 129, 1725–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobbels, A.A.; Michno, J.M.; Campbell, B.W.; Virdi, K.S.; Stec, A.O.; Muehlbauer, G.J.; Naeve, S.L.; Stupar, R.M. An induced chromosomal translocation in soybean disrupts a KASI ortholog and is associated with a high-sucrose and low-oil seed phenotype. G3 2017, 7, 1215–1223. [Google Scholar] [CrossRef]
- Lu, H.F.; Lin, T.; Klein, J.; Wang, S.H.; Qi, J.J.; Zhou, Q.; Sun, J.J.; Zhang, Z.H.; Weng, Y.Q.; Huang, S.W. QTL-seq identifies an early flowering QTL located near flowering locus T in cucumber. Theor. Appl. Genet. 2014, 127, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Illa-Berenguer, E.; van Houten, J.; Huang, Z.; van der Knaap, E. Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor. Appl. Genet. 2015, 128, 1329–1342. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Upadhyaya, H.D.; Bajaj, D.; Kujur, A.; Badoni, S.; Kumar, V.; Tripathi, S.; Gowda, C.L.; Sharma, S.; et al. Deploying QTL-seq for rapid delineation of a potential candidate gene underlying major trait-associated QTL in chickpea. DNA Res. 2015, 22, 193–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.W.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H.; et al. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2013, 8, E58700. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Poland, J.A.; Rife, T.W. Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 2012, 5, 92–102. [Google Scholar] [CrossRef]
- Goff, S.A. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 2002, 296, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Pizza, M. Identification of vccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 2000, 287, 1816–1820. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, J.J.; Win, R.A.; Ganal, M.W.; Tanksley, S.D. Isolation of molecular markers from specific chromosomal intervals using DNA pools from existing mapping populations. Nucleic Acids Res. 1991, 19, 6553–6568. [Google Scholar] [CrossRef] [Green Version]
- Takagi, H.; Uemura, A.; Yaegashi, H.; Tamiru, M.; Abe, A.; Mitsuoka, C.; Utsushi, H.; Natsume, S.; Kanzaki, H.; Matsumura, H.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Fang, N.; Xu, R.; Huang, L.; Zhang, B.; Duan, P.; Li, N.; Luo, Y.; Li, Y. SMALL GRAIN 11 Controls Grain Size, Grain Number and Grain Yield in Rice. Rice 2016, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Megersa, A.; Lee1, D.; Park, J.; Koh, H.-J. Genetic Mapping of a Rice Loose Upper Panicle Mutant. Plant Breed. Biotech. 2015, 3, 366–375. [Google Scholar] [CrossRef] [Green Version]
- Mardis, E.R. Next-Generation DNA Sequencing Method. Annu. Rev. Genom. Hum. Genet. 2008, 9, 387–402. [Google Scholar]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Xu, F.; Sun, X.; Chen, Y.; Huang, Y.; Tong, C.; Bao, J. Rapid Identification of Major QTLs Associated with Rice Grain Weight and Their Utilization. PLoS ONE 2015, 10, e0122206. [Google Scholar] [CrossRef]
- Salvi, S.; Tuberosa, R. To clone or not to clone plant QTLs: Present and future challenges. Trends Plant Sci. 2005, 10, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Ehrenreich, M.I.; Torabi, N.; Jia, Y.; Kent, J.; Martis, S.; Shapiro, A.J.; Gresham, D.; Caudy, A.A.; Kruglyak, L. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 2010, 446, 1039–1042. [Google Scholar] [CrossRef] [PubMed]
- Wenger, W.J.; Schwartz, K.; Sherlock, G. Bulk segregant analysis by high-throughput sequencing reveals a novel xylose utilization gene from Saccharomyces cerevisiae. PLoS Genet. 2010, 6, e1000942. [Google Scholar] [CrossRef] [PubMed]
- Parts, L.; Cubillos, A.F.; Warringer, J.; Jain, K.; Salinas, F.; Bumpstead, S.J.; Molin, M.; Zia, A.; Jared, T.; Simpson, J.T.; Quail, M.A.; et al. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 2011, 21, 1131–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swinnen, S.; Schaerlaekens, K.; Pais, T.; Claesen, J.; Hubmann, G.; Yang, Y.; Demeke, M.; Foulquié-Moreno, M.R.; Goovaerts, A.; Souvereyns, K.; et al. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res. 2012, 22, 975–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magwene, P.M.; Willis, J.H.; Kelly, J.K. The statistics of bulk segregant analysis using next-generation sequencing. PLoS Comput. Biol. 2011, 7, e1002255. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.L.; Wang, B.; Dong, X.; Liu, H.; Ren, L.; Chen, J.; Hauck, A.; Song, W.; Lai, J. An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F2 maize population. BMC Genom. 2014, 15, 433. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Xia, X.; Zhang, Z.; Nong, B.; Zeng, Y.; Xiong, F.; Wu, Y.; Gao, J.; Deng, G.; Li, D. QTL Mapping by Whole Genome Re-sequencing and Analysis of Candidate Genes for Nitrogen Use Efficiency in Rice. Front. Plant Sci. 2017, 8, 1634. [Google Scholar] [CrossRef]
- Qin, Y.; Cheng, P.; Cheng, Y.; Feng, Y.; Huang, D.; Huang, T.; Song, X.; Ying, J. QTL-Seq Identified a Major QTL for Grain Length and Weight in Rice Using Near Isogenic F2 Population. Rice Sci. 2018, 25, 121–131. [Google Scholar]
- Kadambari, G.; Vemireddy, L.R.; Srividhya, A.; Nagireddy, R.; Jena, S.S.; Gandikota, M.; Patil, S.; Veeraghattapu, R.; Deborah, D.A.K.; Reddy, G.E.; et al. QTL-Seq-based genetic analysis identifies a major genomic region governing dwarfness in rice (Oryza sativa L.). Plant Cell Rep. 2018, 37, 677–687. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, D.; Tang, W.; Zheng, Y.; Liang, K.; Cutler, A.J.; Wu, W. Mapping of Quantitative Trait Loci Underlying Cold Tolerance in Rice Seedlings via High Throughput Sequencing of Pooled Extremes. PLoS ONE 2013, 8, e68433. [Google Scholar] [CrossRef] [PubMed]
- Misra1, G.; Badoni1, S.; Anacleto, R.; Graner, A.; Alexandrov, N. Whole genome sequencing-based association study to unravel genetic architecture of cooked grain width and length traits in rice. Sci. Rep. 2017, 7, 1247. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.; Kumar, V.; Singh, B.; Rao, A.; Mithra, S.V.A.; Rai, V.; Singh, A.K.; Singh, N.K.; Sl, K. Mapping QTLs for Salt Tolerance in Rice (Oryza sativa L.) by Bulked Segregant Analysis of Recombinant Inbred Lines Using 50K SNP Chip. PLoS ONE 2016, 11, e0153610. [Google Scholar] [CrossRef] [PubMed]
- Frouin, J.; Filloux, D.; Taillebois, J.; Grenier, C.; Montes, F.; de Lamotte, F.; Verdeil, J.-L.; Courtois, B.; Ahmadi, N. Positional cloning of the rice male sterility gene ms- IR36 widely used in the inter-crossing phase of recurrent selection schemes. Mol. Breed. 2013, 33, 555–567. [Google Scholar] [CrossRef]
- Jeonghwan, S.; Yogendra, B.; Chanmi, L.; Koh, H.-J. Fine Mapping and Candidate Gene Analysis of Small Round Grain Mutant in Rice. Plant Breed. Biotech. 2017, 5, 354–362. [Google Scholar] [Green Version]
- Liu, L.; Meng, F.; He, Y.; Zhu, M.; Shen, Y.; Zhang, Z. Fine Mapping and Candidate Gene Analysis of the Tiller Suppression Gene ts1 in Rice. PLoS ONE 2017, 12, e0170574. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.-L.; Vanavichit, A.; Amornsilpa, S.; Trangoonrung, S. Mapping of Rice Rf Gene by Bulked Line Analysis. DNA Res. 1998, 5, 15–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gh, A.; Nematzadeh, N.; Khush, G.S. Mapping the Gene for Aroma in Rice (Oryza sativa L.) by Bulk Segregant Analysis via RAPD Markers. J. Agric. Sci. Technol. 2004, 6, 129–137. [Google Scholar]




| Mapping Methods | Throughput | Pooled Samples | Population Type | Backcrossing to the Wild Type Parent | Presence or Absence of Unmapped Reads | Use of SNP-Index | Δ (SNP-Index) Plots |
|---|---|---|---|---|---|---|---|
| Conventional BSA | Low | Extreme tails | All | Yes | Absence | No | No |
| MutMap | High | Mutant-type phenotype | F2 | Yes | Absence | Yes | No |
| MutMap Gap | High | Mutant-type phenotype | F2 | Yes | Presence | Yes | No |
| MutMap+ | High | Mutant and wild-type phenotype | M3 | No | Absence | Yes | Yes |
| Modified MutMap | High | Mutant and wild-type phenotype | F2:3 | Yes | Absence | Yes | Yes |
| QTL-Seq | High | Extreme tails | Segregating population | Yes | Absence | Yes | Yes |
| Sequencing | Traits | Gene or QTL | Population Type | Population Size | Pooled Samples Size | References |
|---|---|---|---|---|---|---|
| DNA-seq | Pale green leaves and semidwarfism | osCAO1 | F2 | - | 20, 20 | [21] |
| DNA-seq | Cold tolerance | qCTSS-1, qCTSS-2b and qCTSS-8 | F3 | 10,800 | 430, 385 | [61] |
| DNA-seq | Blast disease and seedling vigour | qPHS3-2 | F2 | 531 | 50, 50 | [45] |
| DNA-seq | Early stage lethality | OsNAP6 | M3 | 223 | 40, 40 | [24] |
| DNA-seq | Blast disease | Pii | F2 | 112 and 78 | 20, 20 | [25] |
| DNA-seq | Salt tolerance | OsRR22 | F2 | 67 | 20, 20 | [26] |
| DNA-seq | Loose Upper Panicle | LUP | F2 | 750 | 20, 20 | [47] |
| DNA-seq | Small grain size | smg11 | F2 | - | 50, 50 | [46] |
| DNA-seq | Endosperm development | WB1 | F2:3 | 1000 | 50, 50 | [30] |
| DNA-seq | Nitrogen Use Efficiency in Rice | qNUE6 | F2 | 280 | 30, 30 | [58] |
| DNA-seq | Cooked grain width and length | GWi11.1 | landrace accessions | 591 | - | [62] |
| DNA-seq | Grain length & Weight | qTGW5.3 | NIL F2 | 176 | 35, 35 | [58] |
| SLAF-Seq | Grain Weight | qTGW3.2, qTGW3.1 and qTGW3.3 | RILs | 234 | 30, 30 | [50] |
| DNA-seq | Dwarfness | asd1 | F2 | 181 | 20, 20 | [60] |
| 50K SNP Chip | Salt tolerance | Saltol | RILs | 216 | 30, 30 | [63] |
| Conventional BSA/individual markers | Male sterility | pms1 | 946 | - | [64] | |
| Conventional BSA/individual markers | Small Round Grain | dep2-3 | F2 and F3 | 980 | 8, 8 | [65] |
| Conventional BSA/individual markers | Tiller Suppression | ts1 | F2 | 279 | 10, 10 | [66] |
| Conventional BSA/individual markers | CMS-WA cytoplasm | Rf | BC1 & F2 | 319 | 6, 15 | [67] |
| Conventional BSA/individual markers | Aroma | AG8-AR | F2 | 125 | 8, 8 | [68] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zegeye, W.A.; Zhang, Y.; Cao, L.; Cheng, S. Whole Genome Resequencing from Bulked Populations as a Rapid QTL and Gene Identification Method in Rice. Int. J. Mol. Sci. 2018, 19, 4000. https://doi.org/10.3390/ijms19124000
Zegeye WA, Zhang Y, Cao L, Cheng S. Whole Genome Resequencing from Bulked Populations as a Rapid QTL and Gene Identification Method in Rice. International Journal of Molecular Sciences. 2018; 19(12):4000. https://doi.org/10.3390/ijms19124000
Chicago/Turabian StyleZegeye, Workie Anley, Yingxin Zhang, Liyong Cao, and Shihua Cheng. 2018. "Whole Genome Resequencing from Bulked Populations as a Rapid QTL and Gene Identification Method in Rice" International Journal of Molecular Sciences 19, no. 12: 4000. https://doi.org/10.3390/ijms19124000




