Chloroplasts at the Crossroad of Photosynthesis, Pathogen Infection and Plant Defense
Abstract
:1. Introduction
2. The Chloroplast is a Major Synthesis Site for Many Plant Hormones
2.1. The Chloroplast is a Major Site of ABA Biosynthesis
2.2. Methionine, the Precursor for ET Biosynthesis, is Made in the Chloroplast
2.3. The Chloroplast is also a Site of JA Synthesis
2.4. The Chloroplast is Involved in the Synthesis of SA
3. The Chloroplast is a Major Site of Free Radical Production
3.1. The Photosynthetic Electron Transport Chain is a Major Site of ROS Production
3.2. The Chloroplast is a ⋅NO Generator
4. The Chloroplast is a Site for Ca2+ Signaling
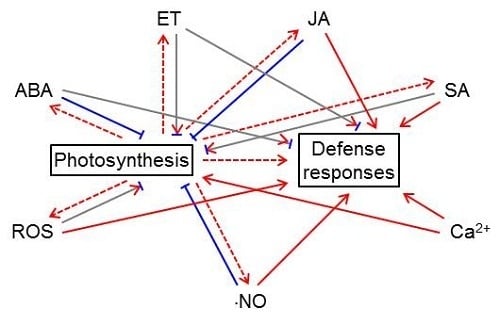
5. The Complex Relationship between Photosynthesis and Defense-related Signals
6. The Participation of Different Plant Signals in Plant Defense against Pathogens
7. PAMP Perception Induces Transcriptional Reprogramming of Nuclear-encoded Chloroplast-targeted Protein Genes
8. Chloroplasts are Targeted by Pathogen Effectors
8.1. AvrRps4 and HopK1
8.2. HopO1-1, HopO1-2, and HopR1
8.3. HopI1 and HopN1
8.4. HopBB1 and HopM1
9. Chloroplast Structure and Functions are Manipulated by Phytotoxins
10. Summary: The Chloroplast Plays a Central Role in the Interplay between Photosynthesis, Pathogen infection, and Plant Defense
11. Future Perspectives and Outstanding Questions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gohre, V. Photosynthetic defence. Nature Plants 2015, 1, 15079. [Google Scholar] [CrossRef] [PubMed]
- Coruzzi, G.M.; Last, R.L. Amino acids. In Biochemistry and Molecular Biology of Plants; Buchanan, R.B., Gruissem, W., Jones, R., Eds.; American Society of Plant Biologists: Rockville, MD, USA, 2000; pp. 358–410. [Google Scholar]
- Rawsthorne, S. Carbon flux and fatty acid synthesis in plants. Prog. Lipid Res. 2002, 41, 182–196. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R. Abscisic acid synthesis and response. Arabidopsis Book 2013, 11, e0166. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.L.-C.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14, 1311. [Google Scholar] [CrossRef]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, Z.; Huang, J.; Lai, Z.; Fan, B. Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 2009, 4, 493–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, D.M.A.; Vlot, A.C.; Wildermuth, M.C.; Klessig, D.F. Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 2011, 9, e0156. [Google Scholar] [CrossRef] [PubMed]
- Nafisi, M.; Goregaoker, S.; Botanga, C.J.; Glawischnig, E.; Olsen, C.E.; Halkier, B.A.; Glazebrook, J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 2007, 19, 2039–2052. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 2006, 141, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Pospíšil, P. Production of reactive oxygen species by Photosystem II. Biochim. Biophys. Acta 2009, 1787, 1151–1160. [Google Scholar] [CrossRef]
- Bechtold, U.; Karpinski, S.; Mullineaux, P.M. The influence of the light environment and photosynthesis on oxidative signalling responses in plant–biotrophic pathogen interactions. Plant Cell Environ. 2005, 28, 1046–1055. [Google Scholar] [CrossRef]
- Buchanan, B.B.; Balmer, Y. Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 2005, 56, 187–220. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Oxidant and antioxidant signalling in plants: A re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 2005, 28, 1056–1071. [Google Scholar] [CrossRef]
- Torres, M.A. ROS in biotic interactions. Physiol. Plant 2010, 138, 414–429. [Google Scholar] [CrossRef]
- Delprato, M.L.; Krapp, A.R.; Carrillo, N. Green light to plant responses to pathogens: The role of chloroplast light-dependent signaling in biotic stress. Photochem. Photobiol. 2015, 91, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Nath, K.; Lu, Y. A paradigm of reactive oxygen species and programmed cell death in plants. J. Cell. Sci. Ther. 2015, 6, 202. [Google Scholar]
- Sewelam, N.; Kazan, K.; Schenk, P.M. Global plant stress signaling: Reactive oxygen species at the cross-road. Front. Plant Sci. 2016, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Collmer, A.; Schneider, D.J.; Lindeberg, M. Lifestyles of the effector rich: Genome-enabled characterization of bacterial plant pathogens. Plant Physiol. 2009, 150, 1623–1630. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, F.; Thilmony, R.; He, S.Y. The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 2002, 1, e0039. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006, 126, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Melotto, M.; Underwood, W.; He, S.Y. Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 2008, 46, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Bender, C.L.; Alarcón-Chaidez, F.; Gross, D.C. Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 1999, 63, 266–292. [Google Scholar]
- Xin, X.F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 2013, 51, 473–498. [Google Scholar]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [PubMed]
- Truman, W.; de Zabala, M.T.; Grant, M. Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J. 2006, 46, 14–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [PubMed]
- de Torres Zabala, M.; Littlejohn, G.; Jayaraman, S.; Studholme, D.; Bailey, T.; Lawson, T.; Tillich, M.; Licht, D.; Bölter, B.; Delfino, L.; et al. Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nature Plants 2015, 1, 15074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, L.A.; Polanski, K.; de Torres-Zabala, M.; Jayaraman, S.; Bowden, L.; Moore, J.; Penfold, C.A.; Jenkins, D.J.; Hill, C.; Baxter, L.; et al. Transcriptional dynamics driving MAMP-triggered immunity and pathogen effector-mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. Plant Cell 2015, 27, 3038–3064. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.; Brown, I.; Knox, P.; Mansfield, J. The role of cell wall-based defences in the early restriction of non-pathogenic hrp mutant bacteria in Arabidopsis. Phytochemistry 2015, 112, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Jin, L.; Shimada, M.; Kim, M.G.; Mackey, D. The phytotoxin coronatine is a multifunctional component of the virulence armament of Pseudomonas syringae. Planta 2014, 240, 1149–1165. [Google Scholar] [CrossRef]
- Feys, B.; Benedetti, C.E.; Penfold, C.N.; Turner, J.G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 1994, 6, 751–759. [Google Scholar] [CrossRef]
- Zhao, Y.; Thilmony, R.; Bender, C.L.; Schaller, A.; He, S.Y.; Howe, G.A. Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 2003, 36, 485–499. [Google Scholar]
- Li, L.; Zhao, Y.; McCaig, B.C.; Wingerd, B.A.; Wang, J.; Whalon, M.E.; Pichersky, E.; Howe, G.A. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 2004, 16, 126–143. [Google Scholar] [CrossRef]
- Katsir, L.; Schilmiller, A.L.; Staswick, P.E.; He, S.Y.; Howe, G.A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Nat. Acad. Sci. USA 2008, 105, 7100–7105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boller, T.; He, S.Y. Innate immunity in plants: An arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009, 324, 742–744. [Google Scholar] [CrossRef] [PubMed]
- Gohre, V.; Jones, A.M.; Sklenar, J.; Robatzek, S.; Weber, A.P. Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Mol. Plant Microbe Interact. 2012, 25, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Hardham, A.R.; Jones, D.A.; Takemoto, D. Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 2007, 10, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Higaki, T.; Kurusu, T.; Hasezawa, S.; Kuchitsu, K. Dynamic intracellular reorganization of cytoskeletons and the vacuole in defense responses and hypersensitive cell death in plants. J. Plant Res. 2011, 124, 315–324. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell. Death Differ. 2011, 18, 1247–1256. [Google Scholar] [CrossRef] [Green Version]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Kangasjärvi, S.; Neukermans, J.; Li, S.; Aro, E.-M.; Noctor, G. Photosynthesis, photorespiration, and light signalling in defence responses. J. Exp. Bot. 2012, 63, 1619–1636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballaré, C.L. Light regulation of plant defense. Annu. Rev. Plant Biol. 2014, 65, 335–363. [Google Scholar] [CrossRef] [PubMed]
- Taiz, L.; Zeiger, E.; Moller, I.M.; Murphy, A. (Eds.) Photosynthesis: The Light Reactions. In Plant Physiology and Development, 6 ed.; Sinauer Associates: Sunderland, MA, USA, 2014; pp. 171–202. [Google Scholar]
- Piotrowska, A.; Bajguz, A. Conjugates of abscisic acid, brassinosteroids, ethylene, gibberellins, and jasmonates. Phytochemistry 2011, 72, 2097–2112. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Piao, H.L.; Kim, H.Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.J. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell. 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012, 35, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Loveys, B.; Kriedemann, P. Internal control of stomatal physiology and photosynthesis. I. Stomatal regulation and associated changes in endogenous levels of abscisic and phaseic acids. Funct. Plant Biol. 1974, 1, 407–415. [Google Scholar] [CrossRef]
- Hornberg, C.; Weiler, E.W. High-affinity binding sites for abscisic acid on the plasmalemma of Vicia faba guard cells. Nature 1984, 310, 321. [Google Scholar] [CrossRef]
- Fedina, I.S.; Tsonev, T.D.; Guleva, E.I. ABA as a modulator of the response of Pisum sativum to salt stress. J. Plant Physiol. 1994, 143, 245–249. [Google Scholar] [CrossRef]
- Fischer, E.; Raschke, K.; Stitt, M. Effects of abscisic acid on photosynthesis in whole leaves: Changes in CO2 assimilation, levels of carbon-reduction-cycle intermediates, and activity of ribulose-1,5-bisphosphate carboxylase. Planta 1986, 169, 536–545. [Google Scholar] [CrossRef]
- Seemann, J.R.; Sharkey, T.D. The effect of abscisic acid and other inhibitors on photosynthetic capacity and the biochemistry of CO2 assimilation. Plant Physiol. 1987, 84, 696–700. [Google Scholar] [CrossRef]
- Popova, L.P.; Tsonev, T.D.; Lazova, G.N.; Stoinova, Z.G. Drought- and ABA-induced changes in photosynthesis of barley plants. Physiol. Plant 1996, 96, 623–629. [Google Scholar] [CrossRef]
- Yamburenko, M.V.; Zubo, Y.O.; Borner, T. Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C-dependent activation of nuclear genes: Repression by guanosine-3'-5'-bisdiphosphate and activation by sigma factor 5. Plant J. 2015, 82, 1030–1041. [Google Scholar] [CrossRef]
- Atkinson, G.C.; Tenson, T.; Hauryliuk, V. The RelA/SpoT homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 2011, 6, e23479. [Google Scholar] [CrossRef]
- Field, B. Green magic: Regulation of the chloroplast stress response by (p)ppGpp in plants and algae. J. Exp. Bot. 2018, 69, 2797–2807. [Google Scholar] [CrossRef] [PubMed]
- Baba, K.; Nakano, T.; Yamagishi, K.; Yoshida, S. Involvement of a nuclear-encoded basic helix-loop-helix protein in transcription of the light-responsive promoter of psbD. Plant Physiol. 2001, 125, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Tsunoyama, Y.; Ishizaki, Y.; Morikawa, K.; Kobori, M.; Nakahira, Y.; Takeba, G.; Toyoshima, Y.; Shiina, T. Blue light-induced transcription of plastid-encoded psbD gene is mediated by a nuclear-encoded transcription initiation factor, AtSig5. Proc. Nat. Acad. Sci. USA 2004, 101, 3304–3309. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Baek, W.; Jung, J.; Kim, J.-H.; Lee, S.C. Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 2015, 16, 15251–15270. [Google Scholar] [CrossRef]
- Mustilli, A.C.; Merlot, S.; Vavasseur, A.; Fenzi, F.; Giraudat, J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef]
- Watkins, J.M.; Chapman, J.M.; Muday, G.K. Abscisic acid-induced reactive oxygen species are modulated by flavonols to control stomata aperture. Plant Physiol. 2017, 175, 1807–1825. [Google Scholar] [CrossRef]
- Sirichandra, C.; Gu, D.; Hu, H.C.; Davanture, M.; Lee, S.; Djaoui, M.; Valot, B.; Zivy, M.; Leung, J.; Merlot, S.; et al. Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett. 2009, 583, 2982–2986. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wang, J.; Li, X.; Xia, X.J.; Zhou, Y.H.; Shi, K.; Chen, Z.; Yu, J.Q. H2O2 mediates the crosstalk of brassinosteroid and abscisic acid in tomato responses to heat and oxidative stresses. J. Exp. Bot. 2014, 65, 4371–4383. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, Y.; Tawaratsumida, T.; Kondo, K.; Kasa, S.; Sakamoto, M.; Aoki, N.; Zheng, S.-H.; Yuasa, T.; Iwaya-Inoue, M. Reactive oxygen species are involved in gibberellin/abscisic acid signaling in barley aleurone cells. Plant Physiol. 2012, 158, 1705–1714. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.; Zhu, G.; Liu, Y.; Zhang, A.; Li, Y.; Liu, R.; Shi, L.; Jia, L.; Zhang, J. Ascorbic acid and reactive oxygen species are involved in the inhibition of seed germination by abscisic acid in rice seeds. J. Exp. Bot. 2012, 63, 1809–1822. [Google Scholar] [CrossRef] [PubMed]
- Desikan, R.; Griffiths, R.; Hancock, J.; Neill, S. A new role for an old enzyme: Nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Nat. Acad. Sci. USA 2002, 99, 16314–16318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Mata, C.; Gay, R.; Sokolovski, S.; Hills, A.; Lamattina, L.; Blatt, M.R. Nitric oxide regulates K+ and Cl− channels in guard cells through a subset of abscisic acid-evoked signaling pathways. Proc. Nat. Acad. Sci. USA 2003, 100, 11116–11121. [Google Scholar] [CrossRef] [PubMed]
- Neill, S.J.; Desikan, R.; Clarke, A.; Hancock, J.T. Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiol. 2002, 128, 13–16. [Google Scholar] [CrossRef]
- She, X.; Song, X. Ethylene inhibits abscisic acid-induced stomatal closure in Vicia faba via reducing nitric oxide levels in guard cells. New Zeal. J. Bot. 2012, 50, 203–216. [Google Scholar] [CrossRef]
- Webb, A.A.; Larman, M.G.; Montgomery, L.T.; Taylor, J.E.; Hetherington, A.M. The role of calcium in ABA-induced gene expression and stomatal movements. Plant J. 2001, 26, 351–362. [Google Scholar] [CrossRef] [Green Version]
- Desikan, R.; Cheung, M.K.; Bright, J.; Henson, D.; Hancock, J.T.; Neill, S.J. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 2004, 55, 205–212. [Google Scholar] [CrossRef]
- Cao, F.Y.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant-pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- De Torres-Zabala, M.; Truman, W.; Bennett, M.H.; Lafforgue, G.; Mansfield, J.W.; Rodriguez Egea, P.; Bogre, L.; Grant, M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007, 26, 1434–1443. [Google Scholar] [PubMed]
- Goritschnig, S.; Weihmann, T.; Zhang, Y.; Fobert, P.; McCourt, P.; Li, X. A novel role for protein farnesylation in plant innate immunity. Plant Physiol. 2008, 148, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Henfling, J.; Bostock, R.; Kuc, J. Effect of abscisic acid on rishitin and lubimin accumulation and resistance to Phytophthora infestans and Cladosporium cucumerinum in potato tuber tissue slices. Phytopathology 1980, 70, 1074–1078. [Google Scholar] [CrossRef]
- Ward, E.W.B.; Cahill, D.M.; Bhattacharyya, M.K. Abscisic acid suppression of phenylalanine ammonia-lyase activity and mRNA, and resistance of soybeans to Phytophthora megasperma f.sp. glycinea. Plant Physiol. 1989, 91, 23–27. [Google Scholar] [CrossRef] [PubMed]
- McDonald, K.L.; Cahill, D.M. Influence of abscisic acid and the abscisic acid biosynthesis inhibitor, norflurazon, on interactions between Phytophthora sojae and soybean (Glycine max). Eur. J. Plant Pathol. 1999, 105, 651–658. [Google Scholar] [CrossRef]
- Mohr, P.G.; Cahill, D.M. Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct. Plant Biol. 2003, 30, 461–469. [Google Scholar] [CrossRef]
- Thaler, J.S.; Bostock, R.M. Interactions between abscisic-acid-mediated responses and plant resistance to pathogens and insects. Ecology 2004, 85, 48–58. [Google Scholar]
- Jiang, C.-J.; Shimono, M.; Sugano, S.; Kojima, M.; Yazawa, K.; Yoshida, R.; Inoue, H.; Hayashi, N.; Sakakibara, H.; Takatsuji, H. Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice–Magnaporthe grisea interaction. Mol. Plant Microbe Interact. 2010, 23, 791–798. [Google Scholar] [CrossRef]
- Lorenzo, O.; Solano, R. Molecular players regulating the jasmonate signalling network. Curr. Opin. Plant Biol. 2005, 8, 532–540. [Google Scholar] [CrossRef]
- Broekgaarden, C.; Caarls, L.; Vos, I.A.; Pieterse, C.M.J.; Van Wees, S.C.M. Ethylene: Traffic controller on hormonal crossroads to defense. Plant Physiol. 2015, 169, 2371–2379. [Google Scholar] [CrossRef]
- Lorenzo, O.; Chico, J.M.; Sanchez-Serrano, J.J.; Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.P.; Badruzsaufari, E.; Schenk, P.M.; Manners, J.M.; Desmond, O.J.; Ehlert, C.; Maclean, D.J.; Ebert, P.R.; Kazan, K. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling Pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 2004, 16, 3460–3479. [Google Scholar] [CrossRef] [PubMed]
- Vos, I.A.; Verhage, A.; Schuurink, R.C.; Watt, L.G.; Pieterse, C.M.J.; Van Wees, S.C.M. Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front. Plant Sci. 2013, 4, 539. [Google Scholar] [CrossRef] [PubMed]
- De Torres Zabala, M.; Bennett, M.H.; Truman, W.H.; Grant, M.R. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 2009, 59, 375–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasuda, M.; Ishikawa, A.; Jikumaru, Y.; Seki, M.; Umezawa, T.; Asami, T.; Maruyama-Nakashita, A.; Kudo, T.; Shinozaki, K.; Yoshida, S.; et al. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 2008, 20, 1678–1692. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Vos, I.A.; Pieterse, C.M.J.; Wees, S.C.M. Costs and benefits of hormone-regulated plant defences. Plant Pathol. 2013, 62, 43–55. [Google Scholar] [CrossRef] [Green Version]
- Adie, B.A.T.; Pérez-Pérez, J.; Pérez-Pérez, M.M.; Godoy, M.; Sánchez-Serrano, J.-J.; Schmelz, E.A.; Solano, R. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 2007, 19, 1665–1681. [Google Scholar] [CrossRef]
- Ravanel, S.; Block, M.A.; Rippert, P.; Jabrin, S.; Curien, G.; Rébeillé, F.; Douce, R. Methionine metabolism in plants: Chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J. Biol. Chem. 2004, 279, 22548–22557. [Google Scholar] [CrossRef]
- Johnson, P.R.; Ecker, J.R. The ethylene gas signal transduction pathway: A molecular perspective. Annu. Rev. Genet. 1998, 32, 227–254. [Google Scholar] [CrossRef] [PubMed]
- Levitt, L.K.; Stein, D.B.; Rubinstein, B. Promotion of stomatal opening by indoleacetic acid and ethrel in epidermal strips of Vicia faba L. Plant Physiol. 1987, 85, 318–321. [Google Scholar] [CrossRef]
- Merritt, F.; Kemper, A.; Tallman, G. Inhibitors of ethylene synthesis Inhibit auxin-induced stomatal opening in epidermis detached from leaves of Vicia faba L. Plant Cell Physiol. 2001, 42, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 2005, 138, 2337–2343. [Google Scholar] [CrossRef] [PubMed]
- Pallas, J.E.; Kays, S.J. Inhibition of photosynthesis by ethylene—a stomatal effect. Plant Physiol. 1982, 70, 598–601. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, S.; Chrominiski, A.; Smith, B.N. Effect of ethylene on stomatal opening in tomato and carnation leaves. Plant Cell Physiol. 1983, 24, 569–572. [Google Scholar]
- Desikan, R.; Last, K.; Harrett-Williams, R.; Tagliavia, C.; Harter, K.; Hooley, R.; Hancock, J.T.; Neill, S.J. Ethylene-induced stomatal closure in Arabidopsis occurs via AtrbohF-mediated hydrogen peroxide synthesis. Plant J. 2006, 47, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Ceusters, J.; Van de Poel, B. Ethylene exerts species-specific and age-dependent control of photosynthesis. Plant Physiol. 2018, 176, 2601–2612. [Google Scholar] [CrossRef]
- Grbić, V.; Bleecker, A.B. Ethylene regulates the timing of leaf senescence in Arabidopsis. Plant J. 1995, 8, 595–602. [Google Scholar] [CrossRef] [Green Version]
- Tholen, D.; Voesenek, L.A.; Poorter, H. Ethylene insensitivity does not increase leaf area or relative growth rate in Arabidopsis, Nicotiana tabacum, and Petunia x hybrida. Plant Physiol. 2004, 134, 1803–1812. [Google Scholar] [CrossRef]
- Tholen, D.; Pons, T.L.; Voesenek, L.A.; Poorter, H. Ethylene insensitivity results in down-regulation of rubisco expression and photosynthetic capacity in tobacco. Plant Physiol. 2007, 144, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Tholen, D.; Pons, T.L.; Voesenek, L.A.; Poorter, H. The role of ethylene perception in the control of photosynthesis. Plant Signal. Behav. 2008, 3, 108–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, S.A.; Park, J.H.; Lee, G.I.; Paek, K.H.; Park, S.K.; Nam, H.G. Identification of three genetic loci controlling leaf senescence in Arabidopsis thaliana. Plant J. 1997, 12, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.F.; Gonzalez-Carranza, Z.H.; Maunders, M.J.; Roberts, J.A. Ethylene and the regulation of senescence processes in transgenic Nicotiana sylvestris plants. Ann. Bot. 2008, 101, 301–310. [Google Scholar] [CrossRef]
- Monteiro, C.C.; Carvalho, R.F.; Gratão, P.L.; Carvalho, G.; Tezotto, T.; Medici, L.O.; Peres, L.E.P.; Azevedo, R.A. Biochemical responses of the ethylene-insensitive Never ripe tomato mutant subjected to cadmium and sodium stresses. Environ. Exp. Bot. 2011, 71, 306–320. [Google Scholar] [CrossRef]
- Zacarias, L.; Reid, M.S. Role of growth regulators in the senescence of Arabidopsis thaliana leaves. Physiol. Plant 1990, 80, 549–554. [Google Scholar] [CrossRef]
- Young, T.E.; Meeley, R.B.; Gallie, D.R. ACC synthase expression regulates leaf performance and drought tolerance in maize. Plant J. 2004, 40, 813–825. [Google Scholar] [CrossRef] [Green Version]
- Pallaghy, C.K.; Raschke, K. No stomatal response to ethylene. Plant Physiol. 1972, 49, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Squier, S.A.; Taylor, G.E.; Selvidge, W.J.; Gunderson, C.A. Effect of ethylene and related hydrocarbons on carbon assimilation and transpiration in herbaceous and woody species. Environ. Sci. Technol. 1985, 19, 432–437. [Google Scholar] [CrossRef]
- Taylor, G.E.; Gunderson, C.A. The response of foliar gas exchange to exogenously applied ethylene. Plant Physiol. 1986, 82, 653–657. [Google Scholar] [CrossRef]
- He, J.; Yue, X.; Wang, R.; Zhang, Y. Ethylene mediates UV-B-induced stomatal closure via peroxidase-dependent hydrogen peroxide synthesis in Vicia faba L. J. Exp. Bot. 2011, 62, 2657–2666. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.-h.; Liu, G.-h.; Hou, L.-x.; Wang, L.-x.; Liu, X. Regulatory function of polyamine oxidase-generated hydrogen peroxide in ethylene-induced stomatal closure in Arabidopsis thaliana. J. Integr. Agr. 2013, 12, 251–262. [Google Scholar] [CrossRef]
- Bent, A.F.; Innes, R.W.; Ecker, J.R.; Staskawicz, B.J. Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol. Plant Microbe Interact. 1992, 5, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Eggermont, K.; Tierens, K.F.; Broekaert, W.F. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 1999, 121, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, T.; Schmidt, J.S.; Zheng, X.; Bent, A.F. Isolation of ethylene-insensitive soybean mutants that are altered in pathogen susceptibility and gene-for-gene disease resistance. Plant Physiol. 1999, 119, 935–950. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, N.; Serizet, C.; Gosti, F.; Giraudat, J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 2000, 12, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Ghassemian, M.; Nambara, E.; Cutler, S.; Kawaide, H.; Kamiya, Y.; McCourt, P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 2000, 12, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Chiwocha, S.D.; Cutler, A.J.; Abrams, S.R.; Ambrose, S.J.; Yang, J.; Ross, A.R.; Kermode, A.R. The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 2005, 42, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Penninckx, I.A.; Thomma, B.P.; Buchala, A.; Metraux, J.P.; Broekaert, W.F. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 1998, 10, 2103–2113. [Google Scholar] [CrossRef]
- Pre, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Van der Ent, S.; Van Wees, S.C.M.; Pieterse, C.M.J. Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 2009, 70, 1581–1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X.; et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 2009, 21, 2527–2540. [Google Scholar] [CrossRef] [PubMed]
- Šašek, V.; Nováková, M.; Jindřichová, B.; Bóka, K.; Valentová, O.; Burketová, L. Recognition of avirulence gene AvrLm1 from hemibiotrophic ascomycete Leptosphaeria maculans triggers salicylic acid and ethylene signaling in Brassica napus. Mol. Plant Microbe Interact. 2012, 25, 1238–1250. [Google Scholar] [CrossRef]
- Kovač, M.; Ravnikar, M. The effect of jasmonic acid on the photosynthetic pigments of potato plants grown in vitro. Plant Sci. 1994, 103, 11–17. [Google Scholar] [CrossRef]
- Zhang, L.; Xing, D. Methyl jasmonate induces production of reactive oxygen species and alterations in mitochondrial dynamics that precede photosynthetic dysfunction and subsequent cell death. Plant Cell Physiol. 2008, 49, 1092–1111. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Lyou, S.H.; Yeu, S.; Kim, M.A.; Rhee, S.; Kim, M.; Lee, J.S.; Choi, Y.D.; Cheong, J.J. Microarray-based screening of jasmonate-responsive genes in Arabidopsis thaliana. Plant Cell Rep. 2007, 26, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Shan, X.; Wang, J.; Chua, L.; Jiang, D.; Peng, W.; Xie, D. The role of Arabidopsis Rubisco activase in jasmonate-induced leaf senescence. Plant Physiol. 2011, 155, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; van Wees, S.C.; van Pelt, J.A.; Knoester, M.; Laan, R.; Gerrits, H.; Weisbeek, P.J.; van Loon, L.C. A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 1998, 10, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.J.; Van Der Ent, S.; Van Loon, L.C.; Pieterse, C.M. Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol. 2008, 180, 511–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ton, J.; De Vos, M.; Robben, C.; Buchala, A.; Metraux, J.P.; Van Loon, L.C.; Pieterse, C.M. Characterization of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J. 2002, 29, 11–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandurska, H.; Stroiński, A.; Kubiś, J. The effect of jasmonic acid on the accumulation of ABA, proline and spermidine and its influence on membrane injury under water deficit in two barley genotypes. Acta Physiol. Plant 2003, 25, 279–285. [Google Scholar] [CrossRef]
- De Ollas, C.; Hernando, B.; Arbona, V.; Gomez-Cadenas, A. Jasmonic acid transient accumulation is needed for abscisic acid increase in citrus roots under drought stress conditions. Physiol. Plant 2013, 147, 296–306. [Google Scholar] [CrossRef]
- De Ollas, C.; Arbona, V.; Gomez-Cadenas, A. Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant Cell Environ. 2015, 38, 2157–2170. [Google Scholar] [CrossRef] [Green Version]
- De Ollas, C.; Dodd, I.C. Physiological impacts of ABA–JA interactions under water-limitation. Plant Mol. Biol. 2016, 91, 641–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, S.; Yamada, H.; Setha, S. Effect of jasmonates differed at fruit ripening stages on 1-aminocyclopropane-1-carboxylate (ACC) synthase and ACC oxidase gene expression in pears. J. Amer. Soc. Hort. Sci. 2007, 132, 120–125. [Google Scholar]
- Suhita, D.; Raghavendra, A.S.; Kwak, J.M.; Vavasseur, A. Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol. 2004, 134, 1536–1545. [Google Scholar] [CrossRef] [PubMed]
- Denness, L.; McKenna, J.F.; Segonzac, C.; Wormit, A.; Madhou, P.; Bennett, M.; Mansfield, J.; Zipfel, C.; Hamann, T. Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 2011, 156, 1364–1374. [Google Scholar] [CrossRef]
- Liu, X.; Shi, W.; Zhang, S.; Lou, C. Nitric oxide involved in signal transduction of jasmonic acid-induced stomatal closure of Vicia faba L. Chin. Sci. Bull. 2005, 50, 520–525. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, Y.; Tang, S.; Pan, J.; Yu, Y.; Han, J.; Li, Y.; Du, X.; Nan, Z.; Sun, Q. AtCNGC2 is involved in jasmonic acid-induced calcium mobilization. J. Exp. Bot. 2016, 67, 809–819. [Google Scholar] [CrossRef]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Garcion, C.; Lohmann, A.; Lamodière, E.; Catinot, J.; Buchala, A.; Doermann, P.; Métraux, J.-P. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 2008, 147, 1279–1287. [Google Scholar] [CrossRef]
- Janda, T.; Gondor, O.K.; Yordanova, R.; Szalai, G.; Pál, M. Salicylic acid and photosynthesis: Signalling and effects. Acta Physiol. Plant 2014, 36, 2537–2546. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez-Coronado, M.A.; Trejo-López, C.; Larqué-Saavedra, A. Effects of salicylic acid on the growth of roots and shoots in soybean. Plant Physiol. Biochem. 1998, 36, 563–565. [Google Scholar] [CrossRef]
- Janda, T.; Szalai, G.; Tari, I.; Páldi, E. Hydroponic treatment with salicylic acid decreases the effects of chilling injury in maize (Zea mays L.) plants. Planta 1999, 208, 175–180. [Google Scholar] [CrossRef]
- Fariduddin, Q.; Hayat, S.; Ahmad, A. Salicylic acid influences net photosynthetic rate, carboxylation efficiency, nitrate reductase activity, and seed yield in Brassica juncea. Photosynthetica 2003, 41, 281–284. [Google Scholar] [CrossRef]
- Khan, W.; Prithiviraj, B.; Smith, D.L. Photosynthetic responses of corn and soybean to foliar application of salicylates. J. Plant Physiol. 2003, 160, 485–492. [Google Scholar] [CrossRef]
- Radwan, D.E.M.; Soltan, D.M. The negative effects of clethodim in photosynthesis and gas-exchange status of maize plants are ameliorated by salicylic acid pretreatment. Photosynthetica 2012, 50, 171–179. [Google Scholar] [CrossRef]
- Sasheva, P.; Yordanova, R.; Janda, T.; Szalai, G.; Maslenkova, L. Study of primary photosynthetic reactions in winter wheat cultivars after cold hardening and freezing. effect of salicylic acid. Bulg. J. Agric. Sci. 2013, 19, 45–48. [Google Scholar]
- Tirani, M.M.; Nasibi, F.; Kalantari, K.M. Interaction of salicylic acid and ethylene and their effects on some physiological and biochemical parameters in canola plants (Brassica napus L.). Photosynthetica 2013, 51, 411–418. [Google Scholar] [CrossRef]
- Mateo, A.; Funck, D.; Muhlenbock, P.; Kular, B.; Mullineaux, P.M.; Karpinski, S. Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 2006, 57, 1795–1807. [Google Scholar] [CrossRef] [Green Version]
- Park, S.W.; Kaimoyo, E.; Kumar, D.; Mosher, S.; Klessig, D.F. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 2007, 318, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.C.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Brown, R.; Kazan, K.; et al. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Amornsiripanitch, N.; Dong, X. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2006, 2, e123. [Google Scholar] [CrossRef] [PubMed]
- Tada, Y.; Spoel, S.H.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef]
- Shimono, M.; Sugano, S.; Nakayama, A.; Jiang, C.J.; Ono, K.; Toki, S.; Takatsuji, H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 2007, 19, 2064–2076. [Google Scholar] [CrossRef] [PubMed]
- Leslie, C.A.; Romani, R.J. Inhibition of ethylene biosynthesis by salicylic acid. Plant Physiol. 1988, 88, 833–837. [Google Scholar] [CrossRef] [PubMed]
- Doherty, H.M.; Selvendran, R.R.; Bowles, D.J. The wound response of tomato plants can be inhibited by aspirin and related hydroxy-benzoic acids. Physiol. Mol. Plant Pathol. 1988, 33, 377–384. [Google Scholar] [CrossRef]
- Pena-Cortés, H.; Albrecht, T.; Prat, S.; Weiler, E.W.; Willmitzer, L. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 1993, 191, 123–128. [Google Scholar] [CrossRef]
- Van Wees, S.C.M.; Luijendijk, M.; Smoorenburg, I.; van Loon, L.C.; Pieterse, C.M.J. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 1999, 41, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Leon-Reyes, A.; Van der Does, D.; De Lange, E.S.; Delker, C.; Wasternack, C.; Van Wees, S.C.; Ritsema, T.; Pieterse, C.M. Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 2010, 232, 1423–1432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Does, D.; Leon-Reyes, A.; Koornneef, A.; Van Verk, M.C.; Rodenburg, N.; Pauwels, L.; Goossens, A.; Körbes, A.P.; Memelink, J.; Ritsema, T.; et al. Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 2013, 25, 744–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zander, M.; Thurow, C.; Gatz, C. TGA transcription factors activate the salicylic acid-suppressible branch of the ethylene-induced defense program by regulating ORA59 expression. Plant Physiol. 2014, 165, 1671–1683. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.A.; Brown, E.J. Cytosol as battleground: Ubiquitin as a weapon for both host and pathogen. Trends Cell. Biol. 2010, 20, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Hicks, S.W.; Galan, J.E. Hijacking the host ubiquitin pathway: Structural strategies of bacterial E3 ubiquitin ligases. Curr. Opin. Microbiol. 2010, 13, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Kubori, T.; Nagai, H. Bacterial effector-involved temporal and spatial regulation by hijack of the host ubiquitin pathway. Front. Microbiol. 2011, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Silva, H.; Klessig, D.F. Active oxygen species in the induction of plant systemic acquired resistance by salicylic acid. Science 1993, 262, 1883–1886. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.V.; Paliyath, G.; Ormrod, D.P.; Murr, D.P.; Watkins, C.B. Influence of salicylic acid on H2O2 production, oxidative stress, and H2O2-metabolizing enzymes. Salicylic acid-mediated oxidative damage requires H2O2. Plant Physiol. 1997, 115, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Khokon, A.R.; Okuma, E.; Hossain, M.A.; Munemasa, S.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Murata, Y. Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 2011, 34, 434–443. [Google Scholar] [CrossRef]
- Miura, K.; Okamoto, H.; Okuma, E.; Shiba, H.; Kamada, H.; Hasegawa, P.M.; Murata, Y. SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 2013, 73, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Durner, J.; Klessig, D.F. Inhibition of ascorbate peroxidase by salicylic acid and 2,6-dichloroisonicotinic acid, two inducers of plant defense responses. Proc. Nat. Acad. Sci. USA 1995, 92, 11312–11316. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Tamaoki, M.; Ioki, M.; Ogawa, D.; Sato, Y.; Aono, M.; Kubo, A.; Saji, S.; Saji, H.; Satoh, S.; et al. Ethylene and salicylic acid control glutathione biosynthesis in ozone-exposed Arabidopsis thaliana. Physiol. Plant 2009, 136, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.G.; Park, C.M. Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 2010, 188, 626–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaouch, S.; Queval, G.; Vanderauwera, S.; Mhamdi, A.; Vandorpe, M.; Langlois-Meurinne, M.; Van Breusegem, F.; Saindrenan, P.; Noctor, G. Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 2010, 153, 1692–1705. [Google Scholar] [CrossRef]
- Herrera-Vásquez, A.; Salinas, P.; Holuigue, L. Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front. Plant Sci. 2015, 6, 171. [Google Scholar] [CrossRef]
- Zottini, M.; Costa, A.; De Michele, R.; Ruzzene, M.; Carimi, F.; Lo Schiavo, F. Salicylic acid activates nitric oxide synthesis in Arabidopsis. J. Exp. Bot. 2007, 58, 1397–1405. [Google Scholar] [CrossRef] [Green Version]
- Kawano, T.; Sahashi, N.; Takahashi, K.; Uozumi, N.; Muto, S. Salicylic acid induces extracellular superoxide generation followed by an increase in cytosolic calcium ion in tobacco suspension culture: The earliest events in salicylic acid signal transduction. Plant Cell Physiol. 1998, 39, 721–730. [Google Scholar] [CrossRef]
- Kawano, T.; Muto, S. Mechanism of peroxidase actions for salicylic acid-induced generation of active oxygen species and an increase in cytosolic calcium in tobacco cell suspension culture. J. Exp. Bot. 2000, 51, 685–693. [Google Scholar] [Green Version]
- Turkan, I. ROS and RNS: Key signalling molecules in plants. J. Exp. Bot. 2018, 69, 3313–3315. [Google Scholar] [CrossRef]
- Shapiguzov, A.; Vainonen, J.P.; Wrzaczek, M.; Kangasjärvi, J. ROS-talk—how the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 2012, 3, 292. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.A.; Jones, J.D.G.; Dangl, J.L. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006, 141, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Bienert, G.P.; Moller, A.L.; Kristiansen, K.A.; Schulz, A.; Moller, I.M.; Schjoerring, J.K.; Jahn, T.P. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 2007, 282, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Dynowski, M.; Schaaf, G.; Loque, D.; Moran, O.; Ludewig, U. Plant plasma membrane water channels conduct the signalling molecule H2O2. Biochem. J. 2008, 414, 53–61. [Google Scholar] [CrossRef]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef] [PubMed]
- Mehler, A.H. Studies on reactions of illuminated chloroplasts: I. Mechanism of the reduction of oxygen and other hill reagents. Arch. Biochem. Biophys. 1951, 33, 65–77. [Google Scholar] [CrossRef]
- Mehler, A.H. Studies on reactions of illuminated chloroplasts. II. Stimulation and inhibition of the reaction with molecular oxygen. Arch. Biochem. Biophys. 1951, 34, 339–351. [Google Scholar] [CrossRef]
- Telfer, A.; Bishop, S.M.; Phillips, D.; Barber, J. Isolated photosynthetic reaction center of photosystem II as a sensitizer for the formation of singlet oxygen. Detection and quantum yield determination using a chemical trapping technique. J. Biol. Chem. 1994, 269, 13244–13253. [Google Scholar]
- Hideg, É.; Kálai, T.; Hideg, K.; Vass, I. Photoinhibition of photosynthesis in vivo results in singlet oxygen production detection via nitroxide-induced fluorescence quenching in broad bean leaves. Biochemistry 1998, 37, 11405–11411. [Google Scholar] [CrossRef]
- Asada, K. The water-water cycle as alternative photon and electron sinks. Phil. Trans. R. Soc. B 2000, 355, 1419–1431. [Google Scholar] [CrossRef]
- Foyer, C.H.; Shigeoka, S. Understanding oxidative stress and antioxidant functions to enhance photosynthesis. Plant Physiol. 2011, 155, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2. [Google Scholar] [CrossRef]
- Chen, S.X.; Schopfer, P. Hydroxyl-radical production in physiological reactions. A novel function of peroxidase. Eur. J. Biochem. 1999, 260, 726–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley, D.J.; Kjellbom, P.; Lamb, C.J. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: A novel, rapid defense response. Cell 1992, 70, 21–30. [Google Scholar] [CrossRef]
- Kovtun, Y.; Chiu, W.L.; Tena, G.; Sheen, J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Nat. Acad. Sci. USA 2000, 97, 2940–2945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montillet, J.-L.; Chamnongpol, S.; Rustérucci, C.; Dat, J.; van de Cotte, B.; Agnel, J.-P.; Battesti, C.; Inzé, D.; Van Breusegem, F.; Triantaphylidès, C. Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol. 2005, 138, 1516–1526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thoma, I.; Loeffler, C.; Sinha, A.K.; Gupta, M.; Krischke, M.; Steffan, B.; Roitsch, T.; Mueller, M.J. Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J. 2003, 34, 363–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, M.E.; Pennell, R.I.; Meijer, P.J.; Ishikawa, A.; Dixon, R.A.; Lamb, C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 1998, 92, 773–784. [Google Scholar] [CrossRef]
- Leborgne-Castel, N.; Lherminier, J.; Der, C.; Fromentin, J.; Houot, V.; Simon-Plas, F. The plant defense elicitor cryptogein stimulates clathrin-mediated endocytosis correlated with reactive oxygen species production in Bright Yellow-2 tobacco cells. Plant Physiol. 2008, 146, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Robatzek, S.; Torres, M.; Kombrink, E.; Somssich, I.E.; Robinson, M.; Schulze-Lefert, P. Chemical interference of pathogen-associated molecular pattern-triggered immune responses in Arabidopsis reveals a potential role for fatty-acid synthase type II complex-derived lipid signals. J. Biol. Chem. 2007, 282, 6803–6811. [Google Scholar] [CrossRef] [PubMed]
- Yao, N.; Greenberg, J.T. Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 2006, 18, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ren, D.; Pike, S.; Pallardy, S.; Gassmann, W.; Zhang, S. Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 2007, 51, 941–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zurbriggen, M.D.; Carrillo, N.; Tognetti, V.B.; Melzer, M.; Peisker, M.; Hause, B.; Hajirezaei, M.R. Chloroplast-generated reactive oxygen species play a major role in localized cell death during the non-host interaction between tobacco and Xanthomonas campestris pv. vesicatoria. Plant J. 2009, 60, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Ishiga, Y.; Ishiga, T.; Wangdi, T.; Mysore, K.S.; Uppalapati, S.R. NTRC and chloroplast-generated reactive oxygen species regulate Pseudomonas syringae pv. tomato disease development in tomato and Arabidopsis. Mol. Plant Microbe Interact. 2012, 25, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Maruta, T.; Noshi, M.; Tanouchi, A.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. H2O2-triggered retrograde signaling from chloroplasts to nucleus plays specific role in response to stress. J. Biol. Chem. 2012, 287, 11717–11729. [Google Scholar] [CrossRef] [PubMed]
- Mubarakshina, M.M.; Ivanov, B.N.; Naydov, I.A.; Hillier, W.; Badger, M.R.; Krieger-Liszkay, A. Production and diffusion of chloroplastic H2O2 and its implication to signalling. J. Exp. Bot. 2010, 61, 3577–3587. [Google Scholar] [CrossRef]
- Borisova, M.M.; Kozuleva, M.A.; Rudenko, N.N.; Naydov, I.A.; Klenina, I.B.; Ivanov, B.N. Photosynthetic electron flow to oxygen and diffusion of hydrogen peroxide through the chloroplast envelope via aquaporins. Biochim. Biophys. Acta 2012, 1817, 1314–1321. [Google Scholar] [CrossRef]
- Wagner, D.; Przybyla, D.; Camp, R.O.D.; Kim, C.; Landgraf, F.; Lee, K.P.; Wursch, M.; Laloi, C.; Nater, M.; Hideg, E.; et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 2004, 306, 1183–1185. [Google Scholar] [CrossRef]
- Lee, K.P.; Kim, C.; Landgraf, F.; Apel, K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Nat. Acad. Sci. USA 2007, 104, 10270–10275. [Google Scholar] [CrossRef] [Green Version]
- Kim, C.; Meskauskiene, R.; Zhang, S.; Lee, K.P.; Lakshmanan Ashok, M.; Blajecka, K.; Herrfurth, C.; Feussner, I.; Apel, K. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 2012, 24, 3026–3039. [Google Scholar] [CrossRef] [PubMed]
- Neill, S.; Barros, R.; Bright, J.; Desikan, R.; Hancock, J.; Harrison, J.; Morris, P.; Ribeiro, D.; Wilson, I. Nitric oxide, stomatal closure, and abiotic stress. J. Exp. Bot. 2008, 59, 165–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Jiang, M.; Zhang, A.; Lu, J. Abscisic acid-induced apoplastic H2O2 accumulation up-regulates the activities of chloroplastic and cytosolic antioxidant enzymes in maize leaves. Planta 2005, 223, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, J. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J. Exp. Bot. 2002, 53, 2401–2410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, M.; Zhang, J. Involvement of plasma-membrane NADPH oxidase in abscisic acid- and water stress-induced antioxidant defense in leaves of maize seedlings. Planta 2002, 215, 1022–1030. [Google Scholar] [CrossRef]
- Lu, C.; Han, M.-H.; Guevara-Garcia, A.; Fedoroff, N.V. Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Nat. Acad. Sci. USA 2002, 99, 15812–15817. [Google Scholar] [CrossRef]
- Zhang, A.; Jiang, M.; Zhang, J.; Tan, M.; Hu, X. Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 2006, 141, 475–487. [Google Scholar] [CrossRef]
- Liu, Y.; Ye, N.; Liu, R.; Chen, M.; Zhang, J. H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 2010, 61, 2979–2990. [Google Scholar] [CrossRef]
- Jakubowicz, M.; Galganska, H.; Nowak, W.; Sadowski, J. Exogenously induced expression of ethylene biosynthesis, ethylene perception, phospholipase D, and Rboh-oxidase genes in broccoli seedlings. J. Exp. Bot. 2010, 61, 3475–3491. [Google Scholar] [CrossRef] [Green Version]
- Ke, D.; Sun, G. The effect of reactive oxygen species on ethylene production induced by osmotic stress in etiolated mungbean seedling. Plant Growth Regul. 2004, 44, 199–206. [Google Scholar] [CrossRef]
- Balazadeh, S.; Jaspert, N.; Arif, M.; Mueller-Roeber, B.; Maurino, V.G. Expression of ROS-responsive genes and transcription factors after metabolic formation of H(2)O(2) in chloroplasts. Front. Plant Sci. 2012, 3, 234. [Google Scholar] [CrossRef] [PubMed]
- Straus, M.R.; Rietz, S.; Ver Loren van Themaat, E.; Bartsch, M.; Parker, J.E. Salicylic acid antagonism of EDS1-driven cell death is important for immune and oxidative stress responses in Arabidopsis. Plant J. 2010, 62, 628–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delledonne, M.; Zeier, J.; Marocco, A.; Lamb, C. Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Nat. Acad. Sci. USA 2001, 98, 13454–13459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bright, J.; Desikan, R.; Hancock, J.T.; Weir, I.S.; Neill, S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2006, 45, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Price, A.H.; Taylor, A.; Ripley, S.J.; Griffiths, A.; Trewavas, A.J.; Knight, M.R. Oxidative signals in tobacco increase cytosolic calcium. Plant Cell 1994, 6, 1301–1310. [Google Scholar] [CrossRef]
- McAinsh, M.R.; Clayton, H.; Mansfield, T.A.; Hetherington, A.M. Changes in stomatal behavior and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiol. 1996, 111, 1031–1042. [Google Scholar] [CrossRef]
- Pei, Z.-M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731. [Google Scholar] [CrossRef]
- Bais, H.P.; Vepachedu, R.; Gilroy, S.; Callaway, R.M.; Vivanco, J.M. Allelopathy and exotic plant invasion: From molecules and genes to species interactions. Science 2003, 301, 1377–1380. [Google Scholar] [CrossRef]
- Rőszer, T. Biosynthesis of nitric oxide in plants. In Nitric Oxide in Plants: Metabolism and Role in Stress Physiology; Khan, M.N., Mobin, M., Mohammad, F., Corpas, F.J., Eds.; Springer International Publishing: Cham, Switzerland, 2014; pp. 17–32. [Google Scholar]
- Jasid, S.; Simontacchi, M.; Bartoli, C.G.; Puntarulo, S. Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiol. 2006, 142, 1246–1255. [Google Scholar] [CrossRef]
- Tewari, R.K.; Prommer, J.; Watanabe, M. Endogenous nitric oxide generation in protoplast chloroplasts. Plant Cell Rep. 2013, 32, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Crawford, N.M. Mechanisms for nitric oxide synthesis in plants. J. Exp. Bot. 2006, 57, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.C.; Cataneo, A.C. Nitric oxide in plants: A brief discussion on this multifunctional molecule. Sci. Agric. 2010, 67, 236–243. [Google Scholar] [CrossRef]
- Shapiro, A.D. Nitric oxide signaling in plants. Vitamins and hormones 2005, 72, 339–398. [Google Scholar]
- Besson-Bard, A.; Pugin, A.; Wendehenne, D. New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 2008, 59, 21–39. [Google Scholar] [CrossRef]
- Hill, A.C.; Bennett, J.H. Inhibition of apparent photosynthesis by nitrogen oxides. Atmos. Environ. 1970, 4, 341–348. [Google Scholar] [CrossRef]
- Saxe, H. Stomatal-dependent and stomatal-independent uptake of NOX. New Phytol. 1986, 103, 199–205. [Google Scholar] [CrossRef]
- Wodala, B.; Deák, Z.; Vass, I.; Erdei, L.; Altorjay, I.; Horváth, F. In vivo target sites of nitric oxide in photosynthetic electron transport as studied by chlorophyll fluorescence in pea leaves. Plant Physiol. 2008, 146, 1920–1927. [Google Scholar] [CrossRef]
- Petrouleas, V.; Diner, B.A. Formation by NO of nitrosyl adducts of redox components of the Photosystem II reaction center. I. NO binds to the acceptor-side non-heme iron. Biochim. Biophys. Acta 1990, 1015, 131–140. [Google Scholar] [CrossRef]
- Diner, B.A.; Petrouleas, V. Formation by NO of nitrosyl adducts of redox components of the Photosystem II reaction center. II. Evidence that HCO3−/CO2 binds to the acceptor-side non-heme iron. Biochim. Biophys. Acta 1990, 1015, 141–149. [Google Scholar] [CrossRef]
- Schansker, G.; Goussias, C.; Petrouleas, V.; Rutherford, A.W. Reduction of the Mn cluster of the water-oxidizing enzyme by nitric oxide: Formation of an S-2 State. Biochemistry 2002, 41, 3057–3064. [Google Scholar] [CrossRef] [PubMed]
- Sanakis, Y.; Goussias, C.; Mason, R.P.; Petrouleas, V. NO interacts with the tyrosine radical Y(D). of photosystem II to form an iminoxyl radical. Biochemistry 1997, 36, 1411–1417. [Google Scholar] [CrossRef] [PubMed]
- Vladkova, R.; Dobrikova, A.G.; Singh, R.; Misra, A.N.; Apostolova, E. Photoelectron transport ability of chloroplast thylakoid membranes treated with NO donor SNP: Changes in flash oxygen evolution and chlorophyll fluorescence. Nitric Oxide 2011, 24, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Abat, J.K.; Mattoo, A.K.; Deswal, R. S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata - ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 2008, 275, 2862–2872. [Google Scholar] [CrossRef] [PubMed]
- Delledonne, M.; Xia, Y.; Dixon, R.A.; Lamb, C. Nitric oxide functions as a signal in plant disease resistance. Nature 1998, 394, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Yun, B.-W.; Feechan, A.; Yin, M.; Saidi, N.B.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.-G.; Kwon, E.; Spoel, S.H.; et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011, 478, 264. [Google Scholar] [CrossRef] [PubMed]
- Klepper, L. NOx evolution by soybean leaves treated with salicylic acid and selected derivatives. Pest. Biochem. Physiol. 1991, 39, 43–48. [Google Scholar] [CrossRef]
- Wendehenne, D.; Durner, J.; Klessig, D.F. Nitric oxide: A new player in plant signalling and defence responses. Curr. Opin. Plant Biol. 2004, 7, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mata, C.; Lamattina, L. Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol. 2001, 126, 1196–1204. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Du, Y.; Hou, Y.-J.; Zhao, Y.; Hsu, C.-C.; Yuan, F.; Zhu, X.; Tao, W.A.; Song, C.-P.; Zhu, J.-K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Nat. Acad. Sci. USA 2015, 112, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Bethke, P.C.; Libourel, I.G.; Jones, R.L. Nitric oxide reduces seed dormancy in Arabidopsis. J. Exp. Bot. 2006, 57, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.J.; Laarhoven, L.J.J.; Harren, F.J.M.; Hall, M.A.; Smith, A.R. Nitric oxide interacts with salicylate to regulate biphasic ethylene production during the hypersensitive response. Plant Physiol. 2008, 148, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.; Lloyd, A.J.; Cristescu, S.M.; Harren, F.J.; Hall, M.A.; Smith, A.R. Biphasic ethylene production during the hypersensitive response in Arabidopsis: A window into defense priming mechanisms? Plant Signal. Behav. 2009, 4, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Hebelstrup, K.H.; van Zanten, M.; Mandon, J.; Voesenek, L.A.; Harren, F.J.; Cristescu, S.M.; Moller, I.M.; Mur, L.A. Haemoglobin modulates NO emission and hyponasty under hypoxia-related stress in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 5581–5591. [Google Scholar] [CrossRef] [PubMed]
- Lindermayr, C.; Saalbach, G.; Bahnweg, G.; Durner, J. Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 2006, 281, 4285–4291. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Liu, M.; Zhou, J. Inhibition by nitric oxide of ethylene biosynthesis and lipoxygenase activity in peach fruit during storage. Postharvest Biol. Technol. 2006, 42, 41–48. [Google Scholar] [CrossRef]
- Huang, X.; Stettmaier, K.; Michel, C.; Hutzler, P.; Mueller, M.J.; Durner, J. Nitric oxide is induced by wounding and influences jasmonic acid signaling in Arabidopsis thaliana. Planta 2004, 218, 938–946. [Google Scholar] [CrossRef]
- Palmieri, M.C.; Sell, S.; Huang, X.; Scherf, M.; Werner, T.; Durner, J.; Lindermayr, C. Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: A bioinformatics approach. J. Exp. Bot. 2008, 59, 177–186. [Google Scholar] [CrossRef]
- Mur, L.A.; Sivakumaran, A.; Mandon, J.; Cristescu, S.M.; Harren, F.J.; Hebelstrup, K.H. Haemoglobin modulates salicylate and jasmonate/ethylene-mediated resistance mechanisms against pathogens. J. Exp. Bot. 2012, 63, 4375–4387. [Google Scholar] [CrossRef] [Green Version]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Nat. Acad. Sci. USA 1998, 95, 10328–10333. [Google Scholar] [CrossRef] [Green Version]
- Lindermayr, C.; Durner, J. Interplay of reactive oxygen species and nitric oxide: Nitric oxide coordinates reactive oxygen species homeostasis. Plant Physiol. 2015, 167, 1209–1210. [Google Scholar] [CrossRef] [PubMed]
- McAinsh, M.R.; Pittman, J.K. Shaping the calcium signature. New Phytol. 2009, 181, 275–294. [Google Scholar] [CrossRef]
- Nomura, H.; Komori, T.; Uemura, S.; Kanda, Y.; Shimotani, K.; Nakai, K.; Furuichi, T.; Takebayashi, K.; Sugimoto, T.; Sano, S.; et al. Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 2012, 3, 926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terry, N.; Huston, R.P. Effects of calcium on the photosynthesis of intact leaves and isolated chloroplasts of sugar beets. Plant Physiol. 1975, 55, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Petroutsos, D.; Busch, A.; Janssen, I.; Trompelt, K.; Bergner, S.V.; Weinl, S.; Holtkamp, M.; Karst, U.; Kudla, J.; Hippler, M. The chloroplast calcium sensor CAS is required for photoacclimation in Chlamydomonas reinhardtii. Plant Cell 2011, 23, 2950–2963. [Google Scholar] [CrossRef]
- Liu, Y.-F.; Zhang, G.-X.; Qi, M.-F.; Li, T.-L. Effects of calcium on photosynthesis, antioxidant system, and chloroplast ultrastructure in tomato leaves under low night temperature stress. J. Plant Growth Regul. 2015, 34, 263–273. [Google Scholar] [CrossRef]
- Dolatabadian, A.; Sanavy, S.A.M.M.; Gholamhoseini, M.; Joghan, A.K.; Majdi, M.; Kashkooli, A.B. The role of calcium in improving photosynthesis and related physiological and biochemical attributes of spring wheat subjected to simulated acid rain. Physiol. Mol. Biol. Plants 2013, 19, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Li, X.; Zhang, L. The effect of calcium chloride on growth, photosynthesis, and antioxidant responses of Zoysia japonica under drought conditions. PLoS ONE 2013, 8, e68214. [Google Scholar] [CrossRef]
- Xu, D.; Wang, W.; Gao, T.; Fang, X.; Gao, X.; Li, J.; Bu, H.; Mu, J. Calcium alleviates decreases in photosynthesis under salt stress by enhancing antioxidant metabolism and adjusting solute accumulation in Calligonum mongolicum. Conserv. Physiol. 2017, 5, cox060. [Google Scholar] [CrossRef]
- Raz, V.; Fluhr, R. Calcium requirement for ethylene-dependent responses. Plant Cell 1992, 4, 1123–1130. [Google Scholar] [CrossRef]
- Zielińska, M.; Michniewicz, M. The effect of calcium on the production of ethylene and abscisic acid by fungus Fusarium culmorum and by wheat seedlings infected with that pathogen. Acta Physiol. Plant 2001, 23, 79–85. [Google Scholar] [CrossRef]
- Sun, Q.-P.; Yu, Y.-K.; Wan, S.-X.; Zhao, F.-K.; Hao, Y.-L. Is there crosstalk between extracellular and intracellular calcium mobilization in jasmonic acid signaling. Plant Growth Regul. 2009, 57, 7–13. [Google Scholar] [CrossRef]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.; Poovaiah, B.W. Ca2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Chandra, S.; Low, P.S. Measurement of Ca2+ fluxes during elicitation of the oxidative burst in aequorin-transformed tobacco cells. J. Biol. Chem. 1997, 272, 28274–28280. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Ohura, I.; Kawakita, K.; Yokota, N.; Fujiwara, M.; Shimamoto, K.; Doke, N.; Yoshioka, H. Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 2007, 19, 1065–1080. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Willmann, M.R.; McCormack, M.; Lee, H.; Shan, L.; He, P.; Bush, J.; Cheng, S.H.; Sheen, J. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 2010, 464, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Modolo, L.V.; Cunha, F.Q.; Braga, M.R.; Salgado, I. Nitric oxide synthase-mediated phytoalexin accumulation in soybean cotyledons in response to the Diaporthe phaseolorum f. sp. meridionalis elicitor. Plant Physiol. 2002, 130, 1288–1297. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B.; Carreras, A.; Quirós, M.; León, A.M.; Romero-Puertas, M.C.; Esteban, F.J.; Valderrama, R.; Palma, J.M.; Sandalio, L.M.; et al. Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol. 2004, 136, 2722–2733. [Google Scholar] [CrossRef]
- Attaran, E.; Major, I.T.; Cruz, J.A.; Rosa, B.A.; Koo, A.J.K.; Chen, J.; Kramer, D.M.; He, S.Y.; Howe, G.A. Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol. 2014, 165, 1302–1314. [Google Scholar] [CrossRef]
- Gururani, Mayank A.; Venkatesh, J.; Tran, L.S.P. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef]
- Boller, T. Chemoperception of microbial signals in plant cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 189–214. [Google Scholar] [CrossRef]
- Zipfel, C.; Robatzek, S. Pathogen-associated molecular pattern-triggered immunity: Veni, vidi…? Plant Physiol. 2010, 154, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Mishina, T.E.; Zeier, J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 2007, 50, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Sato, M.; Glazebrook, J.; Cohen, J.D.; Katagiri, F. Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 2008, 53, 763–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deslandes, L.; Rivas, S. Catch me if you can: Bacterial effectors and plant targets. Trends Plant Sci. 2012, 17, 644–655. [Google Scholar] [CrossRef]
- Hicks, S.W.; Galán, J.E. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat. Rev. Microbiol. 2013, 11, 216–326. [Google Scholar] [CrossRef]
- Cunnac, S.; Lindeberg, M.; Collmer, A. Pseudomonas syringae type III secretion system effectors: Repertoires in search of functions. Curr. Opin. Microbiol. 2009, 12, 53–60. [Google Scholar] [CrossRef]
- Oh, H.S.; Park, D.H.; Collmer, A. Components of the Pseudomonas syringae type III secretion system can suppress and may elicit plant innate immunity. Mol. Plant Microbe Interact. 2010, 23, 727–739. [Google Scholar] [CrossRef]
- Block, A.; Alfano, J.R. Plant targets for Pseudomonas syringae type III effectors: Virulence targets or guarded decoys? Curr. Opin. Microbiol. 2011, 14, 39–46. [Google Scholar] [CrossRef]
- Lindeberg, M.; Cunnac, S.; Collmer, A. Pseudomonas syringae type III effector repertoires: Last words in endless arguments. Trends Microbiol. 2012, 20, 199–208. [Google Scholar] [CrossRef]
- Kim, M.G.; da Cunha, L.; McFall, A.J.; Belkhadir, Y.; DebRoy, S.; Dangl, J.L.; Mackey, D. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 2005, 121, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Jayaraman, J.; Segonzac, C.; Park, H.-J.; Park, H.; Han, S.-W.; Sohn, K.H. Pseudomonas syringae pv. actinidiae type III effectors localized at multiple cellular compartments activate or suppress innate immune responses in Nicotiana benthamiana. Front. Plant Sci. 2017, 8, 2157. [Google Scholar] [PubMed]
- Jelenska, J.; Yao, N.; Vinatzer, B.A.; Wright, C.M.; Brodsky, J.L.; Greenberg, J.T. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 2007, 17, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Herva, J.J.; Gonzalez-Melendi, P.; Cuartas-Lanza, R.; Antunez-Lamas, M.; Rio-Alvarez, I.; Li, Z.; Lopez-Torrejon, G.; Diaz, I.; Del Pozo, J.C.; Chakravarthy, S.; et al. A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell. Microbiol. 2012, 14, 669–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Froehlich, J.E.; Elowsky, C.; Msanne, J.; Ostosh, A.C.; Zhang, C.; Awada, T.; Alfano, J.R. Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 2014, 77, 310–321. [Google Scholar] [CrossRef]
- Sohn, K.H.; Zhang, Y.; Jones, J.D. The Pseudomonas syringae effector protein, AvrRPS4, requires in planta processing and the KRVY domain to function. Plant J. 2009, 57, 1079–1091. [Google Scholar] [CrossRef]
- Sperschneider, J.; Catanzariti, A.-M.; DeBoer, K.; Petre, B.; Gardiner, D.M.; Singh, K.B.; Dodds, P.N.; Taylor, J.M. LOCALIZER: Subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017, 7, 44598. [Google Scholar] [CrossRef]
- Sohn, K.H.; Hughes, R.K.; Piquerez, S.J.; Jones, J.D.G.; Banfield, M.J. Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc. Nat. Acad. Sci. USA 2012, 109, 16371–16376. [Google Scholar] [CrossRef]
- Heidrich, K.; Wirthmueller, L.; Tasset, C.; Pouzet, C.; Deslandes, L.; Parker, J.E. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment–specific immune responses. Science 2011, 334, 1401–1404. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Halane, M.K.; Kim, S.H.; Gassmann, W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 2011, 334, 1405–1408. [Google Scholar] [CrossRef]
- Gassmann, W.; Hinsch, M.E.; Staskawicz, B.J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999, 20, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Hinsch, M.; Staskawicz, B. Identification of a new Arabidopsis disease resistance locus, RPs4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant Microbe Interact. 1996, 9, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.J.; Sohn, K.H.; Wan, L.; Bernoux, M.; Sarris, P.F.; Segonzac, C.; Ve, T.; Ma, Y.; Saucet, S.B.; Ericsson, D.J.; et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 2014, 344, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Sarris, P.F.; Duxbury, Z.; Huh, S.U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.S.; Carvunis, A.R.; Dreze, M.; Epple, P.; Steinbrenner, J.; Moore, J.; Tasan, M.; Galli, M.; Hao, T.; Nishimura, M.T.; et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011, 333, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Teixeira, P.J.; Biswas, S.; Finkel, O.M.; He, Y.; Salas-Gonzalez, I.; English, M.E.; Epple, P.; Mieczkowski, P.; Dangl, J.L. Pseudomonas syringae type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell. Host Microbe 2017, 21, 156–168. [Google Scholar] [CrossRef]
- Guttman, D.S.; Vinatzer, B.A.; Sarkar, S.F.; Ranall, M.V.; Kettler, G.; Greenberg, J.T. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 2002, 295, 1722–1726. [Google Scholar] [CrossRef]
- Jelenska, J.; van Hal, J.A.; Greenberg, J.T. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc. Nat. Acad. Sci. USA 2010, 107, 13177–13182. [Google Scholar] [CrossRef]
- Petnicki-Ocwieja, T.; Schneider, D.J.; Tam, V.C.; Chancey, S.T.; Shan, L.; Jamir, Y.; Schechter, L.M.; Janes, M.D.; Buell, C.R.; Tang, X.; et al. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Nat. Acad. Sci. USA 2002, 99, 7652–7657. [Google Scholar] [CrossRef]
- Guo, M.; Tian, F.; Wamboldt, Y.; Alfano, J.R. The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol. Plant Microbe Interact. 2009, 22, 1069–1080. [Google Scholar] [CrossRef]
- Nomura, K.; Mecey, C.; Lee, Y.-N.; Imboden, L.A.; Chang, J.H.; He, S.Y. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Nat. Acad. Sci. USA 2011, 108, 10774–10779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nomura, K.; Debroy, S.; Lee, Y.H.; Pumplin, N.; Jones, J.; He, S.Y. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 2006, 313, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Duran, R.; Bourdais, G.; He, S.Y.; Robatzek, S. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 2014, 202, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Gangadharan, A.; Sreerekha, M.V.; Whitehill, J.; Ham, J.H.; Mackey, D. The Pseudomonas syringae pv. tomato type III effector HopM1 suppresses Arabidopsis defenses independent of suppressing salicylic acid signaling and of targeting AtMIN7. PLoS ONE 2013, 8, e82032. [Google Scholar]
- Lopez-Solanilla, E.; Bronstein, P.A.; Schneider, A.R.; Collmer, A. HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol. Microbiol. 2004, 54, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Joe, A. Pseudomonas syringae mono-ADP-ribosyltransferase type III effectors: Their plant targets and effects on plant immunity. Ph.D. Thesis, University of Nebraska-Lincoln, Lincoln, NB, USA, 2014. [Google Scholar]
- Kvitko, B.H.; Park, D.H.; Velásquez, A.C.; Wei, C.-F.; Russell, A.B.; Martin, G.B.; Schneider, D.J.; Collmer, A. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog 2009, 5, e1000388. [Google Scholar]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Kubori, T.; Galan, J.E. Temporal regulation of Salmonella virulence effector function by proteasome-dependent protein degradation. Cell 2003, 115, 333–342. [Google Scholar] [CrossRef]
- Knodler, L.A.; Winfree, S.; Drecktrah, D.; Ireland, R.; Steele-Mortimer, O. Ubiquitination of the bacterial inositol phosphatase, SopB, regulates its biological activity at the plasma membrane. Cell. Microbiol. 2009, 11, 1652–1670. [Google Scholar] [CrossRef] [Green Version]
- Patel, J.C.; Hueffer, K.; Lam, T.T.; Galan, J.E. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 2009, 137, 283–294. [Google Scholar] [CrossRef]
- Kubori, T.; Shinzawa, N.; Kanuka, H.; Nagai, H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010, 6, e1001216. [Google Scholar] [CrossRef] [PubMed]
- Huh, S.U.; Cevik, V.; Ding, P.; Duxbury, Z.; Ma, Y.; Tomlinson, L.; Sarris, P.F.; Jones, J.D.G. Protein-protein interactions in the RPS4/RRS1 immune receptor complex. PLoS Pathog. 2017, 13, e1006376. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 2015, 27, 2814–2828. [Google Scholar] [CrossRef]
- Yoo, K.S.; Ok, S.H.; Jeong, B.C.; Jung, K.W.; Cui, M.H.; Hyoung, S.; Lee, M.R.; Song, H.K.; Shin, J.S. Single cystathionine beta-synthase domain-containing proteins modulate development by regulating the thioredoxin system in Arabidopsis. Plant Cell 2011, 23, 3577–3594. [Google Scholar] [CrossRef]
- Montrichard, F.; Alkhalfioui, F.; Yano, H.; Vensel, W.H.; Hurkman, W.J.; Buchanan, B.B. Thioredoxin targets in plants: The first 30 years. J. Proteom. 2009, 72, 452–474. [Google Scholar] [CrossRef]
- Dowen, R.H.; Engel, J.L.; Shao, F.; Ecker, J.R.; Dixon, J.E. A family of bacterial cysteine protease type III effectors utilizes acylation-dependent and -independent strategies to localize to plasma membranes. J. Biol. Chem. 2009, 284, 15867–15879. [Google Scholar] [CrossRef]
- Kelley, W.L. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 1998, 23, 222–227. [Google Scholar] [CrossRef]
- Singer, A.U.; Desveaux, D.; Betts, L.; Chang, J.H.; Nimchuk, Z.; Grant, S.R.; Dangl, J.L.; Sondek, J. Crystal structures of the type III effector protein AvrPphF and its chaperone reveal residues required for plant pathogenesis. Structure 2004, 12, 1669–1681. [Google Scholar] [CrossRef]
- Coburn, J.; Wyatt, R.T.; Iglewski, B.H.; Gill, D.M. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J. Biol. Chem. 1989, 264, 9004–9008. [Google Scholar]
- Weβling, R.; Epple, P.; Altmann, S.; He, Y.; Yang, L.; Henz, S.R.; McDonald, N.; Wiley, K.; Bader, K.C.; Glasser, C.; et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell. Host Microbe 2014, 16, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Kesarwani, M.; Yoo, J.; Dong, X. Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 2007, 144, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.H.; Hwang, I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell. 2010, 19, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.G.; Debbage, J.M. Tabtoxin-induced symptoms are associated with the accumulation of ammonia formed during photorespiration. Physiol. Plant Pathol. 1982, 20, 223–233. [Google Scholar] [CrossRef]
- Thomas, M.D.; Langston-Unkefer, P.J.; Uchytil, T.F.; Durbin, R.D. Inhibition of glutamine synthetase from pea by tabtoxinine-β-lactam. Plant Physiol. 1983, 71, 912–915. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.; Stewart, G.R. The deposition of atmospheric ammonia and its effects on plants. New Phytol. 1993, 125, 283–305. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, R.E. Isolation and structure of a chlorosis-inducing toxin of Pseudomonas phaseolicola. Phytochemistry 1976, 15, 1941–1947. [Google Scholar] [CrossRef]
- Mitchell, R.E.; Bieleski, R.L. Involvement of phaseolotoxin in halo blight of beans: Transport and conversion to functional toxin. Plant Physiol. 1977, 60, 723–729. [Google Scholar] [CrossRef]
- Ferguson, A.R.; Johnston, J.S. Phaseolotoxin: Chlorosis, ornithine accumulation and inhibition of ornithine carbamoyltransferase in different plants. Physiol. Plant Pathol. 1980, 16, 269–275. [Google Scholar] [CrossRef]
- Turner, J.G. Effect of phaseolotoxin on the synthesis of arginine and protein. Plant Physiol. 1986, 80, 760–765. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, S.; Chini, A.; Hamberg, M.; Adie, B.; Porzel, A.; Kramell, R.; Miersch, O.; Wasternack, C.; Solano, R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5, 344. [Google Scholar] [CrossRef] [PubMed]
- Sheard, L.B.; Tan, X.; Mao, H.; Withers, J.; Ben-Nissan, G.; Hinds, T.R.; Kobayashi, Y.; Hsu, F.-F.; Sharon, M.; Browse, J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 2010, 468, 400. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Zheng, X.-y.; Spivey, N.W.; Zeng, W.; Liu, P.-P.; Fu, Z.Q.; Klessig, D.F.; He, S.Y.; Dong, X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell. Host Microbe 2012, 11, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mecey, C.; Hauck, P.; Trapp, M.; Pumplin, N.; Plovanich, A.; Yao, J.; He, S.Y. A critical role of STAYGREEN/Mendel’s I locus in controlling disease symptom development during Pseudomonas syringae pv tomato infection of Arabidopsis. Plant Physiol. 2011, 157, 1965–1974. [Google Scholar] [CrossRef]
- Geng, X.; Cheng, J.; Gangadharan, A.; Mackey, D. The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell 2012, 24, 4763–4774. [Google Scholar] [CrossRef]
- Zhao, Y.F.; Jones, W.T.; Sutherland, P.; Palmer, D.A.; Mitchell, R.E.; Reynolds, P.H.S.; Damicone, J.P.; Bender, C.L. Detection of the phytotoxin coronatine by ELISA and localization in infected plant tissue. Physiol. Mol. Plant Pathol. 2001, 58, 247–258. [Google Scholar] [CrossRef]
- Uppalapati, S.R.; Ayoubi, P.; Weng, H.; Palmer, D.A.; Mitchell, R.E.; Jones, W.; Bender, C.L. The phytotoxin coronatine and methyl jasmonate impact multiple phytohormone pathways in tomato. Plant J. 2005, 42, 201–217. [Google Scholar] [CrossRef] [Green Version]
- Serrano, I.; Audran, C.; Rivas, S. Chloroplasts at work during plant innate immunity. J. Exp. Bot. 2016, 67, 3845–3854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petre, B.; Lorrain, C.; Saunders, D.G.O.; Win, J.; Sklenar, J.; Duplessis, S.; Kamoun, S. Rust fungal effectors mimic host transit peptides to translocate into chloroplasts. Cell. Microbiol. 2016, 18, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.M.; Bender, C.L.; Kunkel, B.N. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 2005, 6, 629–639. [Google Scholar] [CrossRef] [PubMed]


| Name | Full Length (aa) | Cleavable Transit Peptide? | Signature Domain | Known and Potential Target Proteins | Function | References |
|---|---|---|---|---|---|---|
| AvrRps4 | 221a | Yes, 133 aa | SSM4 | EDS1 | Disrupts the interactions of EDS1 with its partners, such as RPS4 | [308,309,311,312,313,314,315] |
| WRKY domain of RRS1, WRKY33, WRKY41, WRKY60, WRKY70 | Activates RPS4-RRS1-dependent ETI; interferes with WRKY-dependent defense | [316,317,318] | ||||
| HopBB1 | 280b | Not sure | AvrPphF-ORF2 | JAZ3, TCP14 | Promotes TCP14 and JAZ3 degradation; de-represses TCP14-regulated JA response | [35,318,319] |
| PTF1d | Modulates psbD expression? | [318] | ||||
| TCP15d, TOE2d, UNE12d | Not known yet | [318] | ||||
| HopI1 | 488a | Non-cleavable transit peptide | DnaJ | cytHsp70-1, cpHsp70-1 | Recruits cytHsp70-1 to chloroplasts, forms complexes with cytHsp70-1 and cpHsp70-1, and activates their ATPase activity | [306,320,321] |
| HopK1 | 338a | Yes, 133 aa | Not identified yet | Not identified yet | Not known yet | [308,322,323] |
| HopM1 | 712a | Not sure | Not identified yet | MIN7, MIN10 | Degrades MIN7 and MIN10 | [305,324,325,326] |
| ARR2? | Degrades ARR2 and suppresses PR gene expression? | [327] | ||||
| At3g11720d | Not known yet | [318] | ||||
| HopN1 | 350a | Non-cleavable transit peptide | Cys protease | PsbQ | Degrades PsbQ | [307,328] |
| HopO1-1 | 283 a | Yes, 28–72 aac | ART | Not identified yet | Not known yet | [310,323,329] |
| HopO1-2 | 298a | Yes, 40–87 aac | ART | APC8d, CSN5A, OBE1d, At5g16940d | Not known yet | [35,310,318,323,329] |
| HopR1 | 1957a | Yes, 52–79 aac | AvrE, SMC_N | LSU1d, JAZ3d, TOE2d | Manipulates nuclear gene expression? | [35,305,310,318,330] |
| PTF1d | Modulates psbD expression? | [318] | ||||
| CBSX2d | Regulates redox status of chloroplastic enzymes? | [318] | ||||
| DUT1d, LSU3d, At3g48550d, At4g17680d | Not known yet | [318] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Y.; Yao, J. Chloroplasts at the Crossroad of Photosynthesis, Pathogen Infection and Plant Defense. Int. J. Mol. Sci. 2018, 19, 3900. https://doi.org/10.3390/ijms19123900
Lu Y, Yao J. Chloroplasts at the Crossroad of Photosynthesis, Pathogen Infection and Plant Defense. International Journal of Molecular Sciences. 2018; 19(12):3900. https://doi.org/10.3390/ijms19123900
Chicago/Turabian StyleLu, Yan, and Jian Yao. 2018. "Chloroplasts at the Crossroad of Photosynthesis, Pathogen Infection and Plant Defense" International Journal of Molecular Sciences 19, no. 12: 3900. https://doi.org/10.3390/ijms19123900