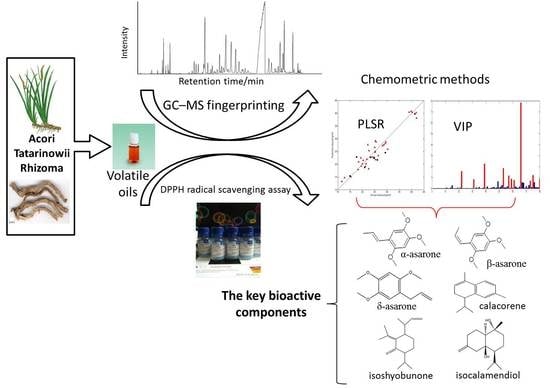
GC-MS Fingerprinting Combined with Chemometric Methods Reveals Key Bioactive Components in Acori Tatarinowii Rhizoma
Abstract
:1. Introduction
2. Results and Discussion
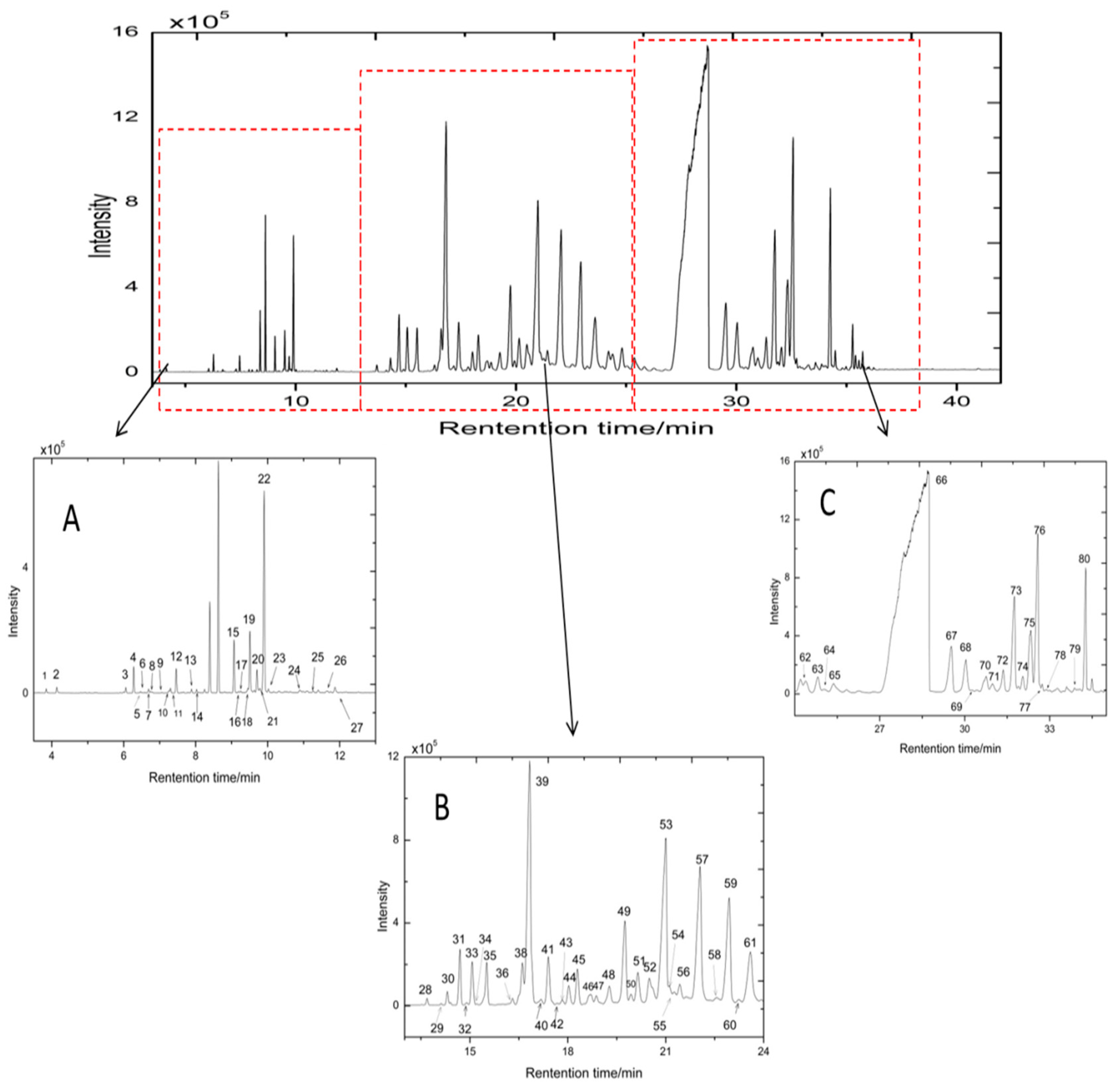
2.1. GC-MS Fingerprinting and DPPH Radical Scavenging Assay
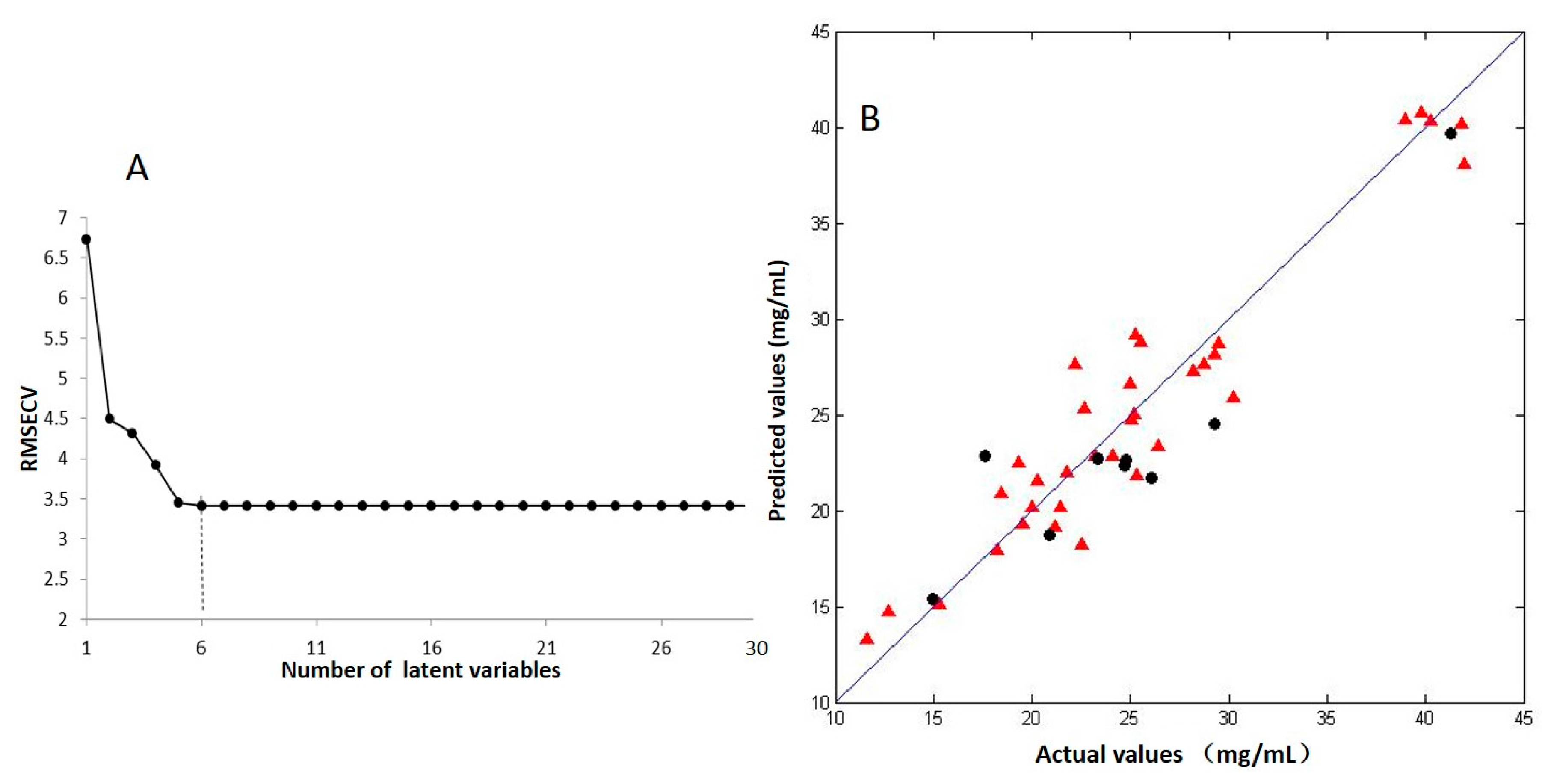
2.2. PLSR Model between the Radical Scavenging Activity and the Contents of Volatile Components
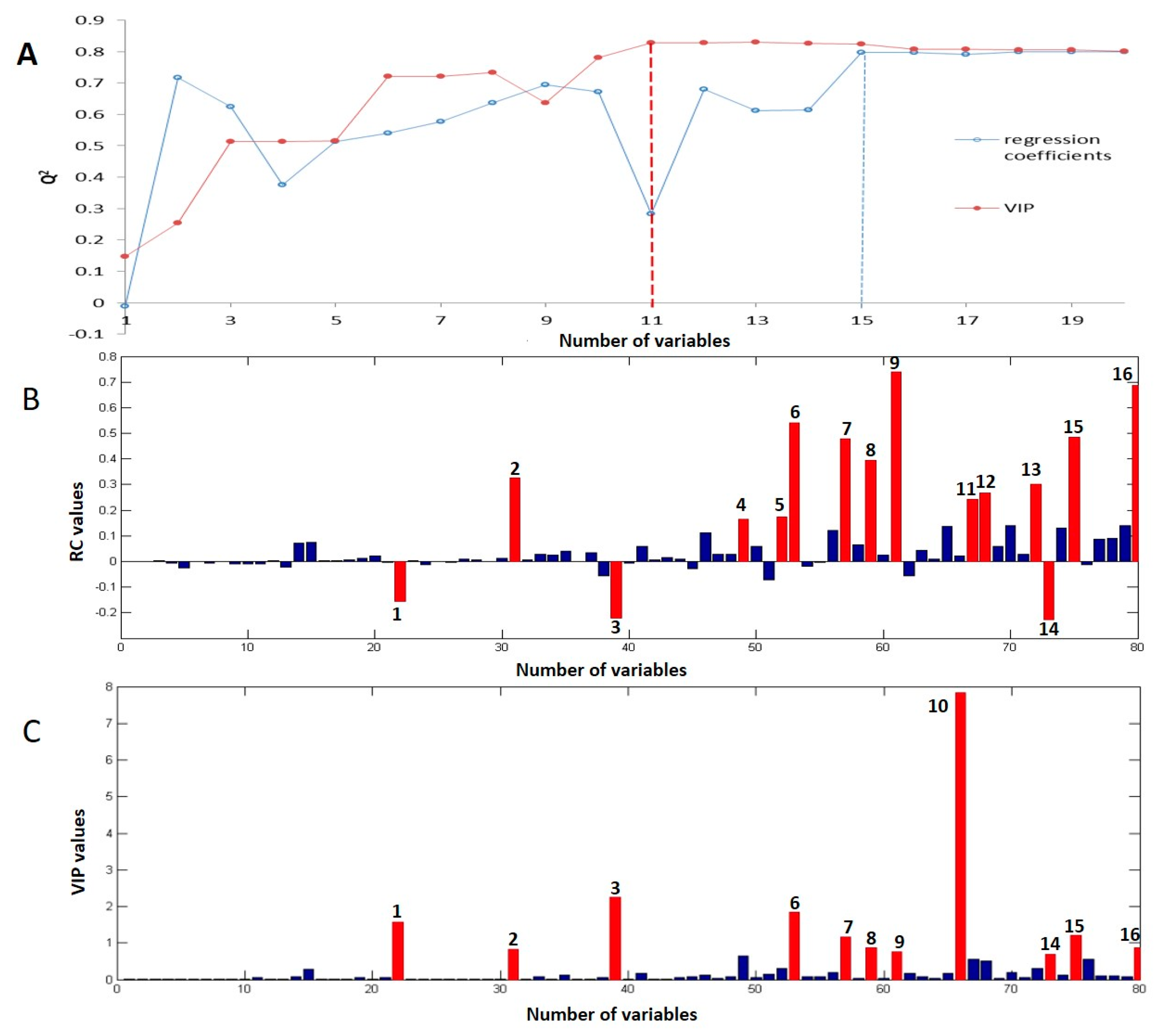
2.3. Key Bioactive Components in ATR for Radical Scavenging Activity
3. Materials and Methods
3.1. Materials
3.2. Extraction of Volatile Oil
3.3. DPPH Radical Scavenging Assay
3.4. GC-MS Analysis
3.5. Statistical Analysis
3.5.1. Partial Least Squares Regression (PLSR)
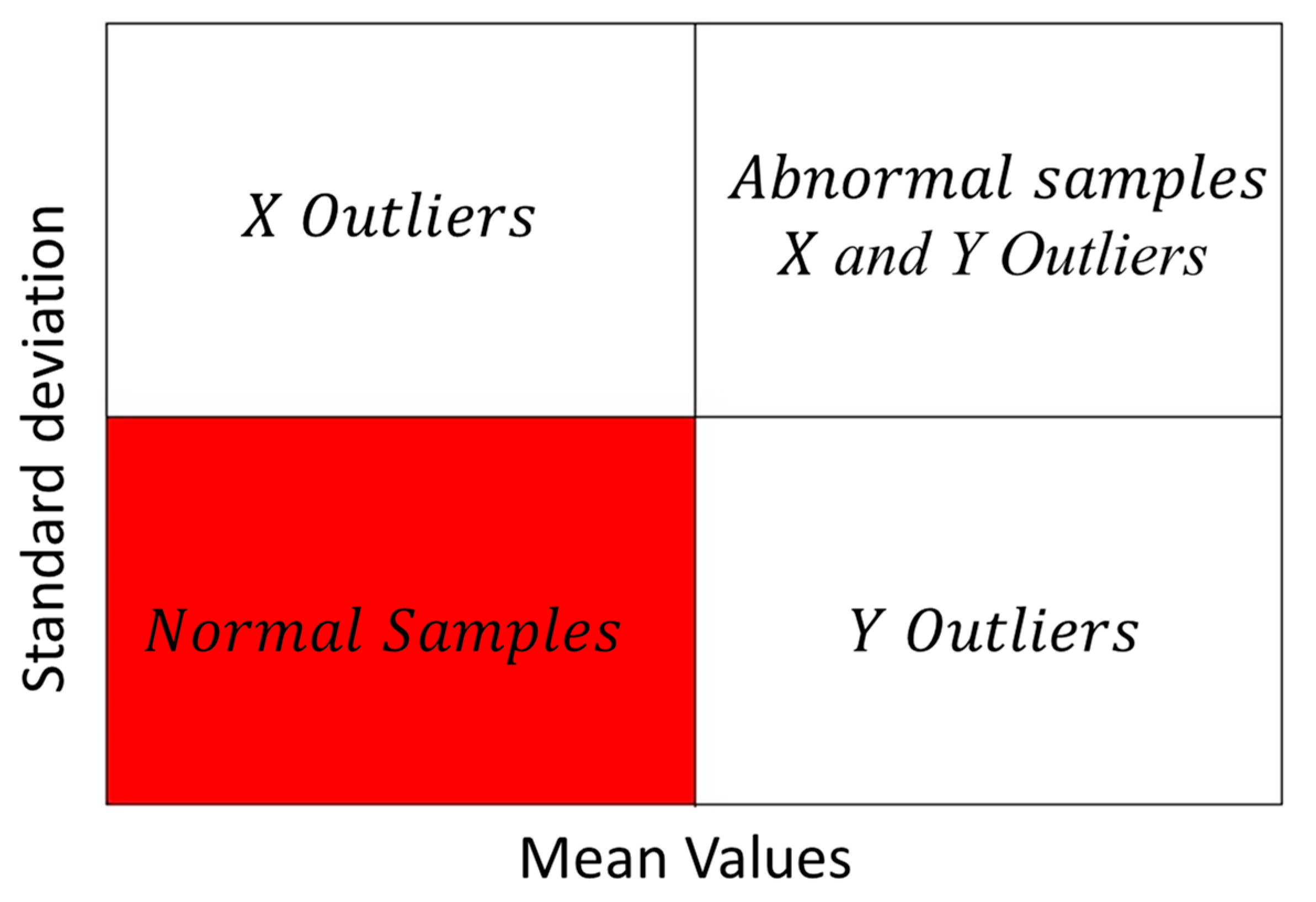
3.5.2. Outlier Identification
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liao, W.P.; Li, C.P.; Yi, Y.H.; Sun, W.W.; Gao, M.M.; Tao, S.; Yang, S.Q. Study of Antiepileptic Effect of Extracts from Acorus tatarinowii Schott. Epilepsia 2005, 46, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, S.; Menom, S.; Shailajan, S. A rapid HPLC-ESI-MS/MS method for determination of β-asarone, a potential anti-epileptic agent, in plasma after oral administration of Acorus calamus extract to rats. Biomed. Chromatogr. 2013, 27, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Kumar, V.; Mal, M.; Houghton, P.J. In vitro acetylcholinesterase inhibitory activity of the essential oil from Acorus calamus and its main constituents. Planta Med. 2007, 73, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Han, T.; Peng, W.; Wang, X.R. Antidepressant-like effects of essential oil and asarone, a major essential oil component from the rhizome of Acorus tatarinowii. Pharm. Boil. 2013, 51, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.; Chen, Y.B.; Chen, D.F.; Lai, X.P.; Liu, D.H.; Deng, R.D.; Zhou, J.H.; Zhang, S.H.; Li, Y.W.; Li, H.; et al. β-Asarone inhibits neuronal apoptosis via the CaMKII/CREB/Bcl-2 signaling pathway in an in vitro model and AbetaPP/PS1 mice. J. Alzheimer’s Dis. 2013, 33, 863–880. [Google Scholar]
- Yang, Y.X.; Chen, Y.T.; Zhou, X.J.; Hong, C.L.; Li, C.Y.; Guo, J.Y. β-asarone, a major component of Acorus tatarinowii Schott, attenuates focal cerebral ischemia induced by middle cerebral artery occlusion in rats. BMC Complement. Altern. Med. 2013, 13, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, L.; Shi, L.; Zhang, Z.; Du, X.; Wang, Z.; Zhang, Z. β-Asarone induces senescence in colorectal cancer cells by inducing lamin B1 expression. Phytomedicine 2013, 20, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Jee, Y.L.; Jung, Y.L.; Bongsik, Y.; Byung, K.H. Antifungal Activity of β-Asarone from Rhizomes of Acorus gramineus. J. Agric. Food Chem. 2004, 52, 776–780. [Google Scholar]
- Zhuang, Y.L.; Ma, Q.Y.; Guo, Y.; Sun, L.P. Protective effects of rambutan (Nephelium lappaceum) peel phenolics on H2O2-induced oxidative damages in HepG2 cells and d-galactose-induced aging mice. Food Chem. Toxicol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Loizzo, M.; Sicari, V.I.; Tenuta, M.C.; Leporini, M.R.; Falco, T.; Pellicanò, T.M.; Menichini, F.; Tundis, R. Phytochemicals content, antioxidant and hypoglycaemic activities of commercial nutmeg mace (Myristica fragrans L.) and pimento (Pimenta dioica(L.) Merr.). Int. J. Food Sci. Tech. 2016, 51, 2057–2063. [Google Scholar] [CrossRef]
- Huang, H.J.; Chen, H.Y.; Chang, Y.S.; Chen, C.C. Insight into two antioxidants binding to the catalase NADPH binding site from traditional Chinese medicines. RSC Adv. 2015, 5, 6625–6635. [Google Scholar] [CrossRef]
- Wang, S.T.; Gao, W.; Fan, Y.X.; Liu, X.G.; Liu, K.; Du, Y.; Wang, L.L.; Li, H.J.; Li, P.; Yang, H. Phenol profiles and antioxidant capacities of Bistort Rhizoma (Polygonum bistorta L.) extracts. RSC Adv. 2016, 6, 27320–27328. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, A.H.; Han, Y.; Lu, S.W.; Sun, H.; Yan, G.L.; Wang, P.; Wang, X.J. Ultra-High Performance Liquid Chromatography Coupled with Time-of-flight Mass Spectrometry Screening and Analyzing the Potential Bioactive Compounds from Traditional Chinese Medicine Kaixin San, Using Multivariate Data Processing Approach and Metabolynx To. RSC Adv. 2014, 5, 85–92. [Google Scholar] [CrossRef]
- Devi, S.A.; Ganjewala, D. Antioxidant Activities of Methanolic Extracts of Sweet-Flag (Acorus calamus) Leaves and Rhizomes. J. Herbs Spices Med. Plants 2011, 17, 1–11. [Google Scholar] [CrossRef]
- Zhang, X.J.; Yi, L.Z.; Deng, B.C.; Chen, L.C.; Shi, S.T.; Zhuang, Y.L.; Zhang, Y. Discrimination of Acori Tatarinowii Rhizoma and Acori Calami Rhizoma based on quantitative gas chromatographic fingerprints and chemometric methods. J. Sep. Sci. 2015, 38, 4078–4085. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.Z.; Dong, N.P.; Liu, S.; Yi, Z.B.; Zhang, Y. Chemical features of Pericarpium Citri Reticulatae and Pericarpium Citri Reticulatae Viride revealed by GC-MS metabolomics analysis. Food Chem. 2014, 186, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sun, G.X.; Luan, J.Y.; Ling, J.H.; Zhang, J.; Yang, F.L. A comprehensive strategy to monitor quality consistency of Weibizhi tablets based on integrated MIR and UV spectroscopic fingerprints, a systematically quantified fingerprint method, antioxidant activities and UPLC-Q-TOF-MS chemical profiling. RSC Adv. 2015, 6, 366–375. [Google Scholar] [CrossRef]
- Liu, X.; Zhan, H.; Qiao, Z.; Zheng, M.; Liu, W.Y.; Feng, F.; Yan, F.R. Chemometric analysis based on HPLC multi-wavelength fingerprints for prediction of antioxidant components in Turpiniae Folium. Chemom. Intell. Lab. 2016, 152, 54–61. [Google Scholar] [CrossRef]
- Fan, X.H.; Cheng, Y.Y.; Ye, Z.L.; Lin, R.C.; Qian, Z.Z. Multiple chromatographic fingerprinting and its application to the quality control of herbal medicines. Anal. Chim. Acta 2006, 555, 217–224. [Google Scholar] [CrossRef]
- Xin, Z.Q.; Ren, D.B.; Zhang, X.J.; Yi, Z.B.; Yi, L.Z. Chromatographic Fingerprints Combined with Chemometric Methods Reveal the Chemical Features of Authentic Radix Polygalae. J. AOAC Int. 2017, 100, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.Z.; Yuan, D.L.; Liang, Y.Z.; Xie, P.S.; Zhao, Y. Quality control and discrimination of pericarpium citri reticulatae and pericarpium citri reticulatae viride based on high-performance liquid chromatographic fingerprints and multivariate statistical analysis. Anal. Chim. Acta 2007, 588, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhao, Y.Y.; Liu, W.Y.; Feng, F.; Xie, N. HPLC with quadrupole TOF-MS and chemometrics analysis for the characterization of Folium Turpiniae from different regions. J. Sep. Sci. 2013, 36, 2552–2561. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Guo, T.; Sui, Y.; Li, F.M. Fingerprint analysis of Flos Carthami by capillary electrophoresis. J. Chromatogr. B 2003, 792, 147–152. [Google Scholar] [CrossRef]
- Chaita, E.; Gikas, E.; Aligiannis, N. Integrated HPTLC-based Methodology for the Tracing of Bioactive Compounds in Herbal Extracts Employing Multivariate Chemometrics. A Case Study on Morus alba. Phytochem. Anal. 2017, 28, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Smilde, A.; Westerhuis, J.A.; De, J. A framework for sequential multiblock component methods. J. Chemometr. 2003, 17, 323–337. [Google Scholar] [CrossRef]
- Reis, M.S. Network-induced supervised learning: Network-induced classification (NI-C) and network-induced regression (NI-R). AichE J. 2013, 59, 1570–1587. [Google Scholar] [CrossRef]
- Vapnik, V. The Nature of Statistical Learning Theory, 2nd ed.; Springer: New York, NY, USA, 2000; Volume 8, pp. 988–999. [Google Scholar]
- Gad, H.A.; El-Ahmady, S.H.; Abou-Shoer, M.I.; Al-Azizi, M.M. Application of chemometrics in authentication of herbal medicines: A review. Phytochem. Anal. 2013, 24, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Kang, Q.; Ru, Q.; Liu, R.; Xu, L.; Liu, J.; Wang, Y.; Zhang, Y.; Li, H.; Zhang, Q.; Wu, Q. On-line monitoring the extract process of Fu-fang Shuanghua oral solution using near infrared spectroscopy and different PLS algorithms. Spectrochim. Acta A 2016, 152, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Lozano, V.; Peña, A.; Durán, M.I.; Mansilla, A.E.; Escandar, G.M. Four-way multivariate calibration using ultra-fast high-performance liquid chromatography with fluorescence excitation–emission detection. Application to the direct analysis of chlorophylls a and b and pheophytins a and b in olive oils. Chemom. Intell. Lab. 2013, 125, 121–131. [Google Scholar] [CrossRef]
- Samadi, N.; Masoum, S.; Mehrara, B.; Hosseini, H. Application of linear multivariate calibration techniques to identify the peaks responsible for the antioxidant activity of Satureja hortensis L. and Oliveria decumbens Vent. essential oils by gas chromatography-mass spectrometry. J. Chromatogr. B 2015, 1001, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.Z.; Dong, N.P.; Shi, S.T.; Deng, B.C.; Yun, Y.H.; Yi, Z.B.; Zhang, Y. Metabolomic identification of novel biomarkers of nasopharyngeal carcinoma. RSC Adv. 2014, 4, 59094–59101. [Google Scholar] [CrossRef]
- Lam, K.Y.; Chen, J.; Lam, C.T.; Wu, Q.; Yao, P.; Dong, T.T.; Lin, H.; Tsim, K.W. Asarone from Acori Tatarinowii Rhizoma Potentiates the Nerve Growth Factor-Induced Neuronal Differentiation in Cultured PC12 Cells: A Signaling Mediated by Protein Kinase A. PLoS ONE 2016, 11, e0163337. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.K.; Kumar, V.; Mal, M.; Houghton, P.J. Scientific Validation of Ayurvedic Tradition from Natural Resources. Pharm. Biol. 2007, 45, 651–666. [Google Scholar] [CrossRef]
- Lubsandorzhieva, P.B.; Boldanova, N.B.; Dashinamzhilov, Z.B. Chemical Composition and In Vitro Antioxidant Activity of Essential Oil from a Hepatoprotective Herbal Mix. Pharm. Chem. J. 2013, 47, 58–61. [Google Scholar] [CrossRef]
- Varma, J.; Tripathi, M.; Ram, V.J.; Pandey, V.B.; Dubey, D.K. γ-Asarone- the fungitoxic principle of the essential oil of Caesulia axillaris. World J. Microbi. Biot. 2002, 18, 277–279. [Google Scholar] [CrossRef]
- Zhu, L.Y.; Lin, Z.; Zhan, L.B.; Lu, X.G.; Peng, J.Y.; Liang, L.N.; Yu, L.; Zheng, L.P.; Zhang, F.L.; Liu, Q.Q. The effects of Zibu Piyin Recipe components on scopolamine-induced learning and memory impairment in the mouse. J. Ethnopharmacol. 2013, 151, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.P.; Yang, K.; Zheng, L.S.; You, C.X.; Cai, Q.; Wang, C.F. Repellant and insecticidal activities of shyobunone and isoshyobunone derived from the essential oil of Acorus calamus rhizomes. Pharmacogn. Mag. 2015, 11, 675. [Google Scholar] [PubMed]
- Huang, Y.Z.; Hua, H.X.; Li, S.G.; Yang, C.J. Contact and fumigant toxicities of calamusenone isolated from Acorus gramineus rhizome against adults of Sitophilus zeamais and Rhizopertha dominica. Insect Sci. 2011, 18, 181–188. [Google Scholar] [CrossRef]
- Scur, M.C.; Pinto, F.G.; Pandini, J.A.; Costa, W.F.; Leite, C.W.; Temponi, L.G. Antimicrobial and antioxidant activity of essential oil and different plant extracts of Psidium cattleianum Sabine. Braz. J. Biol. 2016, 76, 1012–1015. [Google Scholar] [CrossRef] [PubMed]
- Chinese Pharmacopoeia Commission. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2015. [Google Scholar]
- Pérez-Meseguer, J.; Garza-Juárez, A.; Salazar-Aranda, R.; Salazar-Cavazos, M.L.; de La Torre Rodríguez, Y.C.; Rivas-Galindo, V.; Waksman de Torres, N. Development and Validation of an HPLC-DAD Analytical Procedure for Quality Control of Damiana (Turnera diffusa), Using an Antioxidant Marker Isolated from the Plant. J. AOAC Int. 2010, 93, 1161–1168. [Google Scholar] [PubMed]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Saptoro, A.; Tadé, M.O.; Vuthaluru, H. A Modified Kennard-Stone Algorithm for Optimal Division of Data for Developing Artificial Neural Network Models. Chem. Prod. Proc. Model. 2015, 7, 1–14. [Google Scholar] [CrossRef]
- Simpson, M.B. Near-Infrared Spectroscopy for Process Analytical Technology: Theory, Technology and Implementation. In Process Analytical Technology: Spectroscopic Tools and Implementation Strategies for the Chemical and Pharmaceutical Industries; Bakeev, K.A., Ed.; John Wiley & Sons, Ltd.: New York, NY, USA, 2010; Chapter 5; pp. 107–155. [Google Scholar]
- Chen, D.; Shao, X.; Hu, B.; Su, Q. Simultaneous wavelength selection and outlier detection in multivariate regression of near-infrared spectra. Anal. Sci. 2005, 21, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.S.; Liang, L.Z.; Xu, Q.Z.; Li, H.D.; Chen, X. A new strategy of outlier detection for QSAR/QSPR. J. Comput. Chem. 2009, 31, 592–602. [Google Scholar] [CrossRef] [PubMed]

| Matrix | nLV | R2 | Q2 | RMSEC | RMSEP | RMSECV |
|---|---|---|---|---|---|---|
| 49 × 80 a | 8 | 0.8284 | 0.7824 | 3.2597 | 3.2210 | 5.0261 |
| 43 × 80 b | 6 | 0.9090 | 0.8124 | 2.3084 | 3.0996 | 3.4109 |
| 43 × 15 c | 5 | 0.9069 | 0.7969 | 2.3440 | 3.1713 | 2.9716 |
| 43 × 11 d | 6 | 0.8955 | 0.8284 | 2.4745 | 2.9641 | 3.3196 |
| 43 × 10 e | 5 | 0.9050 | 0.7940 | 2.3679 | 3.1932 | 2.9927 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Zhang, B.; Xin, Z.; Ren, D.; Yi, L. GC-MS Fingerprinting Combined with Chemometric Methods Reveals Key Bioactive Components in Acori Tatarinowii Rhizoma. Int. J. Mol. Sci. 2017, 18, 1342. https://doi.org/10.3390/ijms18071342
Liu W, Zhang B, Xin Z, Ren D, Yi L. GC-MS Fingerprinting Combined with Chemometric Methods Reveals Key Bioactive Components in Acori Tatarinowii Rhizoma. International Journal of Molecular Sciences. 2017; 18(7):1342. https://doi.org/10.3390/ijms18071342
Chicago/Turabian StyleLiu, Wenbin, Bingyang Zhang, Zhongquan Xin, Dabing Ren, and Lunzhao Yi. 2017. "GC-MS Fingerprinting Combined with Chemometric Methods Reveals Key Bioactive Components in Acori Tatarinowii Rhizoma" International Journal of Molecular Sciences 18, no. 7: 1342. https://doi.org/10.3390/ijms18071342