Stable Membrane-Association of mRNAs in Etiolated, Greening and Mature Plastids
Abstract
:1. Introduction
2. Results and Discussion
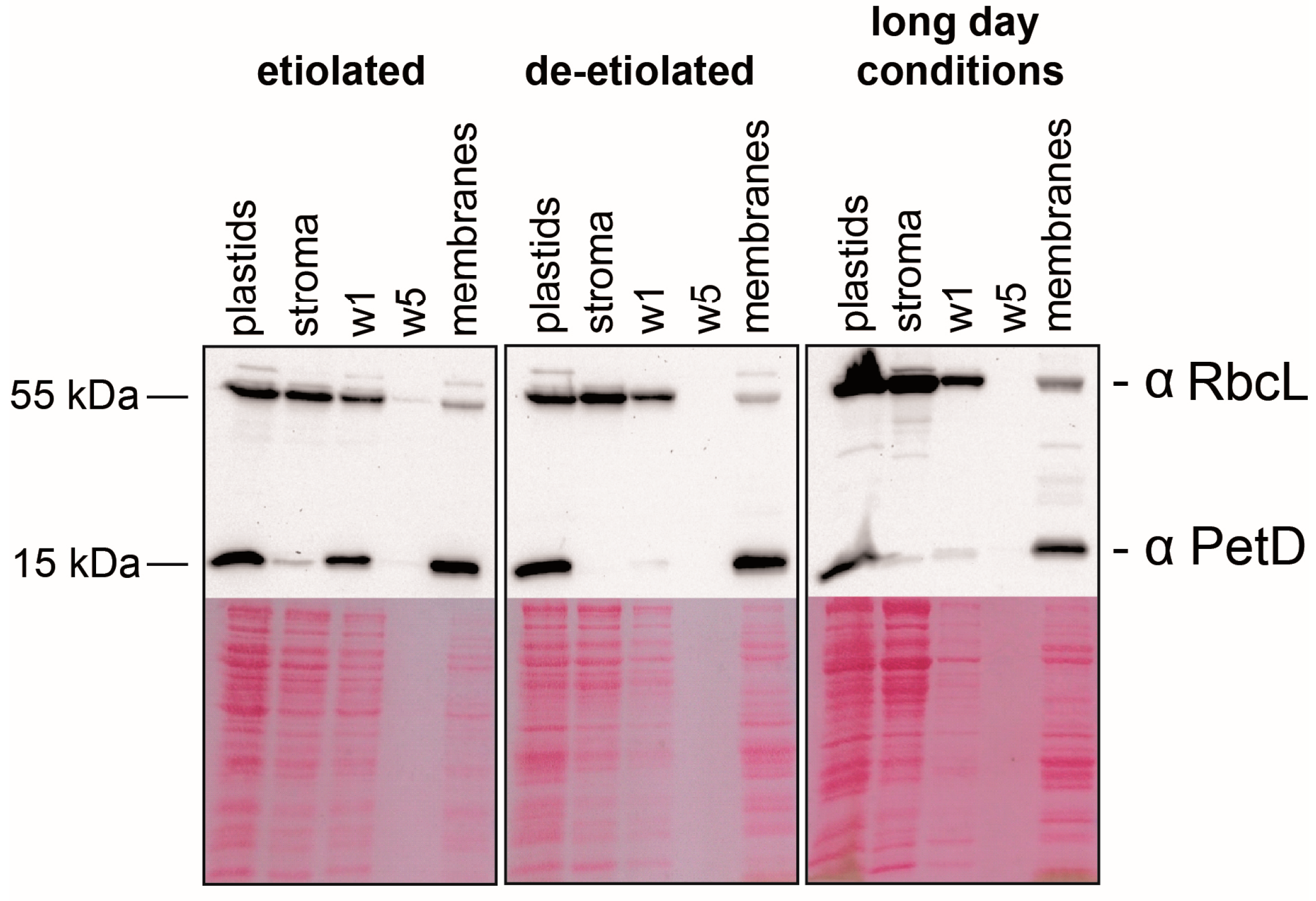
2.1. Preparation of Sub-Organellar Fractions Highly Enriched in Membrane and Stroma Marker Proteins
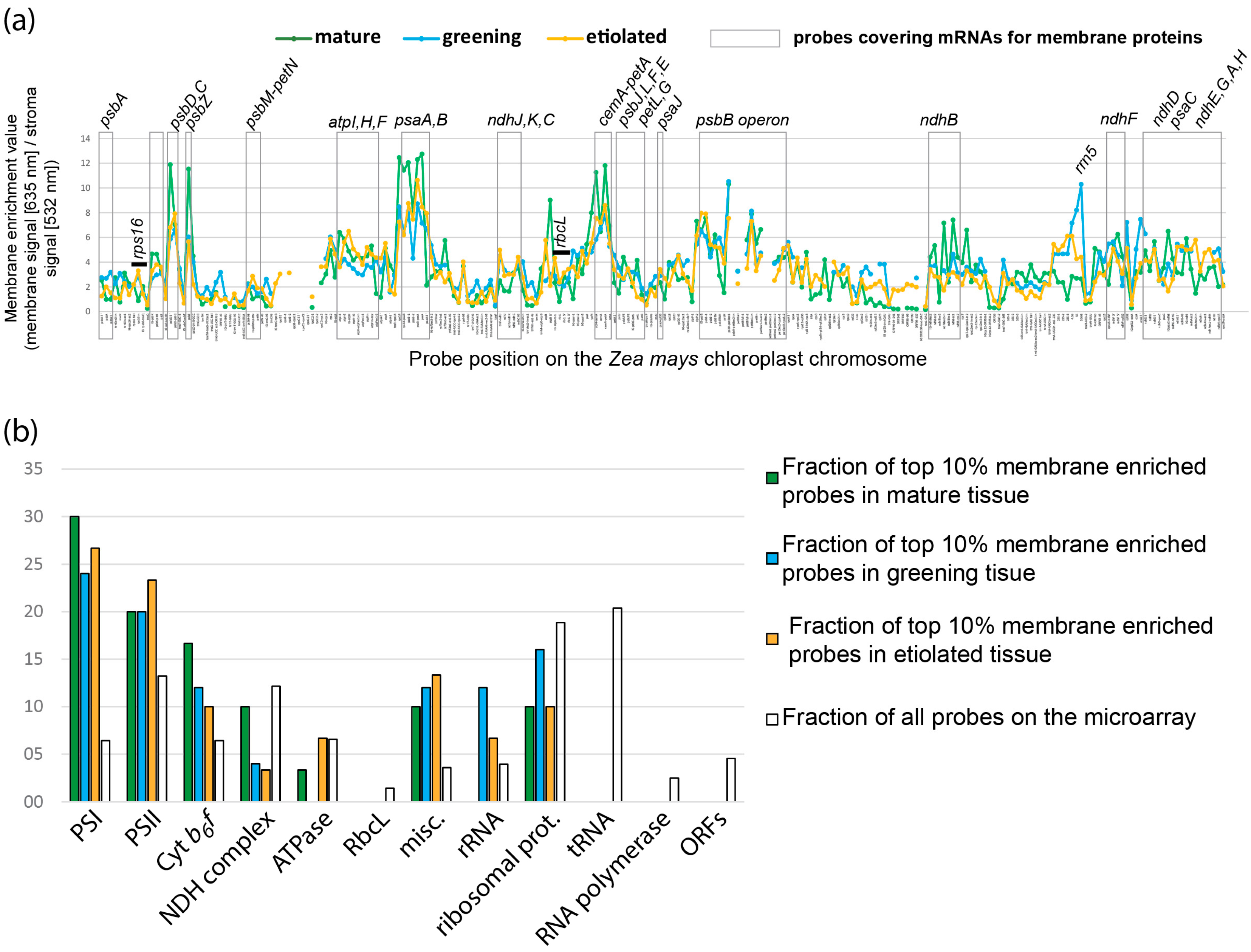
2.2. Global Association of Chloroplast mRNAs for Membrane Proteins with Chloroplast and Etioplast Membranes
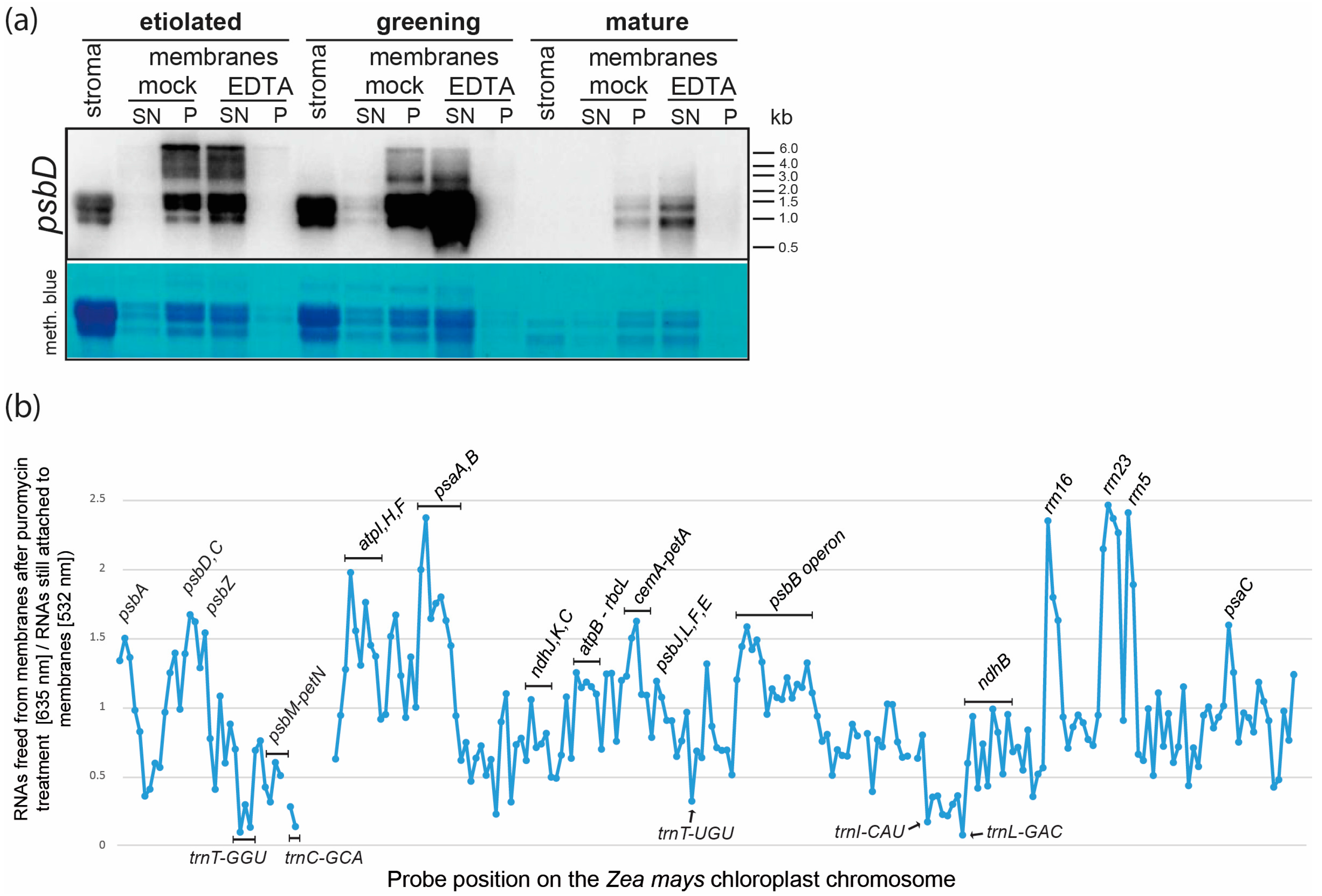
2.3. Membrane Association of Chloroplast RNAs via Ribosomes
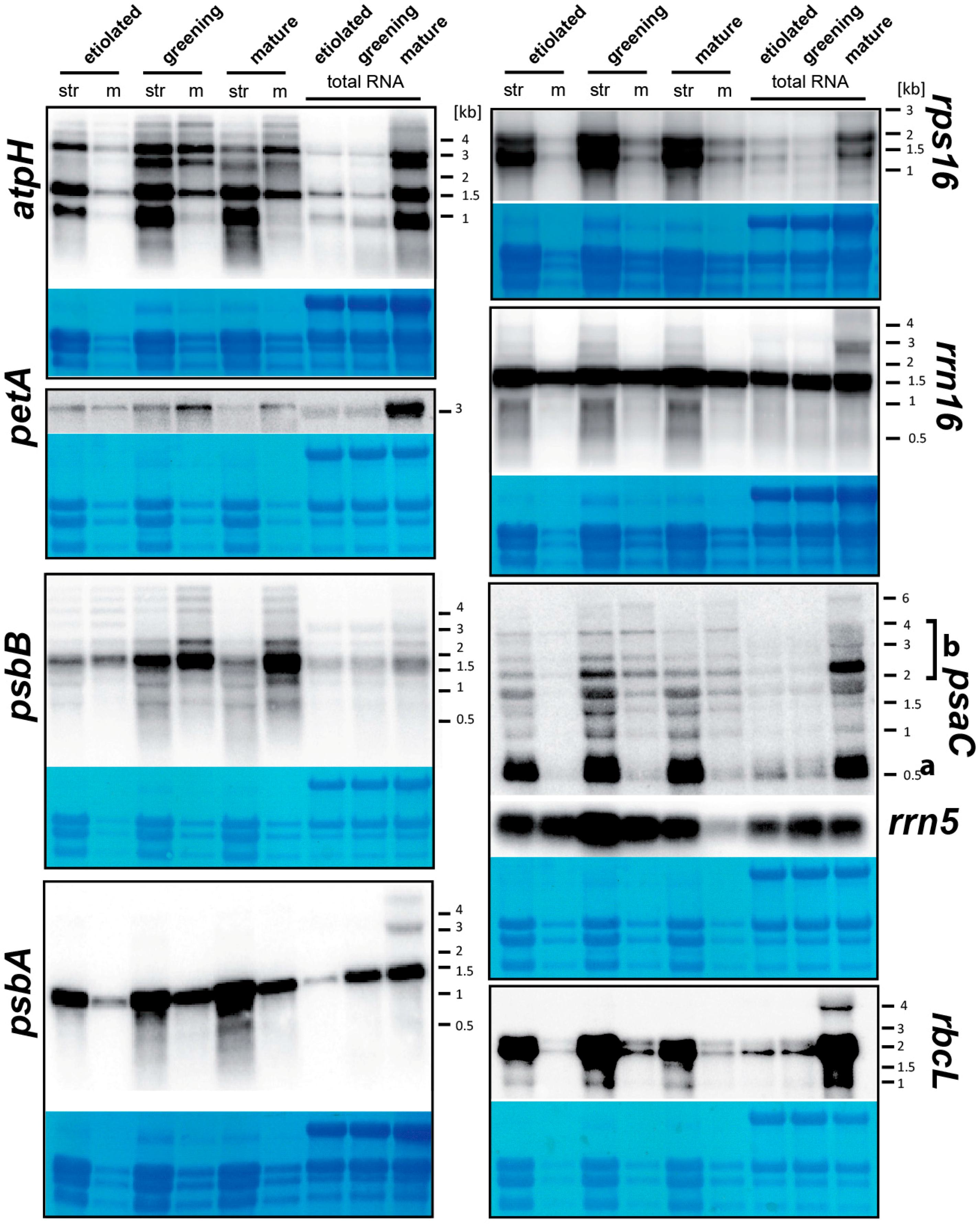
2.4. tRNAs, mRNAs for Stromal Proteins and Short mRNAs for Membrane Proteins Are Enriched in the Soluble Fraction of Plastids
2.5. Etioplasts Display Membrane Enrichment of RNAs Similar to Chloroplasts
2.6. Quantitative Analysis of Membrane-Bound and Stromal mRNAs on a “Per-Chloroplast” Base Uncovers Transcript-Autonomous Localization Patterns
2.6.1. The Analysis of Equal Amounts of Total Cellular RNA from Different Tissues Does Not Accurately Reflect per Organelle RNA Levels for Most Plastid Genes
2.6.2. mRNAs Are Enriched at Chloroplast Membranes in a Transcript-Autonomous Fashion
2.6.3. Differential Membrane-Association of rRNA Species
2.6.4. Constant Association of Plastid RNAs with Chloroplast Membranes Suggests Altered Translational Rates during Chloroplast Greening rather than Increased Accumulation or Improved Recruitment of mRNAs to Membranes
3. Materials and Methods
3.1. Plant Material
3.2. Extraction of Stroma and Membrane Fractions
3.3. EDTA and Puromycin Treatments of Purified Plastid Membranes
3.4. RNA Gel Blot Analyses
3.5. Immunoblot Analyses
3.6. Tiling Microarray Design
3.7. Microarray Hybridisation
3.8. Microarray Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MEV | Membrane Enrichment Value |
| NDH | NADH Dehydrogenase |
References
- Jung, H.; Gkogkas, C.G.; Sonenberg, N.; Holt, C.E. Remote control of gene function by local translation. Cell 2014, 157, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Keiler, K.C. RNA localization in bacteria. Curr. Opin. Microbiol. 2011, 14, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Doroshenk, K.A.; Crofts, A.J.; Morris, R.T.; Wyrick, J.J.; Okita, T.W. Ricerbp: A resource for experimentally identified RNA binding proteins in Oryza sativa. Front. Plant Sci. 2012, 3, 90. [Google Scholar] [CrossRef] [PubMed]
- Uniacke, J.; Zerges, W. Chloroplast protein targeting involves localized translation in Chlamydomonas. Proc. Natl. Acad. Sci. USA 2009, 106, 1439–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uniacke, J.; Zerges, W. Photosystem II assembly and repair are differentially localized in Chlamydomonas. Plant Cell 2007, 19, 3640–3654. [Google Scholar] [CrossRef] [PubMed]
- Schottkowski, M.; Peters, M.; Zhan, Y.; Rifai, O.; Zhang, Y.; Zerges, W. Biogenic membranes of the chloroplast in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2012, 109, 19286–19291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chua, N.-H.; Blobel, G.; Siekevitz, P.; Palade, G.E. Attachment of chloroplast polysomes to thylakoid membranes in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1973, 70, 1554–1558. [Google Scholar] [CrossRef] [PubMed]
- Chua, N.H.; Blobel, G.; Siekevitz, P.; Palade, G.E. Periodic variations in the ratio of free to thylakoid-bound chloroplast ribosomes during the cell cycle of Chlamydomonas reinhardtii. J. Cell Biol. 1976, 71, 497–514. [Google Scholar] [CrossRef] [PubMed]
- Margulies, M.M.; Michaels, A. Ribosomes bound to chloroplast membranes in Chlamydomonas reinhardtii. J. Cell Biol. 1974, 60, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Fish, L.E.; Jagendorf, A.T. Light-induced increase in the number and activity of ribosomes bound to pea chloroplast thylakoids in vivo. Plant Physiol. 1982, 69, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Hurewitz, J.; Jagendorf, A.T. Further characterization of ribosome binding to thylakoid membranes. Plant Physiol. 1987, 84, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Michaels, A.; Margulies, M.M. Amino acid incorporation into protein by ribosomes bound to chloroplast thylakoid membranes: Formation of discrete products. Biochim. Biophys. Acta 1975, 390, 352–362. [Google Scholar] [CrossRef]
- Alscher-Herman, R.; Jagendorf, A.T.; Grumet, R. Ribosome-thylakoid association in peas: Influence of anoxia. Plant Physiol. 1979, 64, 232–235. [Google Scholar] [CrossRef] [PubMed]
- Zoschke, R.; Barkan, A. Genome-wide analysis of thylakoid-bound ribosomes in maize reveals principles of cotranslational targeting to the thylakoid membrane. Proc. Natl. Acad. Sci. USA 2015, 112, E1678–E1687. [Google Scholar] [CrossRef] [PubMed]
- Zoschke, R.; Chotewutmontri, P.; Barkan, A. Translation and co-translational membrane engagement of plastid-encoded chlorophyll-binding proteins are not influenced by chlorophyll availability in maize. Front. Plant Sci. 2017, 8, 385. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A. Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011, 155, 1520–1532. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Kuroda, H.; Yukawa, Y.; Sugiura, M. Translation of partially overlapping psbD–psbC mRNAs in chloroplasts: The role of 5′-processing and translational coupling. Nucleic Acids Res. 2012, 40, 3152–3158. [Google Scholar] [CrossRef] [PubMed]
- Chotewutmontri, P.; Barkan, A. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 2016, 12, e1006106. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988, 7, 2637–2644. [Google Scholar] [PubMed]
- Schmitz-Linneweber, C.; Williams-Carrier, R.; Barkan, A. RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 2005, 17, 2791–2804. [Google Scholar] [CrossRef] [PubMed]
- Zerges, W.; Rochaix, J.D. Low density membranes are associated with RNA-binding proteins and thylakoids in the chloroplast of Chlamydomonas reinhardtii. J. Cell Biol. 1998, 140, 101–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klaff, P.; Gruissem, W. A 43 kd light-regulated chloroplast RNA-binding protein interacts with the psbA 5′ non-translated leader RNA. Photosyn. Res. 1995, 46, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Hotto, A.M.; Huston, Z.E.; Stern, D.B. Overexpression of a natural chloroplast-encoded antisense RNA in tobacco destabilizes 5S rRNA and retards plant growth. BMC Plant Biol. 2010, 10, 213. [Google Scholar] [CrossRef] [PubMed]
- Mulo, P.; Sakurai, I.; Aro, E.M. Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: From transcription to psii repair. Biochim. Biophys. Acta 2012, 1817, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.R.; Mason, H.S.; Mullet, J.E. Light-regulated translation of chloroplast proteins. I. Transcripts of psaA–psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J. Cell Biol. 1988, 106, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.M.; Neckermann, K.; Igloi, G.L.; Kossel, H. Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995, 251, 614–628. [Google Scholar] [CrossRef] [PubMed]
- Blomqvist, L.A.; Ryberg, M.; Sundqvist, C. Proteomic analysis of highly purified prolamellar bodies reveals their significance in chloroplast development. Photosynth. Res. 2008, 96, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Blomqvist, L.A.; Ryberg, M.; Sundqvist, C. Proteomic analysis of the etioplast inner membranes of wheat (Triticum aestivum) by two-dimensional electrophoresis and mass spectrometry. Physiol. Plant. 2006, 128, 368–381. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Murakami, S. Behaviour of the 36,000-dalton protein in the integral membranes of squash etioplasts during greening. Plant Cell Physiol. 1982, 23, 575–583. [Google Scholar]
- Von Zychlinski, A.; Kleffmann, T.; Krishnamurthy, N.; Sjolander, K.; Baginsky, S.; Gruissem, W. Proteome analysis of the rice etioplast: Metabolic and regulatory networks and novel protein functions. Mol. Cell. Proteom. 2005, 4, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Selstam, E.; Sandelius, A.S. A comparison between prolamellar bodies and prothylakoid membranes of etioplasts of dark-grown wheat concerning lipid and polypeptide composition. Plant Physiol. 1984, 76, 1036–1040. [Google Scholar] [CrossRef] [PubMed]
- Kleffmann, T.; von Zychlinski, A.; Russenberger, D.; Hirsch-Hoffmann, M.; Gehrig, P.; Gruissem, W.; Baginsky, S. Proteome dynamics during plastid differentiation in rice. Plant Physiol. 2007, 143, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Kanervo, E.; Singh, M.; Suorsa, M.; Paakkarinen, V.; Aro, E.; Battchikova, N.; Aro, E.M. Expression of protein complexes and individual proteins upon transition of etioplasts to chloroplasts in pea (Pisum sativum). Plant Cell Physiol. 2008, 49, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Lonosky, P.M.; Zhang, X.; Honavar, V.G.; Dobbs, D.L.; Fu, A.; Rodermel, S.R. A proteomic analysis of maize chloroplast biogenesis. Plant Physiol. 2004, 134, 560–574. [Google Scholar] [CrossRef] [PubMed]
- Margulies, M.M.; Tiffany, H.L.; Hattori, T. Photosystem I reaction center polypeptides of spinach are synthesized on thylakoid-bound ribosomes. Arch. Biochem. Biophys. 1987, 254, 454–461. [Google Scholar] [CrossRef]
- Mackender, R.O. Etioplast development in dark-grown leaves of Zea mays L. Plant Physiol. 1978, 62, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.R.; Mullet, J.E. Control of gene expression during higher plant chloroplast biogenesis. Protein synthesis and transcript levels of psbA, psaA–psaB, and rbcL in dark-grown and illuminated barley seedlings. J. Biol. Chem. 1987, 262, 4341–4348. [Google Scholar] [PubMed]
- Laing, W.; Kreuz, K.; Apel, K. Light-dependent, but phytochrome-independent, translational control of the accumulation of the p700 chlorophyll-A protein of photosystem I in barley (Hordeum vulgare L.). Planta 1988, 176, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.R.; Mullet, J.E. Regulation of chloroplast-encoded chlorophyll-binding protein translation during higher plant chloroplast biogenesis. J. Biol. Chem. 1986, 261, 11138–11145. [Google Scholar] [PubMed]
- Ruf, S.; Kossel, H.; Bock, R. Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J. Cell Biol. 1997, 139, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Sugiura, M. Both RNA editing and RNA cleavage are required for translation of tobacco chloroplast NdhD mRNA: A possible regulatory mechanism for the expression of a chloroplast operon consisting of functionally unrelated genes. EMBO J. 1997, 16, 6804–6811. [Google Scholar] [CrossRef] [PubMed]
- Antonkine, M.L.; Jordan, P.; Fromme, P.; Krauss, N.; Golbeck, J.H.; Stehlik, D. Assembly of protein subunits within the stromal ridge of photosystem I. Structural changes between unbound and sequentially PSI-bound polypeptides and correlated changes of the magnetic properties of the terminal iron sulfur clusters. J. Mol. Biol. 2003, 327, 671–697. [Google Scholar] [CrossRef]
- Li, N.; Zhao, J.D.; Warren, P.V.; Warden, J.T.; Bryant, D.A.; Golbeck, J.H. PsaD is required for the stable binding of PsaC to the photosystem I core protein of synechococcus sp. Pcc 6301. Biochemistry 1991, 30, 7863–7872. [Google Scholar] [CrossRef] [PubMed]
- Sharwood, R.E.; Hotto, A.M.; Bollenbach, T.J.; Stern, D.B. Overaccumulation of the chloroplast antisense RNA as5 is correlated with decreased abundance of 5S rRNA in vivo and inefficient 5S rRNA maturation in vitro. RNA 2011, 17, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Breidenbach, E.; Leu, S.; Michaels, A.; Boschetti, A. Synthesis of EF-Tu and distribution of its mRNA between stroma and thylakoids during the cell cycle of Chlamydomonas reinhardii. Biochim. Biophys. Acta 1990, 1048, 209–216. [Google Scholar] [CrossRef]
- Voelker, R.; Barkan, A. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO J. 1995, 14, 3905–3914. [Google Scholar] [PubMed]
- Finster, S.; Eggert, E.; Zoschke, R.; Weihe, A.; Schmitz-Linneweber, C. Light-dependent, plastome-wide association of the plastid-encoded RNA polymerase with chloroplast DNA. Plant J. 2013, 76, 849–860. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Legen, J.; Schmitz-Linneweber, C. Stable Membrane-Association of mRNAs in Etiolated, Greening and Mature Plastids. Int. J. Mol. Sci. 2017, 18, 1881. https://doi.org/10.3390/ijms18091881
Legen J, Schmitz-Linneweber C. Stable Membrane-Association of mRNAs in Etiolated, Greening and Mature Plastids. International Journal of Molecular Sciences. 2017; 18(9):1881. https://doi.org/10.3390/ijms18091881
Chicago/Turabian StyleLegen, Julia, and Christian Schmitz-Linneweber. 2017. "Stable Membrane-Association of mRNAs in Etiolated, Greening and Mature Plastids" International Journal of Molecular Sciences 18, no. 9: 1881. https://doi.org/10.3390/ijms18091881



