Inhibitory Effect of Methotrexate on Rheumatoid Arthritis Inflammation and Comprehensive Metabolomics Analysis Using Ultra-Performance Liquid Chromatography-Quadrupole Time of Flight-Mass Spectrometry (UPLC-Q/TOF-MS)
Abstract
:1. Introduction
2. Results
2.1. Inhibitory Effect of Methotrexate on Murine CIA Paw Swelling
2.2. Downregulation of Methotrexate on NF-κB/NLRP3 Pathways in Murine Model
2.3. Methotrexate Suppressed the Inflammatory Proliferation of Macrophages
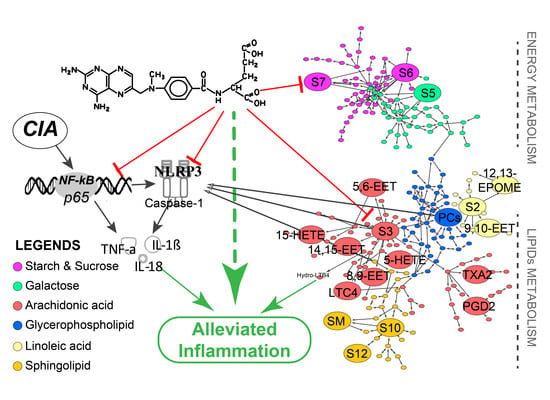
2.4. Methotrexate Inhibited NF-κB and NLRP3/Caspase-1 Pathways in Macrophages
2.5. Multivariate Analysis and Differentiated Metabolic Patterns
2.6. Global Profiles of Distinct Metabolites and Perturbed Metabolic Pathways
2.7. Potential Docking Mode of Methotrexate within Caspase-1 Protein
3. Discussion
4. Materials and Methods
4.1. Experimental Animal and Ethical Approval
4.2. RA induction and Drug Administration
4.3. Assessment on RA Swelling
4.4. Sacrifice and Sample Collection
4.5. Cell Viability Assay with Real Time Cellular Analysis
4.6. Cell Culture and Induction of Inflammation
4.7. qPCR Assay
4.8. Western Blotting and ELISA Assay
4.9. Metabolomic Procedure and Data Processing
4.10. Molecular Docking of Methotrexate within Caspase-1
4.11. Statistic and Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ANOVA | Analysis of Variance |
| ARG | Arginine |
| ATCC | American Type Culture Collection |
| AUC | Area Under Curve |
| CIA | Collagen induced arthritic |
| DCS | Diclofenac sodium |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| ECL | Chemiluminescence |
| EET | Epoxyoctadecenoic acid |
| EpOME | Epoxide of linoleic acid |
| ESI | Electrospray Ionization |
| FBS | Fetal Bovine Serum |
| HETE | Hydroxyeicosatetraenoic acid |
| HMDB | Human Metabolome Database |
| HRP | Horseradish Peroxidase |
| IOD | Integrated optical density |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LPS | Lipopolysaccharide |
| LTC4 | Leukotrienes C4 |
| MS | Mass Spectrometry |
| MS/MS | tandem mass spectrometry |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NLRP3 | Nucleotide binding domain and leucine-rich repeat pyrin 3 domain |
| NSAIDs | Nonsteroidal anti-inflammatory drugs |
| OPLS-DA | Orthogonal Projections to Latent Structures Discriminant Analysis |
| PC | Principle Component |
| PCA | Principle Component Analysis |
| Pc | Phosphatidylcholine |
| PDB | Protein Data Bank |
| PDG2 | Prostaglandin D2 |
| PPM | Mass error ppm |
| PVDF | Polyvinylidene Difluoride |
| Q/TOF | Quadrupole time of flight mass spectrometry |
| QC | Quality Control |
| RA | Rheumatoid arthritis |
| ROC | Receiver Operating Characteristic Curve |
| RSD | Relative Standard Deviation |
| RTCA | Real Time Cellular Analysis |
| sPBS | Stimulated with LPS followed by PBS |
| SM | Sm(d18:0/18:1) |
| TXA2 | Thromboxane A2 |
| UPLC | Ultra-Performance Liquid Chromatography |
| VIP | Variable Importance for the Projection |
References
- Taylor, P.C.; Keystone, E.C.; van der Heijde, D.; Weinblatt, M.E.; Del Carmen Morales, L.; Reyes Gonzaga, J.; Yakushin, S.; Ishii, T.; Emoto, K.; Beattie, S.; et al. Baricitinib versus Placebo or Adalimumab in Rheumatoid Arthritis. N. Engl. J. Med. 2017, 376, 652–662. [Google Scholar] [CrossRef] [PubMed]
- Yousri, N.A.; Bayoumy, K.; Elhaq, W.G.; Mohney, R.P.; Emadi, S.A.; Hammoudeh, M.; Halabi, H.; Masri, B.; Badsha, H.; Uthman, I.; et al. Large Scale Metabolic Profiling identifies Novel Steroids linked to Rheumatoid Arthritis. Sci. Rep. 2017, 7, 9137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vande Walle, L.; Van Opdenbosch, N.; Jacques, P.; Fossoul, A.; Verheugen, E.; Vogel, P.; Beyaert, R.; Elewaut, D.; Kanneganti, T.D.; van Loo, G.; et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature 2014, 512, 69–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [PubMed]
- Engdahl, C.; Bondt, A.; Harre, U.; Raufer, J.; Pfeifle, R.; Camponeschi, A.; Wuhrer, M.; Seeling, M.; Martensson, I.L.; Nimmerjahn, F.; et al. Estrogen induces St6gal1 expression and increases IgG sialylation in mice and patients with rheumatoid arthritis: A potential explanation for the increased risk of rheumatoid arthritis in postmenopausal women. Arthritis Res. Ther. 2018, 20, 84. [Google Scholar] [CrossRef] [PubMed]
- Sohn, D.H.; Rhodes, C.; Onuma, K.; Zhao, X.; Sharpe, O.; Gazitt, T.; Shiao, R.; Fert-Bober, J.; Cheng, D.; Lahey, L.J.; et al. Local Joint inflammation and histone citrullination in a murine model of the transition from preclinical autoimmunity to inflammatory arthritis. Arthritis Rheumatol. 2015, 67, 2877–2887. [Google Scholar] [CrossRef] [PubMed]
- Van Loo, G.; Beyaert, R. Negative regulation of NF-kappaB and its involvement in rheumatoid arthritis. Arthritis Res. Ther. 2011, 13, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Cao, J.; Yu, C.; Yang, Q.; Zhang, Y.; Han, L. Caspase-1 mediated interleukin-18 activation in neutrophils promotes the activity of rheumatoid arthritis in a NLRP3 inflammasome independent manner. Joint Bone Spine 2016, 83, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Choulaki, C.; Papadaki, G.; Repa, A.; Kampouraki, E.; Kambas, K.; Ritis, K.; Bertsias, G.; Boumpas, D.T.; Sidiropoulos, P. Enhanced activity of NLRP3 inflammasome in peripheral blood cells of patients with active rheumatoid arthritis. Arthritis Res. Ther. 2015, 17, 257. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, J.Q.; Xu, J.D.; Zhu, H.; Kong, M.; Zhang, G.H.; Duan, S.M.; Li, X.Y.; Li, G.F.; Liu, L.F.; et al. UPLC/ESI-QTOF-MS-based metabolomics survey on the toxicity of triptolide and detoxication of licorice. Chin. J. Nat. Med. 2017, 15, 474–480. [Google Scholar] [CrossRef]
- Li, R.; Guo, L.X.; Li, Y.; Chang, W.Q.; Liu, J.Q.; Liu, L.F.; Xin, G.Z. Dose-response characteristics of Clematis triterpenoid saponins and clematichinenoside AR in rheumatoid arthritis rats by liquid chromatography/mass spectrometry-based serum and urine metabolomics. J. Pharm. Biomed. Anal. 2017, 136, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Che, N.; Xu, L.; Zhang, Q.; Wang, Q.; Tan, W.; Zhang, M. LC-MS-based serum metabolomics reveals a distinctive signature in patients with rheumatoid arthritis. Clin. Rheumatol. 2018, 37, 1493–1502. [Google Scholar] [CrossRef] [PubMed]
- Van Walsem, A.; Pandhi, S.; Nixon, R.M.; Guyot, P.; Karabis, A.; Moore, R.A. Relative benefit-risk comparing diclofenac to other traditional non-steroidal anti-inflammatory drugs and cyclooxygenase-2 inhibitors in patients with osteoarthritis or rheumatoid arthritis: A network meta-analysis. Arthritis Res. Ther. 2015, 17, 66. [Google Scholar] [CrossRef] [PubMed]
- Huffman, K.M.; Jessee, R.; Andonian, B.; Davis, B.N.; Narowski, R.; Huebner, J.L.; Kraus, V.B.; McCracken, J.; Gilmore, B.F.; Tune, K.N.; et al. Molecular alterations in skeletal muscle in rheumatoid arthritis are related to disease activity, physical inactivity, and disability. Arthritis Res. Ther. 2017, 19, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Tong, H.F.; Li, Y.; Qian, J.C.; Wang, J.X.; Wang, Z.; Ruan, J.C. Effect of the Polymorphism of Folylpolyglutamate Synthetase on Treatment of High-Dose Methotrexate in Pediatric Patients with Acute Lymphocytic Leukemia. Med. Sci. Monit. 2016, 22, 4967–4973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peres, R.S.; Liew, F.Y.; Talbot, J.; Carregaro, V.; Oliveira, R.D.; Almeida, S.L.; Franca, R.F.; Donate, P.B.; Pinto, L.G.; Ferreira, F.I.; et al. Low expression of CD39 on regulatory T cells as a biomarker for resistance to methotrexate therapy in rheumatoid arthritis. Proc. Natl. Acad. Sci. USA 2015, 112, 2509–2514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, B.; Zheng, H.; Wu, F.; Wu, J.; Liu, X.; Tang, C.; Lu, S.; Chen, Z.; Song, F.; Ruan, J.; et al. Metabolomics analysis of Danggui Sini decoction on treatment of collagen-induced arthritis in rats. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1061–1062, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Schett, G. Review: Immune cells and mediators of inflammatory arthritis. Autoimmunity 2008, 41, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Bauernfeind, F.G.; Horvath, G.; Stutz, A.; Alnemri, E.S.; MacDonald, K.; Speert, D.; Fernandes-Alnemri, T.; Wu, J.; Monks, B.G.; Fitzgerald, K.A.; et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 2009, 183, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Hayden, M.S. New regulators of NF-kappaB in inflammation. Nat. Rev. Immunol. 2008, 8, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Mathews, R.J.; Robinson, J.I.; Battellino, M.; Wong, C.; Taylor, J.C.; Eyre, S.; Churchman, S.M.; Wilson, A.G.; Isaacs, J.D.; Hyrich, K.; et al. Evidence of NLRP3-inflammasome activation in rheumatoid arthritis (RA); genetic variants within the NLRP3-inflammasome complex in relation to susceptibility to RA and response to anti-TNF treatment. Ann. Rheum. Dis. 2014, 73, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cai, F.; Zhang, Y.; Yang, A.; Liu, L. Celastrol, an NF-kappaB inhibitor, ameliorates hypercalciuria and articular cartilage lesions in a mouse model of secondary osteoporosis. J. Pharmacol. Sci. 2016, 130, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.X.; Liu, Y.; Zhou, W.; Li, H.W.; Yang, J.; Chen, Z.B. Shikonin inhibits TNF-alpha production through suppressing PKC-NF-kappaB-dependent decrease of IL-10 in rheumatoid arthritis-like cell model. J. Nat. Med. 2017, 71, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.X.; Xiong, W.; Zhao, G.D.; Peng, Y.; Zhong, Z.F.; Xu, L.; Duan, R.; Tsim, K.W.K.; Yu, H.; Wang, Y.T. Discrimination of three Siegesbeckiae Herba species using UPLC-QTOF/MS-based metabolomics approach. Food Chem. Toxicol. 2018, 119, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Gromovsky, A.D.; Brown, J.M.; Chung, S. Gamma-tocotrienol attenuates the aberrant lipid mediator production in NLRP3 inflammasome-stimulated macrophages. J. Nutr. Biochem. 2018, 58, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Camandola, S.; Leonarduzzi, G.; Musso, T.; Varesio, L.; Carini, R.; Scavazza, A.; Chiarpotto, E.; Baeuerle, P.A.; Poli, G. Nuclear factor kB is activated by arachidonic acid but not by eicosapentaenoic acid. Biochem. Biophys. Res. Commun. 1996, 229, 643–647. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Gris, D.; Lei, Y.; Jha, S.; Zhang, L.; Huang, M.T.; Brickey, W.J.; Ting, J.P. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 2011, 12, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Morisseau, C.; Hammock, B.D. Impact of soluble epoxide hydrolase and epoxyeicosanoids on human health. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 37–58. [Google Scholar] [CrossRef] [PubMed]
- Node, K.; Huo, Y.; Ruan, X.; Yang, B.; Spiecker, M.; Ley, K.; Zeldin, D.C.; Liao, J.K. Anti-inflammatory properties of cytochrome P450 epoxygenase-derived eicosanoids. Science 1999, 285, 1276–1279. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.C. Lipidomic profiling of bioactive lipids by mass spectrometry during microbial infections. Seminars Immunol. 2013, 25, 240–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishizaki, T.; Shigemori, K.; Nakai, T.; Miyabo, S.; Ozawa, T.; Chang, S.W.; Voelkel, N.F. Leukotoxin, 9,10-epoxy-12-octadecenoate causes edematous lung injury via activation of vascular nitric oxide synthase. Am. J. Physiol. 1995, 269, L65–L70. [Google Scholar] [CrossRef] [PubMed]
- De Lima-Salgado, T.M.; Alba-Loureiro, T.C.; do Nascimento, C.S.; Nunes, M.T.; Curi, R. Molecular mechanisms by which saturated fatty acids modulate TNF-alpha expression in mouse macrophage lineage. Cell Biochem. Biophys. 2011, 59, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.M.; Roche, H.M. Conjugated linoleic acid and inflammatory cell signalling. Prostaglandins Leukot. Essent. Fatty Acids 2010, 82, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Luczaj, W.; Gindzienska-Sieskiewicz, E.; Jarocka-Karpowicz, I.; Andrisic, L.; Sierakowski, S.; Zarkovic, N.; Waeg, G.; Skrzydlewska, E. The onset of lipid peroxidation in rheumatoid arthritis: Consequences and monitoring. Free Radic. Res. 2016, 50, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.M.; Segovia, S.A.; Zhang, X.D.; Gray, C.; Vickers, M.H. Maternal high-fat diet-induced programing of gut taste receptor and inflammatory gene expression in rat offspring is ameliorated by CLA supplementation. Physiol. Rep. 2015, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aggarwal, N.; Korenbaum, E.; Mahadeva, R.; Immenschuh, S.; Grau, V.; Dinarello, C.A.; Welte, T.; Janciauskiene, S. α-Linoleic acid enhances the capacity of alpha-1 antitrypsin to inhibit lipopolysaccharide-induced IL-1beta in human blood neutrophils. Mol. Med. 2016, 22. [Google Scholar] [CrossRef] [PubMed]
- Fijlstra, M.; Tissing, W.J.; Stellaard, F.; Verkade, H.J.; Rings, E.H. Reduced absorption of long-chain fatty acids during methotrexate-induced gastrointestinal mucositis in the rat. Clin. Nutr. 2013, 32, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Shen, J.; Pan, W.; Shen, S.; Das, U.N. Effects of polyunsaturated fatty acids on the growth of gastric cancer cells in vitro. Lipids Health Dis. 2013, 12, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adada, M.; Luberto, C.; Canals, D. Inhibitors of the sphingomyelin cycle: Sphingomyelin synthases and sphingomyelinases. Chem. Phys. Lipids 2016, 197, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Meshcheryakova, A.; Mechtcheriakova, D.; Pietschmann, P. Sphingosine 1-phosphate signaling in bone remodeling: Multifaceted roles and therapeutic potential. Expert Opin. Ther. Targets 2017, 21, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Luberto, C.; Yoo, D.S.; Suidan, H.S.; Bartoli, G.M.; Hannun, Y.A. Differential effects of sphingomyelin hydrolysis and resynthesis on the activation of NF-kappa B in normal and SV40-transformed human fibroblasts. J. Biol. Chem. 2000, 275, 14760–14766. [Google Scholar] [CrossRef] [PubMed]
- Maceyka, M.; Spiegel, S. Sphingolipid metabolites in inflammatory disease. Nature 2014, 510, 58–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Won, J.S.; Singh, A.K.; Singh, I. Lactosylceramide: A lipid second messenger in neuroinflammatory disease. J. Neurochem. 2007, 103 (Suppl. 1), 180–191. [Google Scholar] [CrossRef] [PubMed]
- Kosinska, M.K.; Liebisch, G.; Lochnit, G.; Wilhelm, J.; Klein, H.; Kaesser, U.; Lasczkowski, G.; Rickert, M.; Schmitz, G.; Steinmeyer, J. Sphingolipids in human synovial fluid--a lipidomic study. PLoS ONE 2014, 9, e91769. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.Y.; Park, S.H.; Lee, H.; Kwon, H.H.; Kim, S.K. Resting energy expenditure is not associated with disease activity in women with rheumatoid arthritis: Cross-sectional study. Korean J. Intern. Med. 2014, 29, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Echigo, R.; Shimohata, N.; Karatsu, K.; Yano, F.; Kayasuga-Kariya, Y.; Fujisawa, A.; Ohto, T.; Kita, Y.; Nakamura, M.; Suzuki, S.; et al. Trehalose treatment suppresses inflammation, oxidative stress, and vasospasm induced by experimental subarachnoid hemorrhage. J. Transl. Med. 2012, 10, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, B.; Schiller, J.; Wagner, U.; Hantzschel, H.; Arnold, K. The phosphatidylcholine/ lysophosphatidylcholine ratio in human plasma is an indicator of the severity of rheumatoid arthritis: Investigations by 31P NMR and MALDI-TOF MS. Clin. Biochem. 2005, 38, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Yun, X.; Dong, S.; Hu, Q.; Dai, Y.; Xia, Y. 1H NMR-based metabolomics approach to investigate the urine samples of collagen-induced arthritis rats and the intervention of tetrandrine. J. Pharm. Biomed. Anal. 2018, 154, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Semerano, L.; Minichiello, E.; Bessis, N.; Boissier, M.C. Novel Immunotherapeutic Avenues for Rheumatoid Arthritis. Trends Mol. Med. 2016, 22, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Tanwar, A.; Chawla, R.; Ansari, M.M.; Thakur, P.; Chakotiya, A.S.; Goel, R.; Ojha, H.; Asif, M.; Basu, M.; Arora, R.; et al. In vivo anti-arthritic efficacy of Camellia sinensis (L.) in collagen induced arthritis model. Biomed. Pharmacother. 2017, 87, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, N.; Wang, Z.; Qi, Z.; Zhu, H.; Zheng, B.; Li, P.; Liu, J. Nontargeted Metabolomic Analysis of Four Different Parts of Platycodon grandiflorum Grown in Northeast China. Molecules 2017, 22, 1280. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Guijas, C.; Montenegro-Burke, J.R.; Domingo-Almenara, X.; Palermo, A.; Warth, B.; Hermann, G.; Koellensperger, G.; Huan, T.; Uritboonthai, W.; Aisporna, A.E.; et al. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]












| NO | tR/min | Mass/Da | ESI § | VIP | Identified Compound | KEGG ID | PPM | MSE Fragmentation † | Pathways |
|---|---|---|---|---|---|---|---|---|---|
| S1 * | 17.26 | 295.2306 | − | 1.47 | 9,10- & 12,13-EpOME | C14825 ▲ | 9 | 134.8979, 152.9976, 167.1678, 183.0139, 195.8134, 197.2099, 223.1726, 277.2354, 295.2302 | Linoleic acid |
| S2 | 18.15 | 303.2299 | + | 2.47 | Linoleic acid | C01595 | 1 | 105.1102, 146.9803, 149.0211, 165.0605, 185.0739, 221.9592, 281.1889 | Linoleic acid |
| S3 | 22.98 | 303.2358 | − | 1.43 | Arachidonic acid | C00219 | 9 | 100.9381, 160.8434, 219.8489, 243.9024, 255.2361, 259.2451, 303.2365 | Arachidonic acid |
| S4 * | 18.26 | 319.2309 | − | 1.02 | EETs # & HETEs ## | C14768 ▲▲ | 8 | 114.9385, 127.4831, 136.9957, 163.1423, 174.9591, 219.1486, 235.1270, 255.6255, 257.6209, 319.2300, etc. | Arachidonic acid |
| S5 | 29.05 | 341.1083 | − | 2.26 | Alpha-lactose | C00243 | 2 | 116.9300, 145.1682, 160.8460, 265.1563, 277.4372, 281.2538, 311.1749 | Starch and Galactose |
| S6 | 29.24 | 341.1122 | − | 2.01 | Sucrose | C00089 | 2 | 101.0466, 146.9683, 160.9446, 249.5477, 341.1111 | Starch and Galactose |
| S7 | 29.23 | 387.1167 | − | 1.25 | Trehalose | C01083 | 6 | 101.0466, 146.9683, 160.9446, 179.0878, 341.1111 | Starch and Galactose |
| S8 * | 19.10 | 397.2239 | − | 1.15 | Prostanoids | C02198 ▲▲▲ | 2 | 115.1325, 134.9978, 166.8685, 183.0141, 205.5410, 207.1378, 279.2340, 303.2343, 351.1919 | Arachidonic acid |
| S9 | 18.69 | 624.3016 | − | 1.12 | Leukotriene C4 | C02166 | 9 | 146.1692, 167.0458, 225.0720, 508.3372, 562.3419 | Arachidonic acid |
| S10 * | 2.55 | 648.6284 | − | 1.13 | Ceramide (d18:1/24:0) | C00195 | 2 | 248.9640, 316.9509, 452.9229, 649.8531 | Sphingolipid |
| S11 | 24.76 | 730.5941 | + | 1.97 | Sm(d18:0/18:1) | C00550 | 6 | 149.0213, 183.0713, 282.2614, 310.2769, 561.3276 | Sphingolipid |
| S12 | 25.75 | 850.5526 | − | 1.82 | Lactosylceramide (d18:1/12:0) | C01290 | 1 | 146.9697, 179.0561, 198.2738, 480.3100, 626.3899 | Sphingolipid |
| S13 | 28.68 | 734.5693 | + | 2.97 | Pc (16:0/16:0) | C00157 | 0 | 184.0714, 496.2644, 551.5518, 734.5715 | Linoleic acid & Glycerophospholipid |
| S14 | 26.62 | 742.5342 | - | 3.87 | Pc (15:0/18:2) | C00157 | 7 | 141.0705, 279.2360, 281.2520, 655.4543, 742.5328 | |
| S15 | 27.63 | 752.5248 | + | 2.78 | Pc (16:1/16:1) | C00157 | 6 | 184.0159, 237.0634, 279.2367, 281.0246, 464.3080, 474.2838 | |
| S16 | 24.79 | 756.5538 | + | 1.85 | Pc (14:0/20:3) | C00157 | 0 | 183.0151, 247.2348, 271.2355, 468.3029, 628.3584 | |
| S17 | 23.84 | 758.5713 | + | 2.89 | Pc (18:1/16:1) | C00157 | 2 | 184.0157, 255.2365, 590.5201, 655.4091, 740.5466 | |
| S18 | 26.38 | 782.5714 | + | 3.71 | Pc (20:4/16:0) | C00157 | 3 | 184.0706, 257.2607, 478.3263, 679.5030 | |
| S19 | 27.41 | 786.6048 | + | 18.77 | Pc (18:1/18:1) | C00157 | 5 | 184.0718, 283.2617, 339.3408, 604.5342, 786.6055 | |
| S20 | 27.08 | 798.5323 | − | 2.59 | Pc (20:4/14:0) | C00157 | 4 | 227.2040, 257.2359, 303.2356, 439.2255, 669.4767 | |
| S21 | 26.90 | 804.5745 | − | 1.42 | Pc (14:1/20:0) | C00157 | 2 | 181.1628, 265.2368, 307.2393, 758.5609 | |
| S22 | 28.72 | 810.6019 | + | 23.25 | Pc (20:3/18:1) | C00157 | 1 | 184.0713, 221.0816, 283.2609, 307.2764, 627.3745 | |
| S23 | 25.79 | 832.5876 | + | 6.78 | Pc (18:2/20:2) | C00157 | 6 | 184.0728, 309.2785, 627.3777, 684.4404, 752.5748 | |
| S24 | 26.87 | 854.5839 | − | 4.99 | Pc (20:4/18:0) | C00157 | 9 | 255.2352, 279.2362, 283.2639, 303.2360, 725.5103 |
| Items | Potential Energy-OPLS3 | Docking Score | XP GScore | H-Bond |
|---|---|---|---|---|
| Values | 209.403 | −4.683 | −5.953 | 6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, Z.; Wang, G.; Ran, N.; Lin, H.; Wang, Z.; Guan, X.; Yuan, Y.; Fang, K.; Liu, J.; Wang, F. Inhibitory Effect of Methotrexate on Rheumatoid Arthritis Inflammation and Comprehensive Metabolomics Analysis Using Ultra-Performance Liquid Chromatography-Quadrupole Time of Flight-Mass Spectrometry (UPLC-Q/TOF-MS). Int. J. Mol. Sci. 2018, 19, 2894. https://doi.org/10.3390/ijms19102894
Pang Z, Wang G, Ran N, Lin H, Wang Z, Guan X, Yuan Y, Fang K, Liu J, Wang F. Inhibitory Effect of Methotrexate on Rheumatoid Arthritis Inflammation and Comprehensive Metabolomics Analysis Using Ultra-Performance Liquid Chromatography-Quadrupole Time of Flight-Mass Spectrometry (UPLC-Q/TOF-MS). International Journal of Molecular Sciences. 2018; 19(10):2894. https://doi.org/10.3390/ijms19102894
Chicago/Turabian StylePang, Zhiqiang, Guoqiang Wang, Nan Ran, Hongqiang Lin, Ziyan Wang, Xuewa Guan, Yuze Yuan, Keyong Fang, Jinping Liu, and Fang Wang. 2018. "Inhibitory Effect of Methotrexate on Rheumatoid Arthritis Inflammation and Comprehensive Metabolomics Analysis Using Ultra-Performance Liquid Chromatography-Quadrupole Time of Flight-Mass Spectrometry (UPLC-Q/TOF-MS)" International Journal of Molecular Sciences 19, no. 10: 2894. https://doi.org/10.3390/ijms19102894