Red-Shifted Aminated Derivatives of GFP Chromophore for Live-Cell Protein Labeling with Lipocalins
Abstract
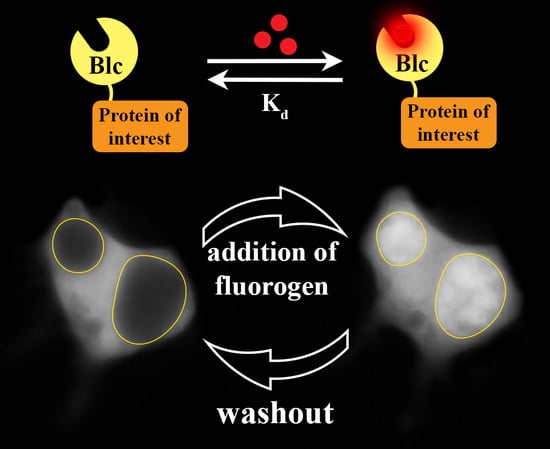
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Chemical Section
3.2. Synthetic procedures
3.2.1. (Z)-4-(4-(diethylamino)-2-(difluoroboryl)benzylidene)-1-methyl-2-(styryl)-1H-imidazol-5(4H)-ones (1) (general procedure)
(Z)-4-(4-(diethylamino)-2-(difluoroboryl)benzylidene)-1-methyl-2-((E)-2-(4-methoxyphenyl)vinyl)-1H-imidazol-5(4H)-one (1a)
(Z)-4-(4-(diethylamino)-2-(difluoroboryl)benzylidene)-1-methyl-2-((E)-2-(thiophen-2-yl)vinyl)-1H-imidazol-5(4H)-one (1b)
(Z)-4-(4-(diethylamino)-2-(difluoroboryl)benzylidene)-2-((4-(4-methoxyphenyl)buta-1,3-dien-1-yl)-1-methyl-1H-imidazol-5(4H)-one (1c)
(Z)-4-(4-(diethylamino)-2-(difluoroboryl)benzylidene)-1-methyl-2-((E)-2-(pyridin-4-yl)vinyl)-1H-imidazol-5(4H)-one (1d)
3.2.2. (Z)-4-(2-(Difluoroboryl)-benzylidene)-1-methyl-2-aminoethenyl-1H-imidazol-5(4H)-ones (2) (general procedure)
(Z)-4-(4-(diethylamino)-2-(difluoroboryl)benzylidene)-2-((E)-2-(dimethylamino)vinyl)-1-methyl-1H-imidazol-5(4H)-one (2a)
(Z)-4-(2-(difluoroboryl)-4-(diethylamino)-benzylidene)-1-methyl-2-((E)-2-piperidin-1-ylethenyl)-1H-imidazol-5(4H)-one (2b)
(Z)-4-(2-(difluoroboryl)-4-(diethylamino)-benzylidene)-1-methyl-2-((E)-2-pyrrolidin-1-ylethenyl)-1H-imidazol-5(4H)-one (2c)
(Z)-4-(4-(diethylamino)-2-(difluoroboryl)benzylidene)-1-methyl-2-((E)-2-morpholinovinyl)-1H-imidazol-5(4H)-one (2d)
3.2.3. (Z)-4-(4-(Diethylamino)-2-(difluoroboryl)-benzylidene)-1H-imidazo [1,2-a]pyridin-5(4H)-one (3)
3.2.4. 7-(Diethylamino)-3H-phenoxazin-3-one (4)
3.2.5. (Z)-2-acetyl-4-(4-(diethylamino)benzylidene)-1-methyl-1H-imidazol-5(4H)-one (5)
3.2.6. (Z)-4-(4-(diethylamino)-2-hydroxybenzylidene)-1,2-dimethyl-1H-imidazol-5(4H)-one (6)
3.2.7. (Z)-2-(4-(diethylamino)benzylidene)-6,7-dihydroimidazo[1,2-a] pyrazine-3,8(2H,5H)-dione (7)
3.3. Plasmids
3.4. Chromophore Titration
3.5. Analysis of Fluorescence Titration Data
3.6. Cell Culture
3.7. Fluorescence Microscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef] [PubMed]
- Szent-Gyorgyi, C.; Schmidt, B.F.; Schmidt, B.A.; Creeger, Y.; Fisher, G.W.; Zakel, K.L.; Adler, S.; Fitzpatrick, J.A.J.; Woolford, C.A.; Yan, Q.; et al. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat. Biotechnol. 2008, 26, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Plamont, M.-A.; Billon-Denis, E.; Maurin, S.; Gauron, C.; Pimenta, F.M.; Specht, C.G.; Shi, J.; Quérard, J.; Pan, B.; Rossignol, J.; et al. Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Bozhanova, N.G.; Baranov, M.S.; Klementieva, N.V.; Sarkisyan, K.S.; Gavrikov, A.S.; Yampolsky, I.V.; Zagaynova, E.V.; Lukyanov, S.A.; Lukyanov, K.A.; Mishin, A.S. Protein labeling for live cell fluorescence microscopy with a highly photostable renewable signal. Chem. Sci. 2017, 8, 7138–7142. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Vorobieva, A.A.; Sheffler, W.; Doyle, L.A.; Park, H.; Bick, M.J.; Mao, B.; Foight, G.W.; Lee, M.Y.; Gagnon, L.A.; et al. De novo design of a fluorescence-activating β-barrel. Nature 2018, 561, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Raue, R. Methine Dyes and Pigments. In Ullmann’s Encyclopedia of Industrial Chemistry; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2000; Volume 18, p. 77. ISBN 9783527306732. [Google Scholar]
- Paul, W. Xanthene Dyes. In Kirk-Othmer Encyclopedia of Chemical Technology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2000; Volume 4, p. 558. ISBN 9780471238966. [Google Scholar]
- Loudet, A.; Burgess, K. BODIPY dyes and their derivatives: Syntheses and spectroscopic properties. Chem. Rev. 2007, 107, 4891–4932. [Google Scholar] [CrossRef] [PubMed]
- Christie, R.M. Fluorescent dyes. Rev. Prog. Color. Relat. Top. 2008, 23, 1–18. [Google Scholar] [CrossRef]
- Walker, C.L.; Lukyanov, K.A.; Yampolsky, I.V.; Mishin, A.S.; Bommarius, A.S.; Duraj-Thatte, A.M.; Azizi, B.; Tolbert, L.M.; Solntsev, K.M. Fluorescence imaging using synthetic GFP chromophores. Curr. Opin. Chem. Biol. 2015, 27, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Feng, G.; Luo, C.; Yi, H.; Yuan, L.; Lin, B.; Luo, X.; Hu, X.; Wang, H.; Lei, C.; Nie, Z.; et al. DNA mimics of red fluorescent proteins (RFP) based on G-quadruplex-confined synthetic RFP chromophores. Nucleic Acids Res. 2017, 45, 10380–10392. [Google Scholar] [CrossRef] [PubMed]
- Baranov, M.S.; Solntsev, K.M.; Baleeva, N.S.; Mishin, A.S.; Lukyanov, S.A.; Lukyanov, K.A.; Yampolsky, I.V. Red-Shifted Fluorescent Aminated Derivatives of a Conformationally Locked GFP Chromophore. Chem.-A Eur. J. 2014, 20, 13234–13241. [Google Scholar] [CrossRef] [PubMed]
- Baleeva, N.S.; Zaitseva, S.O.; Gorbachev, D.A.; Smirnov, A.Y.; Zagudaylova, M.B.; Baranov, M.S. The Role of N -Substituents in Radiationless Deactivation of Aminated Derivatives of a Locked GFP Chromophore. Eur. J. Org. Chem. 2017, 2017, 5219–5224. [Google Scholar] [CrossRef]
- Grabowski, Z.R.; Rotkiewicz, K.; Rettig, W. Structural changes accompanying intramolecular electron transfer: Focus on twisted intramolecular charge-transfer states and structures. Chem. Rev. 2003, 103, 3899–4032. [Google Scholar] [CrossRef] [PubMed]
- Subach, F.V.; Piatkevich, K.D.; Verkhusha, V.V. Directed molecular evolution to design advanced red fluorescent proteins. Nat. Methods 2011, 8, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Filonov, G.S.; Kim, H.; Hirsch, M.; Li, X.; Moon, J.D.; Jaffrey, S.R. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex. Nat. Chem. Biol. 2017, 13, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Baleeva, N.S.; Myannik, K.A.; Yampolsky, I.V.; Baranov, M.S. Bioinspired Fluorescent Dyes Based on a Conformationally Locked Chromophore of the Fluorescent Protein Kaede. Eur. J. Org. Chem. 2015, 2015, 5716–5721. [Google Scholar] [CrossRef]
- Otsuki, S.; Taguchi, T. Solvent-dependent photophysical properties of aminophenoxazone dyes as optical probes. J. Photochem. Photobiol. A Chem. 1997, 104, 189–195. [Google Scholar] [CrossRef]
- Greenspan, P.; Mayer, E.P.; Fowler, S.D. Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 1985, 100, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Baleeva, N.S.; Levina, E.A.; Baranov, M.S. Synthesis of 2-arylidene-6,7-dihydroimidazo[1,2-a]pyrazine-3,8(2H,5H)-diones by oxidation of 4-arylidene-2-methyl-1H-imidazol-5(4H)-ones with selenium dioxide. Chem. Heterocycl. Compd. 2017, 53, 930–933. [Google Scholar] [CrossRef]
- Yampolsky, I.V.; Remington, S.J.; Martynov, V.I.; Potapov, V.K.; Lukyanov, S.; Lukyanov, K.A. Synthesis and properties of the chromophore of the asFP595 chromoprotein from Anemonia sulcata. Biochemistry 2005, 44, 5788–5793. [Google Scholar] [CrossRef] [PubMed]
- Baldridge, A.; Feng, S.; Chang, Y.-T.; Tolbert, L.M. Recapture of GFP chromophore fluorescence in a protein host. ACS Comb. Sci. 2011, 13, 214–217. [Google Scholar] [CrossRef] [PubMed]
- Bindels, D.S.; Haarbosch, L.; van Weeren, L.; Postma, M.; Wiese, K.E.; Mastop, M.; Aumonier, S.; Gotthard, G.; Royant, A.; Hink, M.A.; et al. mScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 2017, 14, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Würth, C.; Grabolle, M.; Pauli, J.; Spieles, M.; Resch-Genger, U. Relative and absolute determination of fluorescence quantum yields of transparent samples. Nat. Protoc. 2013, 8, 1535–1550. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Zhang, Y.; Li, Y. Copper-catalyzed coupling reaction of C-OMe bonds adjacent to a nitrogen atom with terminal alkynes. J. Org. Chem. 2010, 75, 4554–4561. [Google Scholar] [CrossRef] [PubMed]
- Ovesný, M.; Křížek, P.; Borkovec, J.; Svindrych, Z.; Hagen, G.M. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 2014, 30, 2389–2390. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Plamont, M.-A.; Sladitschek, H.L.; Rodrigues, V.; Aujard, I.; Neveu, P.; Le Saux, T.; Jullien, L.; Gautier, A. Dynamic multicolor protein labeling in living cells. Chem. Sci. 2017, 8, 5598–5605. [Google Scholar] [CrossRef] [PubMed]







| Fluorescent Tag | λabs, nm | λem, nm | EC | Kd, µM | FQY |
|---|---|---|---|---|---|
| DiB1:3 | 605 | 625 | 55 | 0.09 | 0.29 |
| DiB2:3 | 600 | 625 | 55 | 0.54 | - |
| DiB3:3 | 622 | 637 | 55 | 2.39 | - |
| Blc-L141Q:3 | 607 | 630 | 55 | 0.12 | - |
| Blc-V74F:3 | 605 | 634 | 55 | 0.48 | - |
| DiB1:M739 [4] | 513 | 542 | 45.8 | 0.1 | 0.32 |
| DiB2:M739 [4] | 510 | 539 | 51 | 4 | 0.32 |
| DiB3:M739 [4] | 546 | 565 | 43 | 9 | 0.15 |
| mCherry, data from Ref. [23] | 586 | 610 | 88 | N/A | 0.23 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bozhanova, N.G.; Baranov, M.S.; Baleeva, N.S.; Gavrikov, A.S.; Mishin, A.S. Red-Shifted Aminated Derivatives of GFP Chromophore for Live-Cell Protein Labeling with Lipocalins. Int. J. Mol. Sci. 2018, 19, 3778. https://doi.org/10.3390/ijms19123778
Bozhanova NG, Baranov MS, Baleeva NS, Gavrikov AS, Mishin AS. Red-Shifted Aminated Derivatives of GFP Chromophore for Live-Cell Protein Labeling with Lipocalins. International Journal of Molecular Sciences. 2018; 19(12):3778. https://doi.org/10.3390/ijms19123778
Chicago/Turabian StyleBozhanova, Nina G., Mikhail S. Baranov, Nadezhda S. Baleeva, Alexey S. Gavrikov, and Alexander S. Mishin. 2018. "Red-Shifted Aminated Derivatives of GFP Chromophore for Live-Cell Protein Labeling with Lipocalins" International Journal of Molecular Sciences 19, no. 12: 3778. https://doi.org/10.3390/ijms19123778
APA StyleBozhanova, N. G., Baranov, M. S., Baleeva, N. S., Gavrikov, A. S., & Mishin, A. S. (2018). Red-Shifted Aminated Derivatives of GFP Chromophore for Live-Cell Protein Labeling with Lipocalins. International Journal of Molecular Sciences, 19(12), 3778. https://doi.org/10.3390/ijms19123778