Use of Yarrowia lipolytica Lipase Immobilized in Cell Debris for the Production of Lipolyzed Milk Fat (LMF)
Abstract
:1. Introduction
2. Results and Discussion
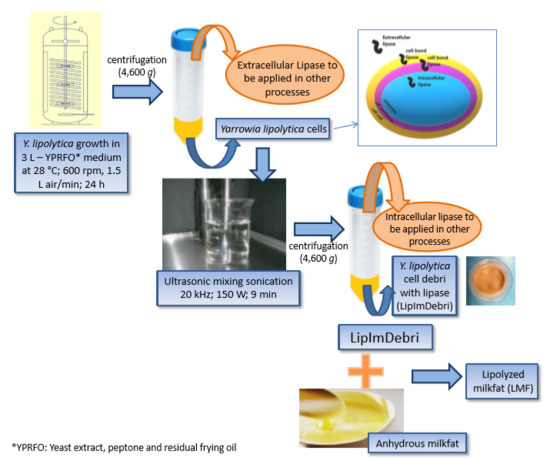
2.1. Production of Lipase from Y. lipolytica Immobilized in Cell Debris Induced by Residual Frying Oil
2.2. Characterization of Lipase Immobilized in Cell Debris
2.3. Anhydrous Milk Fat Production
2.4. Lipolysis of Milk Fat
3. Materials and Methods
3.1. Materials
3.2. Strain, Media, and Inoculum Preparation
3.3. Production of Lipases (Fermentation Process)
3.4. Preparation of Lipase Immobilized in Cell Debris
3.5. Lipase Activity
3.6. Effect of Temperature and pH on Lipase Immobilized in Cell Debris
3.7. Thermal Stability and Stability in Different Solvents of Lipase Immobilized in Cell Debris
3.8. Repeated Use of Lipase Immobilized in Cell Debris
3.9. Preparation of Anhydrous Milk Fat
3.10. Milk Fat Hydrolysis Reactions
3.11. Monitoring Lipid Composition during Milk Fat Hydrolysis Reactions
3.11.1. High Performance Liquid Chromatography (HPLC)
3.11.2. Thin Layer Chromatography (TLC)
3.11.3. Analysis of Fatty Acid Composition by Gas Chromatography (GC)
3.12. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HPLC | High-performance liquid chromatography |
| GC | Gas chromatography |
| TLC | Thin layer chromatography |
| LMF | Lipolyzed milk fat |
| LipImDebri | Lipase immobilized in cell debris |
References
- Cao, Z.M.; Li, J.; Wang, W.; Ge, T.; Yue, R.; Li, V.L.; Colvin, W.; Yu, W. Food related applications of magnetic iron oxide nanoparticles: Enzyme immobilization, protein purification, and food analysis. Trends Food Sci. Technol. 2012, 27, 47–56. [Google Scholar] [CrossRef]
- Kumar, A.; Venkatesu, P. Overview of the stability of α-chymotrypsin in different solvent media. Chem. Rev. 2002, 112, 4283–4307. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Thakur, S.; Bhatt, A.K. Microbial lipases: Industrial applications and properties (a review). Int. Res. J. Biol. Sci. 2012, 1, 88–92. [Google Scholar] [CrossRef]
- de Oliveira, U.M.; de Matos, L.J.L.; de Souza, M.C.M.; Pinheiro, B.B.; dos Santos, J.C.; Gonçalves, L.R. Effect of the presence of surfactants and immobilization conditions on catalysts’ properties of Rhizomucor miehei lipase onto chitosan. Appl. Biochem. Biotechnol. 2018, 184, 1263–1285. [Google Scholar] [CrossRef] [PubMed]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- DiCosimo, R.; McAuliffe, J.; Poulose, A.J.; Bohlmann, G. Industrial use of immobilized enzymes. Chem. Soc. Rev. 2013, 42, 6437. [Google Scholar] [CrossRef] [PubMed]
- Galvão, W.S.; Pinheiro, B.B.; Golçalves, L.R.B.; de Mattos, M.C.; Fonseca, T.S.; Regis, T.; Zampieri, D.; dos Santos, J.C.S.; Costa, L.S.; Correa, M.A.; et al. Novel nanohybrid biocatalyst: Application in the kinetic resolution of secondary alcohols. J. Mater. Sci. 2018, 53, 14121–14137. [Google Scholar] [CrossRef]
- Zdarta, J.; Meyer, A.S.; Jesionowski, T.; Pinelo, M. Developments in support materials for immobilization of oxidoreductases: A comprehensive review. Adv. Colloid Interface Sci. 2018, 258, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Cantone, S.; Ferrario, V.; Corici, L.; Ebert, C.; Fattor, D.; Spizzo, P.; Gardossi, L. Efficient immobilisation of industrial biocatalysts: Criteria and constraints for the selection of organic polymeric carriers and immobilisation methods. Chem. Soc. Rev. 2013, 42, 6262–6276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, H.; Hama, S.; Tamalampudi, S.; Noda, H. Whole-cell biocatalysts for biodiesel fuel production. Trends Biotechnol. 2008, 26, 668–673. [Google Scholar] [CrossRef] [PubMed]
- Adamczak, M.; Bednarski, W. Enhanced activity of intracellular lipases from Rhizomucor miehei and Yarrowia lipolytica by immobilization on biomass support particles. Process Biochem. 2004, 39, 1347–1361. [Google Scholar] [CrossRef]
- Barth, G.; Gaillardin, C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 1977, 19, 219–237. [Google Scholar] [CrossRef]
- Fickers, P.; Nicaud, J.M.; Gaillardin, C.; Destain, J.; Thonart, P. Carbon and nitrogen sources modulate lipase production in the yeast Yarrowia lipolytica. J. Appl. Microbiol. 2004, 96, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Nunes, P.; Martins, A.B.; Santa Brigida, A.I.; Miguez Da Rocha Leao, M.H.; Amaral, P. Intracellular lipase production by Yarrowia lipolytica using different carbon sources. Chem. Eng. Trans. 2014, 38, 421–426. [Google Scholar] [CrossRef]
- Ota, Y.; Gomi, K.; Kato, S.; Sugiura, T.; Minoda, Y. Purification and some properties of cell-bond lipase from Saccharomycopsis lipolytica. Agric. Biol. Chem. 1982, 46, 2885–2893. [Google Scholar] [CrossRef]
- Kilcawley, K.N. The enzyme effect. Dairy Ind. Int. 2001, 66, 26–28. [Google Scholar]
- Regado, M.A.; Cristóvão, B.M.; Moutinho, C.G.; Balcão, V.M.; Aires-Barros, R.; Ferreira, J.P.M.; Malcata, F.X. Lipolysis of milkfats for flavour. Int. J. Food Sci. Technol. 2007, 42, 961–968. [Google Scholar] [CrossRef]
- Pereira, A.S.; Fraga, J.L.; Diniz, M.M.; Fontes-Sant’Ana, G.F.; Amaral, P.F.F. High catalytic activity of lipase from Yarrowia lipolytica immobilized by microencapsulation. Int. J. Mol. Sci. under review.
- Choe, E.; Min, D.B. Chemistry of deep-fat frying oils. J. Food Sci. 2007, 72, R77–R86. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Qin, S.; Tan, T. Purification and characterization of the extracellular lipase Lip2 from Yarrowia lipolytica. Process Biochem. 2007, 42, 384–391. [Google Scholar] [CrossRef]
- Kumari, A.; Gupta, R. Purification and Biochemical Characterization of a Novel Magnesium Dependent Lipase from Trichosporon asahii MSR 54 and its Application in Biodiesel Production. Asian J. Biotechnol. 2012, 4, 70–82. [Google Scholar] [CrossRef]
- Brígida, A.I.; Amaral, P.F.F.; Gonçalves, L.R.; Rocha-Leão, M.H.M.; Coelho, M.A.Z. Yarrowia lipolytica IMUFRJ 50682: Lipase production in a multiphase bioreactor. Curr. Biochem. Eng. 2014, 1, 65–74. [Google Scholar] [CrossRef]
- Kumari, A.; Verma, V.V.; Gupta, R. Comparative biochemical characterization and in silico analysis of novel lipases Lip11 and Lip12 with Lip2 from Yarrowia lipolytica. World J. Microbiol. Biotechnol. 2012, 28, 3103–3111. [Google Scholar] [CrossRef] [PubMed]
- Castro-Ochoa, L.D.; Rodríguez-Gómez, C.; Valerio-Alfaro, G.; Ros, R.O. Screening, purification and characterization of the thermoalkalophilic lipase produced by Bacillus thermoleovorans CCR11. Enzym. Microb. Technol. 2005, 37, 648–654. [Google Scholar] [CrossRef]
- Brígida, A.I.; Pinheiro, Á.D.; Ferreira, A.L.; Pinto, G.A.; Gonçalves, L.R. Immobilization of Candida antarctica lipase B by covalent attachment to green coconut fiber. Appl. Biochem. Biotecnol. 2007, 146, 173–187. [Google Scholar] [CrossRef] [PubMed]
- Stolarzewicz, I.A.; Zaborniak, P.; Fabiszewska, A.U.; Białecka-Florjańczyka, E. Study on the Properties of Immobilized Biocatalysts with Lipase Activity Produced by Yarrowia lipolytica in Batch Culture. Chem. Biochem. Eng. Q. 2017, 31, 251–259. [Google Scholar] [CrossRef]
- Fickers, P.; Fudalej, F.; Nicaud, J.; Destain, J.E.; Thonart, P. Selection of new over-producing derivates for the improvement of extracellular lipase production by the nonconventional yeast Yarrowia lipolytica. J. Appl. Microb. 2005, 115, 379–386. [Google Scholar] [CrossRef]
- Brígida, A.I.; Pinheiro, Á.D.; Ferreira, A.L.; Gonçalves, L.R. Immobilization of Candida antarctica lipase B by adsorption to green coconut fiber. Biotech. Fuels Chem. 2007. [Google Scholar] [CrossRef]
- Kakugawa, K.; Shobayashi, M.; Suzuki, O.; Miyakawa, T. Purification and characterization of a lipase from the glycolipid-producing yeast Kurtzmanomyces sp. I-11. Biosci. Biotechnol. Biochem. 2002, 66, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Balan, A.; Ibrahim, D.; Rahim, R.A. Organic-solvent and surfactant tolerant thermostable lipase, isolated from a thermophilic bacterium, Geobacillus thermodenitrificans IBRL-NRA. Adv. Stud. Biol. 2013, 5, 389–401. [Google Scholar] [CrossRef]
- Syal, P.; Gupta, R. Cloning, expression, and biochemical characterization of an enantioselective lipase, YLIP9, from Yarrowia lipolytica MSR80. Appl. Biochem. Biotechnol. 2015, 176, 110–124. [Google Scholar] [CrossRef] [PubMed]
- FAO; WHO. Standard for Butter. Codex Stan 279, Revised 1999, Amended 2003 and 2006; Food Agriculture Organization of the United Nations: Rome, Italy, 2007. [Google Scholar]
- Månsson, H.L. Fatty acids in bovine milk fat. Food Nutr. Res. 2008, 52. [Google Scholar] [CrossRef]
- Torres, A.G.; Trugo, N.M.; Trugo, L.C. Mathematical method for the prediction of retention times of fatty acid methyl esters in temperature-programmed capillary gas chromatography. J. Agric. Food Chem. 2002, 50, 4156–4163. [Google Scholar] [CrossRef] [PubMed]
- Lindmark-Månsson, H.; Fondén, R.; Pettersson, H.E. Composition of Swedish dairy milk. Int. Dairy J. 2003, 13, 409–425. [Google Scholar] [CrossRef]
- Akil, E.; Carvalho, T.; Bárea, B.; Finotelli, P.; Lecomte, J.; Torres, A.G.; Amaral, P.; Villeneuve, P. Accessing regio-and typo-selectivity of Yarrowia lipolytica lipase in its free form and immobilized onto magnetic nanoparticles. Biochem. Eng. J. 2016, 109, 101–111. [Google Scholar] [CrossRef]
- Chen, J.Y.P.; Yang, B.A. Enhancement of release of short-chain fatty acids from milk fat with immobilized microbial lipase. J. Food Sci. 1992, 57, 781–782. [Google Scholar] [CrossRef]
- Lubary, M.; Hofland, G.W.; Ter Horst, J.H. The potential of milk fat for the synthesis of valuable derivatives. Eur. Food Res. Technol. 2011, 232, 1–8. [Google Scholar] [CrossRef]
- Haegler, A.N.; Mendonça Haegler, L.C. Yeast from marine and estuarine waters with different levels of pollution in the State of Rio de Janeiro, Brazil. Appl. Environ. Microbiol. 1981, 41, 173–178. [Google Scholar]
- Kalo, P.; Huotari, H.; Antila, M. Pseudomonas fluorescens lipase-catalysed interesterification of butterfat in the absence of a solvent. Milchwiss. Milk Sci. Int. 1990, 45, 281–285. [Google Scholar]
- Balcão, V.M.; Malcata, F.X. Lipase-catalyzed modification of butterfat via acidolysis with oleic acid. J. Mol. Catal. B Enzym. 1997, 3, 161–169. [Google Scholar] [CrossRef] [Green Version]
- Tan, B.; Brzuskiewicz, L. Separation of tocopherol and tocotrienol isomers using normal- and reverse-phase liquid chromatography. Anal. Biochem. 1989, 180, 368–373. [Google Scholar] [CrossRef]
- Lepage, G.; Roy, C.C. Direct transterification of all classes of lipids in a one-step reaction. J. Lipid Res. 1986, 27, 114–120. [Google Scholar] [PubMed]
- Mhøs, S.; Grahl-Nielsen, O. Prediction of gas chromatographic retention of polyunsaturated fatty acid methyl esters. J. Chromatogr. A 2006, 11, 171–180. [Google Scholar] [CrossRef]





| Fatty Acids | Contents (%, mol/mol) | Fatty Acids | Contents (%, mol/mol) |
|---|---|---|---|
| Saturated | Monounsaturated | ||
| 4:0 | 0.63 ± 0.45 | 14:1 | 0.66 ± 0.02 |
| 6:0 | 0.48 ± 0.23 | 15:1 | 0.28 ± 0.01 |
| 8:0 | 1.35 ± 0.33 | 16:1 | 1.30 ± 0.06 |
| 10:0 | 0.11 ± 0.03 | 17:1 | 0.84 ± 0.01 |
| 11:0 | 0.16 ± 0.06 | 18:1n-9 | 28.5 ± 2.03 |
| 12:0 | 1.88 ± 0.17 | Polyunsaturated | |
| 13:0 | 0.06 ± 0.00 | 18:2n-6 | 2.17 ± 0.03 |
| 14:0 | 9.13 ± 0.26 | 18:3n-6 | 0.43 ± 0.02 |
| 15:0 | 0.94 ± 0.03 | 18:3n-3 | 0.44 ± 0.02 |
| 16:0 | 31.9 ± 0.79 | 20:1n-9 | 0.75 ± 0.02 |
| 17:0 | 0.50 ± 0.01 | 20:3n-6 | 0.03 ± 0.00 |
| 18:0 | 16.0 ± 0.15 | 20:3n-3 | 0.10 ± 0.01 |
| 20:0 | 0.24 ± 0.01 | Conjugated | |
| 21:0 | 0.06 ± 0.01 | CLA * | 0.35 ± 0.02 |
| 22:0 | 0.12 ± 0.02 | ||
| Branched-chain | |||
| i14:0 | 0.08 ± 0.00 | ||
| i15:0 | 0.26 ± 0.01 | ||
| i18:0 | 0.29 ± 0.00 |
| Time of Lipolysis (h) | Lipid Classes | |
|---|---|---|
| 500 mg LipImDebri | 750 mg LipImDebri | |
| Free Fatty Acid (FFA) + Monoacylglycerol (MAG) Fraction (%) | ||
| 0 | 0.00 a,C | 0,00 a,C |
| 1.5 | 0.98 ± 0.29 a,C | 5.48 ± 2.53 a,B,C |
| 3.0 | 11.02 ± 1.00 a,B,C | 19.54 ±0.94 b,A,B,C |
| 4.5 | 17.86 ± 5.84 a,A,B | 11.79 ± 4.58 a,A,B |
| 6.0 | 28.23 ± 3.45 a,A | 14.30 ± 5.34 a,A,B |
| Diacylglycerol (DAG) Fraction (%) | ||
| 0 | 1.11 ± 0.02 a,A | 1.11 ± 0.02 a,A |
| 1.5 | 0.31 ± 0.34 a,A | 0.57 ± 0.15 a,A |
| 3.0 | 1.57 ± 0.94 a,A | 1.27 ± 0.02 a,A |
| 4.5 | 1.18 ± 0.52 a,A | 0.62 ± 0.20 a,A |
| 6.0 | 0.56 ± 0.05 a,A | 1.41 ± 0.48 a,A |
| Triacylglycerol (TAG) Fraction (%) | ||
| 0 | 98.96 ± 0.11 a,A | 98.96 ± 0.11 a,A |
| 1.5 | 98.71 ± 0.63 a,A | 93.95 ± 2.68 a,A,B |
| 3.0 | 87.41 ± 1.95 a,B | 79.19 ± 0.96 b,C |
| 4.5 | 80.96 ± 5.30 a,B | 87.60 ±4.37 a,AB,C |
| 6.0 | 72.71 ± 5.52 a,C | 84.29 ± 4.86 a,B,C |
| Fatty Acid | LipImDebri (mg), Lipolysis Time (h) | ||
|---|---|---|---|
| 500, 4.5 | 500, 6 | 750, 3 | |
| Saturated | |||
| 4:0 | 0.74 ± 0.29 a,D | 0.51 ± 0.13 a,D | 0.76 ± 0.53 a,E |
| 6:0 | 0.45 ± 0.09 a,D | 0.27 ± 0.02 a,D | 0.65 ± 0.49 a,E |
| 8:0 | 4.40 ± 1.98 a,C,D | 6.79 ± 5.30 a,D | 3.63 ± 3.46 a,E,F |
| 10:0 | 0.74 ± 0.02 a,C,D | 0.56 ± 0.15 a,D | 1.44 ± 1.16 a,E |
| 12:0 | 1.99 ± 0.83 a,C,D | 2.42 ± 1.57 a,D | 2.74 ± 0.16 a,D,E |
| 14:0 | 8.17 ± 0.03 a,C | 6.03 ± 0.99 a,D | 8.14 ± 2.20 a,D |
| 15:0 | 0.90 ± 0.05 a,C,D | 0.79 ± 0.12 a,D | 0.81 ± 0.01 a,E |
| 16:0 | 28.80 ± 3.00 a,A | 24.97 ± 6.94 a,B | 24.67 ± 0.71 a,B |
| 18:0 | 13.94 ± 1.60 a,B | 15.36 ± 3.25 a,C | 12.79 ± 1.60 a,C |
| Branched-Chain | |||
| i15:0 | 0.22 ± 0.01 a,D | 0.22 ± 0.01 a,D | 0.21 ± 0.03 a,E |
| a15:0 | 0.34 ± 0.03 a,C,D | 0.29 ± 0.05 a,D | 0.31 ± 0.06 a,E |
| Monounsaturated | |||
| 14:1 | 0.56 ± 0.09 a,C,D | 0.40 ± 0.06 a,D | 0.69 ± 0.41 a,E |
| 16:1 | 1.19 ± 0.27 a,C,D | 1.07 ± 0.08 a,D | 1.32 ± 0.39 a,E |
| 17:1 | 0.69 ± 0.13 a,C,D | 0.64 ± 0.02 a,D | 0.56 ± 0.08 a,E |
| 18:1n-9 | 31.27 ± 5.16 a,A | 35.18 ± 1.83 a,A | 35.31 ± 1.05 a,A |
| Polyunsaturated | |||
| 18:2n-6 | 4.29 ± 1.24 a,C,D | 3.84 ± 0.54 a,D | 4.82 ± 0.47 a,D,E |
| 20:1n-9 | 1.33 ± 0.32 a,C,D | 0.67 ± 0.07 a,D | 0.85 ± 0.18 a,E |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fraga, J.L.; Penha, A.C.B.; Da S. Pereira, A.; Silva, K.A.; Akil, E.; Torres, A.G.; Amaral, P.F.F. Use of Yarrowia lipolytica Lipase Immobilized in Cell Debris for the Production of Lipolyzed Milk Fat (LMF). Int. J. Mol. Sci. 2018, 19, 3413. https://doi.org/10.3390/ijms19113413
Fraga JL, Penha ACB, Da S. Pereira A, Silva KA, Akil E, Torres AG, Amaral PFF. Use of Yarrowia lipolytica Lipase Immobilized in Cell Debris for the Production of Lipolyzed Milk Fat (LMF). International Journal of Molecular Sciences. 2018; 19(11):3413. https://doi.org/10.3390/ijms19113413
Chicago/Turabian StyleFraga, Jully L., Adrian C. B. Penha, Adejanildo Da S. Pereira, Kelly A. Silva, Emília Akil, Alexandre G. Torres, and Priscilla F. F. Amaral. 2018. "Use of Yarrowia lipolytica Lipase Immobilized in Cell Debris for the Production of Lipolyzed Milk Fat (LMF)" International Journal of Molecular Sciences 19, no. 11: 3413. https://doi.org/10.3390/ijms19113413