Gene Expression in Osteolysis: Review on the Identification of Altered Molecular Pathways in Preclinical and Clinical Studies
Abstract
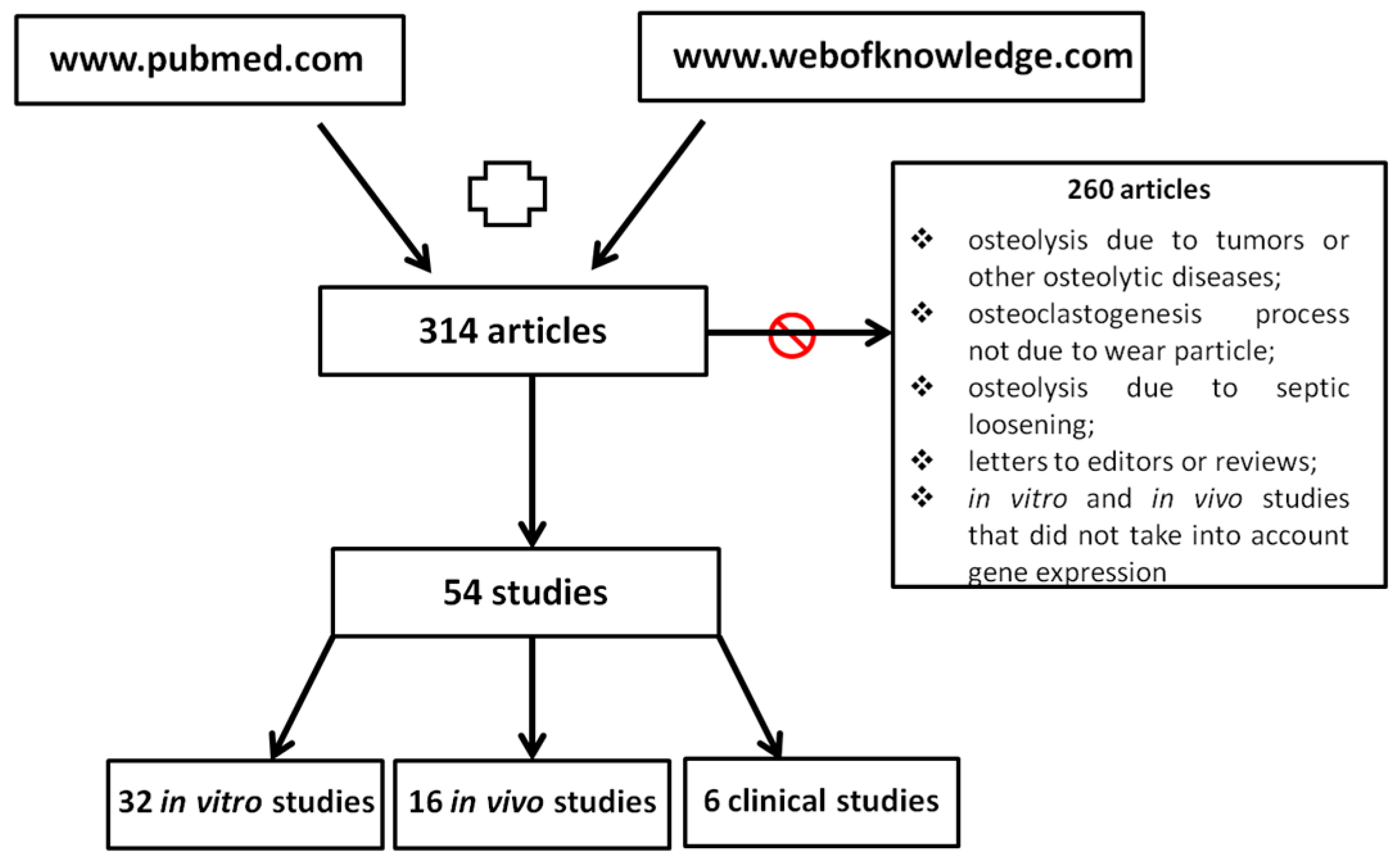
:1. Introduction
2. Gene Expression in Osteolysis
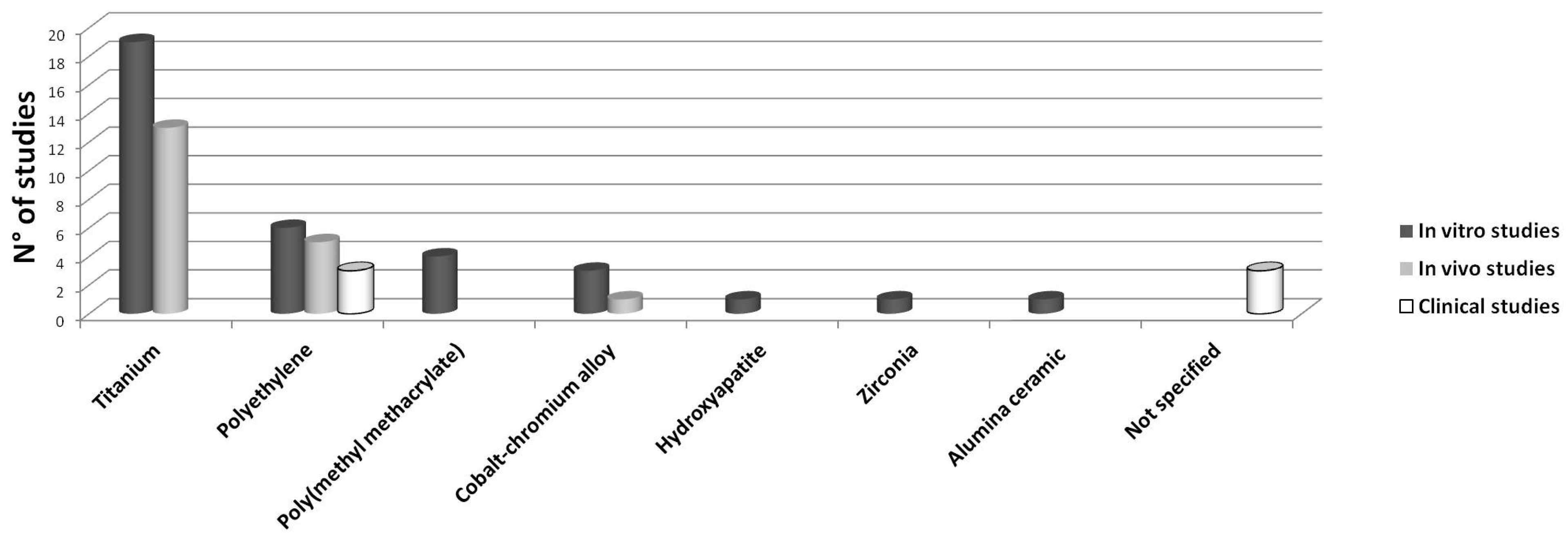
2.1. In Vitro Studies
2.1.1. Ti Particles (19/34 Studies)
2.1.2. PE Particles (7/34 Studies)
2.1.3. PMMA Particles (3/34 Studies)
2.1.4. Co-Cr Alloy (3/34 Studies)
2.1.5. Other Than Genes
2.2. In Vivo Studies
2.2.1. Ti Particles (12/16 Studies)
2.2.2. PE Particles (4/16 Studies)
2.2.3. Other Than Genes
2.3. Clinical Studies
2.4. Other Than Genes
3. Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ø | Diameter |
| AKT | Protein kinase B |
| ALP | Alkaline phosphatase |
| BAD | Bcl-2-associated death promoter |
| TpBCL2 | B-cell lymphoma 2 |
| Bglap | Osteocalcin gene |
| BMD | Bone mineral density |
| BMM | Bone marrow monocytes |
| BMP | Bone morphogenetic protein |
| BS/BV | Bone surface/bone volume |
| BV | Bone volume |
| BV/TV | Bone volume/tissue volume |
| Calcr | Calcitonin receptor gene |
| CAT | Chloramphenicol acetyltransferase |
| Catk | Cathepsin k gene |
| CB | Cannabinoid receptor |
| CCL | C–C motif chemokine ligand |
| CCR | C–C motif chemokine receptor |
| CD | Cluster of differentiation |
| CE | Aluminia ceramic |
| CGRPR1 | Calcitonin gene-related peptide type 1 receptor |
| CHIT1 | Chitinase 1 |
| CHOP | C/EBP homologous protein |
| CM | Conditioned medium |
| Cnr2 | Cannabinoid receptor type 2 gene |
| COLL I | Collagen type I |
| COMP | Cartilage oligomeric matrix protein |
| COX | Cyclooxygenase |
| CREB | cAMP response element-binding protein |
| CSF1 | Colony stimulating factor 1 |
| CXCL | C-X-C motif chemokine ligand |
| CXCR | C-X-C motif chemokine receptor |
| CYS1 | Cystin 1 |
| DAP12 | TYRO protein tyrosine kinase binding protein |
| DC-STAMP | Dendritic cells (DC)-specific transmembrane protein |
| DKK | Dickkopf-related protein |
| E | Estrogen |
| ERK | Extracellular signal–regulated kinases |
| FAM213 | Family with sequence similarity 213 Member A |
| FB | Fibroblasts |
| FGFR | Receptor of fibroblast growth factor |
| Gpx | Glutathione peroxidase |
| GR | Glutathione reductase |
| HA | Hydroxyapatite |
| HDP | High density polyethylene |
| HPRT1 | hypoxanthine phosphoribosyltransferase 1 |
| IGF | Insulin-like growth factor |
| IL | Interleukin |
| Il | Interleukin gene |
| Il1ra | Interleukin 1 receptor antagonist gene |
| iNOS | Inducible nitric oxide synthase |
| IRE-1 | Inositol-requiring kinase 1 |
| KREMEN1 | Kringle containing transmembrane |
| LDH | Lactate dehydrogenase |
| Lrp5 | Low-density lipoprotein receptor-related protein 5 gene |
| MAPK | Mitogen-activated protein kinase |
| MCP-1 | Monocyte chemoattractant protein-1 |
| MD2 | Myeloid differentiation protein-2 |
| MhcII | Major histocompatibility complex class II |
| MIP-1α | Macrophage inflammatory protein-1 alpha |
| MMP | Matrix metalloprotease |
| MRC | Mannose receptor C |
| MSC | Mesenchymal stem cells |
| MSK | Mitogen- and stress-activated protein kinase |
| Myd | Myeloid differentiation primary response gene |
| Nell1 | Neural EGFL like 1 |
| Nfatc1 | Nuclear factor of activated T-cells, cytoplasmic 1 gene |
| Nfκb | Nuclear factor κ-B |
| NO | Nitric oxide |
| NOD | Nodulation factor |
| NP | Nanoparticles |
| OA | Osteoarthritis |
| OB | Osteoblasts |
| OC | Osteoclasts |
| OCN | Osteonectin |
| OPG | Osteoprotegerin |
| OPN | Osteopontin |
| OSCAR | Osteoclast-associated immunoglobulin-like receptor |
| Vegf | Vascular endothelial growth factor gene |
| P2RY1 | Purinergic receptor P2Y1 |
| P2RY6 | Pyrimidinergic receptor P2Y6 |
| Pdgf | Platelet derived growth factor gene |
| PGE2 | Prostaglandin E2 |
| PIK3CB | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta |
| PIK3R2 | Phosphoinositide-3-kinase regulatory subunit |
| PMMA | Poly methyl methacrilate |
| Ptges | Prostaglandin E synthase gene |
| pz | Patients |
| RANK | Receptor activator of nuclear factor κ-B |
| RANKL | Receptor activator of nuclear factor κ-B ligand |
| ROS | Reactive oxygen species |
| Rpl19 | Ribosomal protein L19 gene |
| Runx2 | Runt-related transcription factor 2 gene |
| SF | Synovial fibroblasts |
| SFRP1 | Secreted frizzled related protein |
| SOCS3 | Suppressor of cytokine signaling 3 |
| SOD | Superoxide dismutase |
| SOST | Sclerostin |
| SOX9 | Sry-related HMG box |
| Sp7 | Osterix gene |
| SPP1 | Secreted phosphoprotein 1 |
| SQSTM1 | Sequestosome 1 |
| SSA | Sagittal suture area |
| TAK | Transforming growth factor beta-activated kinase |
| Tb.Th | Trabecular thickness |
| THA | Total hip arthroplasty |
| THR | Total hip replacement |
| Ti | Titanium |
| Ticam | Toll-like receptor adaptor molecule gene |
| TiO2-M | Micro-sized titanium dioxide particles |
| TiO2-N | Nano-sized titanium dioxide particles |
| TIRAP | TIR domain containing adaptor protein |
| Tlr | toll-like receptor gene |
| Tnfrsf | TNF receptor superfamily gene |
| Tnfsf11 | RANKL gene |
| Tnfrsf11b | Osteoprotegerin gene |
| Tnfrsf11a | RANK gene |
| TNFα | Tumor necrosis factor α |
| Tnf | Tumor necrosis factor α gene |
| TKR | Total knee replacement |
| TRAF | TNF receptor associated factor |
| TRAM | Translocation associated membrane protein |
| Trap | Tartrate-resistant acid phosphatase gene |
| Trem | Triggering receptor expressed on myeloid cells gene |
| Trif | TIR-domain-containing adapter-inducing interferon-β gene |
| UHMWPE | Ultra high molecular weight polyethylene |
References
- Drynda, A.; Singh, G.; Buchhorn, G.H.; Awiszus, F.; Ruetschi, M.; Feuerstein, B.; Kliche, S.; Lohmann, S.H. Metallic wear debris may regulate CXCR4 expression in vitro and in vivo. J. Biomed. Mater. Res. A 2015, 103, 1940–1948. [Google Scholar] [CrossRef] [PubMed]
- Chiu, R.; Smith, K.E.; Ma, G.K.; Ma, T.; Smith, R.L.; Goodman, S.B. Polymethylmethacrylate particles impair osteoprogenitor viability and expression of osteogenic transcription factors Runx2, osterix, and Dlx5. J. Orthop. Res. 2010, 28, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Saad, S.; Dharmapatni, A.A.; Crotti, T.N.; Cantley, M.D.; Algate, K.; Findlay, D.M.; Atkins, G.J.; Haynes, D.R. Semaphorin-3a, neuropilin-1 and plexin-A1 in prosthetic-particle induced bone loss. Acta Biomater. 2016, 30, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.P.; Wei, J.L.; Tian, Q.Y.; Liu, A.T.; Yi, Y.S.; Einhorn, T.A.; Liu, C.J. Progranulin suppresses titanium particle induced inflammatory osteolysis by targeting TNFα signaling. Sci. Rep. 2016, 6, 20909. [Google Scholar] [CrossRef] [PubMed]
- Cordova, L.A.; Stresing, V.; Gobin, B.; Rosset, P.; Passuti, N.; Gouin, F.; Trichet, V.; Layrolle, P.; Heymann, D. Orthopaedic implant failure: Aseptic implant loosening—The contribution and future challenges of mouse models in translational research. Clin. Sci. 2014, 127, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Thomas, V.; Halloran, B.A.; Ambalavanan, N.; Catledge, S.A.; Vohra, Y.K. In vitro studies on the effect of particle size on macrophage responses to nanodiamond wear debris. Acta Biomater. 2012, 8, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Cobelli, N.; Scharf, B.; Crisi, G.M.; Hardin, J.; Santambrogio, L. Mediators of the inflammatory response to joint replacement devices. Nat. Rev. Rheumatol. 2011, 7, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, S.D.; Seyler, T.M.; Bennett, D.; Delanois, R.E.; Saleh, K.J.; Thongtrangan, I.; Kuskowski, M.; Cheng, E.Y.; Sharkey, P.F.; Parvizi, J.; et al. Total hip arthroplasties: What are the reasons for revision? Int. Orthop. 2008, 32, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Willert, H.G.; Semlitsch, M. Reactions of the articular capsule to wear products of artificial joint prostheses. J. Biomed. Mater. Res. 1977, 11, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Haynes, D.R.; Rogers, S.D.; Hay, S.; Pearcy, M.J.; Howie, D.W. The differences in toxicity and release of bone-resorbing mediators induced by titanium and cobalt-chromium-alloy wear particles. J. Bone Jt. Surg. Am. 1993, 75, 825–834. [Google Scholar] [CrossRef]
- Ohlin, A.; Johnell, O.; Lerner, U.H. The pathogenesis of loosening of total hip arthroplasties. The production of factors by periprosthetic tissues that stimulate in vitro bone resorption. Clin. Orthop. Relat. Res. 1990, 253, 287–296. [Google Scholar]
- Howie, D.W.; Cornish, B.L.; Vernon-Roberts, B. Resurfacing hip arthroplasty. Classification of loosening and the role of prosthesis wear particles. Clin. Orthop. Relat. Res. 1990, 255, 144–159. [Google Scholar]
- Gallo, J.; Goodman, S.B.; Konttinen, Y.T.; Raska, M. Particle disease: Biologic mechanisms of periprosthetic osteolysis in total hip arthroplasty. Innate Immun. 2013, 19, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Hallab, N.J.; Jacobs, J.J. Biologic effects of implant debris. Bull. NYU Hosp. Jt. Dis. 2009, 67, 182–188. [Google Scholar] [PubMed]
- Nich, C.; Takakubo, Y.; Pajarinen, J.; Ainola, M.; Salem, A.; Sillat, T.; Rao, A.J.; Raska, M.; Tamaki, Y.; Takagi, M.; et al. Macrophages‑Key cells in the response to wear debris from joint replacements. J. Biomed. Mater. Res. A 2013, 101, 3033–3045. [Google Scholar] [CrossRef] [PubMed]
- Geng, D.; Mao, H.; Wang, J.; Zhu, X.; Huang, C.; Chen, L.; Yang, H.; Xu, Y. Protective effects of COX-2 inhibitor on titanium-particle-induced inflammatory osteolysis via the down-regulation of RANK/RANKL. Acta Biomater. 2011, 7, 3216–3221. [Google Scholar] [CrossRef] [PubMed]
- Watters, T.S.; Cardona, D.M.; Menon, K.S.; Vinson, E.N.; Bolognesi, M.P.; Dodd, L.G. Aseptic lymphocyte-dominated vasculitis-associated lesion: A clinicopathologic review of an underrecognized cause of prosthetic failure. Am. J. Clin. Pathol. 2010, 134, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Nich, C.; Langlois, J.; Marchadier, A.; Vidal, C.; Cohen-Solal, M.; Petite, H.; Hamadouche, M. Oestrogen deficiency modulates particle-induced osteolysis. Arthritis Res. Ther. 2011, 13, R100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Fang, Y.; Wang, Q.; Sun, Y.; Xiong, C.; Cao, L.; Wang, B.; Bao, N.; Zhao, J. Tumor necrosis factor-like weak inducer of apoptosis regulates particle-induced inflammatory osteolysis via the p38 mitogen-activated protein kinase signaling pathway. Mol. Med. Rep. 2015, 12, 1499–1505. [Google Scholar] [PubMed]
- Jablonski, H.; Kauther, M.D.; Bachmann, H.S.; Jäger, M.; Wedemeyer, C. Calcitonin gene-related peptide modulates the production of pro-inflammatory cytokines associated with periprosthetic osteolysis by THP-1 macrophage-like cells. Neuroimmunomodulation 2015, 22, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Sundfeldt, M.; Carlsson, L.V.; Johansson, C.B.; Thomsen, P.; Gretzer, C. Aseptic loosening, not only a question of wear: A review of different theories. Acta Orthop. 2006, 77, 177–197. [Google Scholar] [CrossRef] [PubMed]
- Purdue, P.E.; Koulouvaris, P.; Nestor, B.J.; Sculco, T.P. The central role of wear debris in periprosthetic osteolysis. HSS J. 2006, 2, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Della Valle, C.J.; Mesko, N.W.; Quigley, L.; Rosenberg, A.G.; Jacobs, J.J.; Galante, J.O. Primary total hip arthroplasty with a porous-coated acetabular component. A concise follow-up, at minimum of twenty years, of previous reports. J. Bone Jt. Surg. Am. 2009, 91, 1130–1135. [Google Scholar] [CrossRef] [PubMed]
- Dallari, D.; Fini, M.; Giavaresi, G.; del Piccolo, N.; Stagni, C.; Amendola, L.; Rani, N.; Gnudi, S.; Giardino, R. Effect of Pulsed Electromagnetic stimulation on patient undergoing hip revision prosthesis: A randomized prospective double-blind study. Bioelectromagnetics 2009, 30, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Mertens, M.T.; Sing, J.A. Biomarkers in Arthroplasty: A Systematic Review. Open Orthop. J. 2011, 16, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Holt, G.; Murnaghan, C.; Reilly, J.; Meek, R.M. The biology of aseptic osteolysis. Clin. Orthop. Relat. Res. 2007, 460, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhang, J.; Hao, S.; Jin, Q. Adenovirus-mediated small interfering RNA targeting tumor necrosis factor-α inhibits titanium particle-induced osteoclastogenesis and bone resorption. Int. J. Mol. Med. 2013, 32, 296–306. [Google Scholar] [PubMed]
- Lee, H.G.; Hsu, A.; Goto, H.; Nizami, S.; Lee, J.H.; Cadet, E.R.; Tang, P.; Shaji, R.; Chandhanayinyong, C.; Kweon, S.H.; et al. Aggravation of inflammatory response by costimulation with titanium particles and mechanical perturbations in osteoblast- and macrophage-like cells. Am. J. Physiol. Cell Physiol. 2013, 304, C431–C439. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Pan, X.; Zhao, S.; Peng, X.; Cheng, T.; Zhang, X. Protection against Titanium Particle-Induced Inflammatory Osteolysis by the Proteasome Inhibitor Bortezomib In Vivo. Inflammation 2012, 35, 1378–1391. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Li, Z.; Wang, Y.; Wang, H.; Zhang, Z.; Chen, W.; Zhang, Y.; Xiao, Y.; Li, C.; Guo, Y.; et al. Resveratrol protects against titanium particle-induced aseptic loosening through reduction of oxidative stress and inactivation of NF-κB. Inflammation 2016, 39, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Geng, D.C.; Xu, Y.Z.; Yang, H.L.; Zhu, X.S.; Zhu, G.M.; Wang, X.B. Inhibition of titanium particle-induced inflammatory osteolysis through inactivation of cannabinoid receptor 2 by AM630. Biomed. Mater. Res. Part A 2010, 95, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Peng, X.C.; Li, F.F.; Zhang, X.L.; Hu, K.Z.; Zhu, J.F.; Zeng, B.F. Transforming growth factor-b activated kinase 1 signaling pathways regulate TNF-α production by titanium alloy particles in RAW264.7 cells. J. Biomed. Mater. Res. 2010, 93, 1493–1499. [Google Scholar]
- Zhou, F.; Lu, J.; Zhu, X.; Mao, H.; Yang, H.; Geng, D.; Xu, Y. Effects of a Cannabinoid Receptor 2 Selective Antagonist on the Inflammatory Reaction to Titanium Particles In Vivo and In Vitro. J. Int. Med. Res. 2010, 38, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.B.; Ding, Y.; Huang, D.S.; Zeng, W.K.; Guan, Z.P.; Zhang, M.L. RNA interference targeting p110β reduces tumor necrosis factor-alpha production in cellular response to wear particles in vitro and osteolysis in vivo. Inflammation 2013, 36, 1041–1054. [Google Scholar] [CrossRef] [PubMed]
- Cang, D.; Guo, K.; Zhao, F. Dendritic cells enhance UHMWPE wear particle-induced osteoclast differentiation of macrophages. J. Biomed. Mater. Res. Part A 2015, 103, 3349–3354. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Xu, Z.; Wooley, P.H.; Zhang, J.; Yang, S.Y. Therapeutic potentials of naringin on polymethylmethacrylate induced osteoclastogenesis and osteolysis, in vitro and in vivo assessments. Drug Des. Dev. Ther. 2014, 8, 1–11. [Google Scholar]
- Sartori, M.; Vincenzi, F.; Ravani, A.; Cepollaro, S.; Martini, L.; Varani, K.; Fini, M.; Tschon, M. RAW264.7 co-cultured with ultra-high molecular weight polyethylene particles spontaneously differentiate into osteoclasts: An in vitro model of periprosthetic osteolysis. J. Biomed. Mater. Res. Part A 2017, 105, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Zhao, F.; Tang, X.; Pei, F.; Dong, H.; Zhu, L.; Guo, K. Type-2 cannabinoid receptor regulates proliferation, apoptosis, differentiation, and OPG/RANKL ratio of MC3T3-E1 cells exposed to Titanium particles. Mol. Cell. Biochem. 2015, 399, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Minematsu, H.; Kim, K.O.; Celil Aydemir, A.B.; Shin, M.J.; Nizami, S.A.; Chung, K.J.; Hsu, A.C.; Jacobs, C.R.; Lee, F.Y. Actin and ERK1/2-CEBPb signaling mediates phagocytosis-induced innate immune response of osteoprogenitor cells. Biomaterials 2011, 32, 9197–9206. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, K.; Li, F.; Jiang, J.; Jia, T.; Yang, S.Y. Biological responses of preosteoblasts to particulate and ion forms of Co-Cr alloy. Biomed. Mater. Res. Part A 2015, 103, 3564–3571. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Wang, W.; Li, X.; Markel, D.C.; Ren, W. Mitigative Effect of Erythromycin on PMMA Challenged Preosteoblastic MC3T3-E1 Cells. Sci. World J. 2014, 2014, 107196. [Google Scholar] [CrossRef] [PubMed]
- Vallés, G.; Pérez, C.; Boré, A.; Martín-Saavedra, F.; Saldaña, L.; Vilaboa, N. Simvastatin prevents the induction of interleukin-6 gene expression by titanium particles in human osteoblastic cells. Acta Biomater. 2013, 9, 4916–4925. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kauther, M.D.; Hartl, J.; Wedemeyer, C. Effects of α-calcitonin gene-related peptide on osteoprotegerin and receptor activator of nuclear factor-κB ligand expression in MG-63 osteoblastlike cells exposed to polyethylene particles. J. Orthop. Surg. Res. 2010, 5, 83. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, L.; Yu, X.; Gu, Z.; Zhang, X. The inhibitory effect of strontium-doped calcium polyphosphate particles on cytokines from macrophages and osteoblasts leading to aseptic loosening in vitro. Biomed. Mater. 2014, 9, 025010. [Google Scholar] [CrossRef] [PubMed]
- Schoenenberger, A.D.; Schipanski, A.; Malheiro, V.; Kucki, M.; Snedeker, J.G.; Wick, P.; Maniura-Weber, K. Macrophage polarization by titanium dioxide (TiO2) particles: Size matters. ACS Biomater. Sci. Eng. 2016, 2, 908–919. [Google Scholar] [CrossRef]
- Potnis, P.A.; Dutta, D.K.; Wood, S.C. Toll-like receptor 4 signaling pathway mediates proinflammatory immune response to cobalt-alloy particles. Cell. Immunol. 2013, 282, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Fujii, J.; Niida, S.; Yasunaga, Y.; Yamasaki, A.; Ochi, M. Wear debris stimulates bone-resorbing factor expression in the fibroblasts and osteoblasts. Hip Int. 2011, 21, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Kauther, M.D.; Xu, J.; Wedemeyer, C. α-Calcitonin Gene-Related Peptide Can Reverse the Catabolic Influence of UHMWPE Particles on RANKL Expression in Primary Human Osteoblasts. Int. J. Biol. Sci. 2010, 6, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.X.; Wu, C.L.; Zhu, Z.; Li, M.Q.; Mao, Y.Q.; Liu, M.; Wang, X.Q.; Yu, D.G.; Tang, T.T. Calcineurin/NFAT pathway mediates wear particle-induced TNF-α release and osteoclastogenesis from mice bone marrow macrophages in vitro. Acta Pharmacol. Sin. 2013, 34, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Xu, Y.; Zhu, M.; Gu, Y.; Zhang, W.; Shao, H.; Wang, Y.; Ping, Z.; Hu, X.; Wang, L.; et al. Inhibition of titanium-particle-induced inflammatory osteolysis after local administration of dopamine and suppression of osteoclastogenesis via D2-like receptor signaling pathway. Biomaterials 2016, 80, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Jia, T.; Gong, W.; Ning, B.; Wooley, P.H.; Yang, S.Y. Macrophages polarization in IL-10 treatment of particle-induced inflammation and osteolysis. Am. J. Pathol. 2016, 186, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Haleem-Smith, H.; Argintar, E.; Bush, C.; Hampton, D.; Postma, W.F.; Chen, F.H.; Rimington, T.; Lamb, J.; Tuan, R.S. Biological responses of human mesenchymal stem cells to titanium wear debris particles. J. Orthop. Res. 2012, 30, 853–863. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, N.; Shi, T.; Zhou, G.; Wang, Z.; Gan, J.; Guo, T.; Qian, H.; Bao, N.; Zhao, J. ER Stress Mediates TiAl6V4 Particle-Induced Peri-Implant Osteolysis by Promoting RANKL Expression in Fibroblasts. PLoS ONE 2015, 10, e0137774. [Google Scholar] [CrossRef] [PubMed]
- Obando-Pereda, G.A.; Fischer, L.; Stach-Machado, D.R. Titanium and zirconia particle-induced pro-inflammatory gene expression in cultured macrophages and osteolysis, inflammatory hyperalgesia and edema in vivo. Life Sci. 2014, 97, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, A.; Norgard, M.; Andersson, G.; Fahlgren, A. Fluid Pressure Induces Osteoclast Differentiation Comparably to Titanium Particles but Through a Molecular Pathway Only Partly Involving TNFa. J. Cell. Biochem. 2012, 113, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Zou, Y.; Liu, X.; Shang, J.; Cheng, M.; Dai, M. Dose-dependent effects of lanthanum chloride on wear particle-induced aseptic inflammation in a murine air-pouch model. J. Rare Earths 2013, 31, 420–427. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, X.; Guo, Y.; Shi, S.; Mao, X.; Pan, X.; Cheng, T. MMP-9 inhibition suppresses wear debris-induced inflammatory osteolysis through downregulation of RANK/RANKL in a murine osteolysis model. Int. J. Mol. Med. 2012, 30, 1417–1423. [Google Scholar] [PubMed]
- Geng, D.; Xu, Y.; Yang, H.; Wang, J.; Zhu, X.; Zhu, G.; Wang, X. Protection against titanium particle induced osteolysis by cannabinoid receptor 2 selective antagonist. Biomaterials 2010, 31, 1996–2000. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, Y.; Guo, F.; Lu, Z.; Hei, C.; Li, P.; Jin, Q. Protective effect of p38 MAPK inhibitor on wear debris-induced inflammatory osteolysis through downregulating RANK/RANKL in a mouse model. Genet. Mol. Res. 2015, 14, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Guo, Y.; Mao, X.; Zhang, X. Inhibition of p38 Mitogen-Activated Protein Kinase Down-regulates the Inflammatory Osteolysis Response to Titanium Particles in a Murine Osteolysis Model. Inflammation 2012, 35, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Córdova, L.A.; Trichet, V.; Escriou, V.; Rosset, P.; Amiaud, J.; Battaglia, S.; Charrier, C.; Berreur, M.; Brion, R.; Gouin, F.; et al. Inhibition of osteolysis and increase of bone formation after local administration of siRNA-targeting RANK in a polyethylene particle-induced osteolysis model. Acta Biomater. 2015, 13, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Onodera, S.; Tohyama, H.; Kwon, H.J.; Honma, K.I.; Yasuda, K. In Vivo Imaging of Particle-Induced Inflammation and Osteolysis in the Calvariae of NFκB/Luciferase Transgenic Mice. J. Biomed. Biotechnol. 2011, 2011, 727063. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Peng, J.; Wang, Y.; Wang, A.; Zhang, X.; Yuan, M.; Zhang, L.; Zhao, B.; Liu, B.; Fan, M.; et al. NELL1 Promotes Bone Regeneration in Polyethylene Particle-Induced Osteolysis. Tissue Eng. Part A 2012, 18, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhu, S.; Cui, J.; Shao, H.; Zhang, W.; Yang, H.; Xu, Y.; Geng, D.; Yu, L. Strontium ranelate inhibits titanium-particle-induced osteolysis by restraining inflammatory osteoclastogenesis in vivo. Acta Biomater. 2014, 10, 4912–4918. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Jia, T.; Gong, W.; Wooley, P.H.; Yang, S.Y. Titanium particle-challenged osteoblasts promote osteoclastogenesis and osteolysis in a murine model of periprosthestic osteolysis. Acta Biomater. 2013, 9, 7564–7572. [Google Scholar] [CrossRef] [PubMed]
- MacInnes, S.J.; del Vescovo, E.; Kiss-Toth, E.; Ollier, W.E.; Kay, P.R.; Gordon, A.; Greenfield, E.M.; Wilkinson, M.J. Genetic variation in inflammatory and bone turnover pathways and risk of osteolytic responses to prosthetic materials. J. Orthop. Res. 2015, 33, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Mao, X.; Cheng, T.; Peng, X.; Zhang, X.; Liu, Z.; Wang, Q.; Chen, Y. Up-regulated expression of MIF by interfacial membrane fibroblasts and macrophages around aseptically loosened implants. J. Surg. Res. 2012, 176, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Tomankova, T.; Kriegova, E.; Fillerova, R.; Luzna, P.; Ehrmann, J.; Gallo, J. Comparison of periprosthetic tissues in knee and hip joints: Differential expression of CCL3 and DC-STAMP in total knee and hip arthroplasty and similar cytokine profiles in primary knee and hip osteoarthritis. Osteoarthr. Cartil. 2014, 22, 1851–1860. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.; Greenfield, E.M.; Eastell, R.; Kiss-Toth, E.; Wilkinson, J.M. Individual susceptibility to periprosthetic osteolysis is associated with altered patterns of innate immune gene expression in response to pro-inflammatory stimuli. J. Orthop. Res. 2010, 28, 1127–1135. [Google Scholar] [CrossRef] [PubMed]
- Alias, E.; Dharmapatni, A.S.; Holding, A.C.; Atkins, G.J.; Findlay, D.M.; Howie, D.W.; Crotti, T.N.; Haynes, D.R. Polyethylene particles stimulate expression of ITAM-related molecules in peri-implant tissues and when stimulating osteoclastogenesis in vitro. Acta Biomater. 2012, 8, 3104–3112. [Google Scholar] [CrossRef] [PubMed]
- Michaëlsson, K. Surgeon volume and early complications after primary total hip arthroplasty. BMJ 2014, 348, g3433. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.H.; Tamaki, Y.; Pajarinen, J.; Waters, H.A.; Woo, D.K.; Yao, Z.; Goodman, S.B. Chronic inflammation in biomaterial-induced periprosthetic osteolysis: NF-κB as a therapeutic target. Acta Biomater. 2014, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yarilina, A.; Xu, K.; Chen, J.; Ivashkiv, L.B. TNF activates calcium-nuclear factor of activated T cells (NFAT)c1 signaling pathways in human macrophages. Proc. Natl. Acad. Sci. USA 2011, 108, 1573–1578. [Google Scholar] [CrossRef] [PubMed]
- Gibon, E.; Ma, T.; Ren, P.G.; Fritton, K.; Biswal, S.; Yao, Z.; Smith, L.; Goodman, S.B. Selective inhibition of the MCP-1-CCR2 ligand-receptor axis decreases systemic trafficking of macrophages in the presence of UHMWPE particles. J. Orthop. Res. 2012, 30, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Mine, Y.; Makihira, S.; Nikawa, H.; Murata, H.; Hosokawa, R.; Hiyama, A.; Mimura, S. Impact of Titanium ions on osteoblast-, osteoclast- and gingival epithelial-like cells. J. Prosthodont. Res. 2010, 54, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhang, X.; Zeng, B. Locally administered lentivirus-mediated siRNA inhibits wear debris-induced inflammation in murine air pouch model. Biotechnol. Lett. 2008, 30, 1923–1929. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Yang, S.; Wooley, P. A novel murine model of orthopaedic wear-debris associated osteolysis. Scand. J. Rehabil. Med. 2004, 33, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, E.M.; Benz, E.B.; Lu, A.P.; Goater, J.J.; Mollano, A.V.; Rosier, R.N.; Puzas, J.E.; O’Keefe, R.J. Quantitative small-animal surrogate to evaluate drug efficacy in preventing wear debris-induced osteolysis. J. Orthop. Res. 2000, 18, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yang, X.; Nestor, B.; Bostrom, M.; Camacho, N.; Li, G.; Purdue, E.; Sculco, T. Histologic and FTIR studies on long term effect of PMMA particle in a murine intramedullary osteolysis model. Trans. Orthop. Res. Soc. 2007, 53, 217. [Google Scholar]
- Merriam, F.V.; Wang, Z.Y.; Guerios, S.D.; Bjorling, D.E. Cannabinoid receptor 2 is increased in acutely and chronically inflamed bladder of rats. Neurosci. Lett. 2008, 445, 130–134. [Google Scholar] [CrossRef] [PubMed]
- Park-Min, K.H.; Ji, J.D.; Antoniv, T.; Reid, A.C.; Silver, R.B.; Humphrey, M.B.; Nakamura, M.; Ivashkiv, L.B. IL-10 suppresses calcium-mediated costimulation of receptor activator NF-κB signaling during human osteoclast differentiation by inhibiting TREM-2 expression. J. Immunol. 2009, 183, 2444–2455. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Zhu, T.; Craft, C.S.; Broekelmann, T.J.; Mecham, R.P.; Teitelbaum, S.L. Cytoskeletal dysfunction dominates in DAP12-deficient osteoclasts. J. Cell Sci. 2010, 123, 2955–2963. [Google Scholar] [CrossRef] [PubMed]
- Galvin, A.L.; Tipper, J.L.; Jennings, L.M.; Stone, M.H.; Jin, Z.M.; Ingham, E.; Fisher, I. Wear and biological activity of highly crosslinked polyethylene in the hip under low serum protein concentrations. Proc. Inst. Mech. Eng. Part H 2007, 221, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Doorn, P.F.; Campbell, P.A.; Worrall, J.; Benya, P.D.; McKellop, H.A.; Amstutz, H.C. Metal wear particle characterization from metal on metal total hip replacements: Transmission electron microscopy study of periprosthetic tissues and isolated particles. J. Biomed. Mater. Res. 1998, 42, 103–111. [Google Scholar] [CrossRef]
- Nishihira, J.; Mitsuyama, K. Overview of the role of macrophage migration inhibitory factor (MIF) in inflammatory bowel disease. Curr. Pharm. Des. 2009, 15, 2104–2109. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, K.; Schoppet, M.; Al-Fakhri, N.; Helas, S.; Jessberger, R.; Hofbauer, L.C.; Goettsch, C. The role of osteoclast-associated receptor in osteoimmunology. J. Immunol. 2011, 186, 13–18. [Google Scholar] [CrossRef] [PubMed]
| Type of Wear Particles | Cell Type | Evaluations | Results: Osteolysis vs. Control | Ref. |
|---|---|---|---|---|
| 100 mg/mL Ti alloy particles (ø < 8 µm) | Raw 264.7 | Gene expression (Tnf) Protein levels (TNFα, p-TAK1, TAK1, p-P38, p-38, cytosolic and nuclear p50 and p65) | ↑ TAK1, Tnf, TNFα, nuclear and cytosolic p50, nuclear p65, p-38 | [32] |
| 1 mg/mL ZrO2 or Ti alloy particles (ø 5 µm) | Mice peritoneal macrophages | Gene expression (Tlr2, Tlr3, Tlr4, Tlr9, Myd88, Ticam2, Tnfrsf11a, Tnf, Il1, Il6) Protein levels (IL1β, IL6, TNFα) | ↑ Tlr2, Tlr3, Tlr4, Tlr9, MyD88, Ticam2, Nfκb, Tnf, Il1, Il6, TNFα. ↑ Tlr2, Tlr9; ↓ Tlr3, Tlr4, MyD88, Ticam2, Nfκb, Tnf, Il1, Il6 in ZrO2 than Ti | [54] |
| 0.05, 0.5, 1 mg/mL Ti alloy particles (ø 0.52 µm) | SFs from OA pz | Protein levels (IRE1-α, CHOP, RANKL, OPG, sRANKL) Gene expression (TNFSF11, TNFRSF11B) OCs number | ↑ CHOP, TNFSF11, RANKL, sRANKL, IRE1α (dose-dependent manner) | [53] |
| 0.1 mg/mL Ti particles (ø < 10 µm) | RAW264.7 | Cell viability Gene expression (Tnf, Il1, Il6, Il10, Ccl2, Ccl3, Nos2, Cox2) Protein levels (TNFα, IL1β, IL6, IL10, MCP-1, MIP-1α, iNOS, COX2) | ↑ Tnf, Il1, Il6, Il10, Ccl2, Ccl3, Nos2, Cox2, TNFα, IL1β, IL6, MCP1, MIP1α, iNOS, COX2 ↓ IL10 | [29] |
| 0.1 mg/mL Ti particles (ø 2.6 mm) | Raw264.7 | NO production Intracellular ROS Lipid peroxidation Gene expression (Nos2, Nox1, Nox2, Sod1, Nos3, Gpx, Gr, Catk, Tnf, Nfκb) SOD, CAT, GR activities Protein levels (TNFα, p-NFκB -p65, NFκB-p65) | ↑ NO, ROS, Nos2, Nox1, Nox2, Tnf, Nfκb, TNFα, p- NFκB -p65, GR, SOD, CAT ↓ Sod1, Nos3, Gpx | [30] |
| 0.1 mg/mL Ti particles (ø 4.5 µm) | RAW264.7 | CB2 activity Bone resorption pit assay OCs number Protein levels (IL1β, TNFα) Gene expression (Cnr2, Tnfrsf11a, Ctsk) | ↑ OCs number, Cnr2, Tnfrsf11a, Ctsk, TNFα, IL1β, CB2 activity, resorption area | [31] |
| 0.1 mg/mL Ti particles (ø 4.50 µm) | RAW264.7 | Protein levels (IL1β, TNFα) Gene expression (Il1, Tnf) | ↑ Il1, Tnf, IL1β, TNFα | [33] |
| Calvaria with CM of RAW264.7 stimulated with 1 mg/mL Ti particles RAW264.7 with 1% Ti particles (ø < 3.6 µm) | Calvaria from mice (5-day old) and RAW264.7 | OCs number Gene expression (Trap, Cstk, Calcr, Nfκb, Tnf, Il1, Il6, Cox2, Nos2) Protein levels (TNFα, IL1β, IL6, cytosol and nuclear p65) | Calvaria: ↑ OCs number, Trap, Cstk, Calcr, Nfκb. RAW264.7: ↑ Tnf, Il1, Il6, Cox2, Nos2, NOS2, IL1β, IL6, TNFα, nucleus p65; ↓ cytosol p65 | [4] |
| 4x104–4x106 particles/mL TiO2-N (ø <0.1 µm) or TiO2-M (ø <5 µm) | THP-1 | Protein levels (TNFα, IL-10) Gene expression (TNF, IL10, CXCL10, CCR7, MRC1) | TiO2-M: ↑ TNFα, IL-10, TNF, IL10, CXCL10, CCR7 (at the highest concentration) | [45] |
| 0.1 mg/mL Ti particles (ø 3.6 µm) | Mice BMM | OCs number Bone resorption assay Gene expression (Ctsk, Trap, Nfatc1, Mmp9, Il1, Il6, Tnf) Protein levels (TNFα, IL1β, IL6) | ↑ OCs number, Trap, Nfatc1, Ctsk, Mmp9, Tnf, Il1, Il6, TNFα, IL1β, IL6, resorbed area | [50] |
| TiO2 (ø 8 nm) | RAW264.7 | Cell viability Gene expression (Tnf, Il1, Cxcl2, Ccl2, Vegfa, Pdgfa) | ↓ cell viability, Cxcl2, ↑ Ccl2 | [6] |
| 2.5 mg/mL Ti particles (ø 4.5 µm) | MC3T3-E1 | Gene expression (Cnr2, Tnfrsf11b, Tnfsf11) Caspase-3 activity Protein level (CB2, ALP, OCN, COLL I); Cell viability Apoptosis Mineralization | ↑ Cnr2, CB2, apoptosis, caspase-3 activity, Tnfsf11 ↓ cell viability, ALP, COLL I, OCN, mineralization, Tnfrsf11b/Tnfsf11 | [38] |
| 0.05% (v/v) Ti particles (ø 0.3–1 µm) | MC3T3-E1, RAW264.7, mice primary OBs and BMMs | Gene expression (Il6, Cox2, Il1, Tnf, Fam213A) Protein levels (p-ERK) Cell membrane damage and death | MC3T3-E1:↑ Il6, Cox2, Fam213A, membrane damage ↓ viability. RAW264.7: ↑ Il6, Cox2, Tnf, Il1 membrane damage ↓ viability OBs, BMMs: ↑ Il6, Cox2, Fam213A, Il1, Tnf, p-ERK | [28] |
| 0.1 mg/mL Ti particles (ø < 10000 µm) | MC3T3-E1, RAW264.7 | Gene expression (Tnf, Il6, Il1, Trap, Nfatc1, Csf1, Tnfsf11, Tnfrsf11b) Protein levels (TNFα, NFATc1, BMP2) Cell viability OCs number; Bone pit resorption assay | RAW264.7: ↑ Tnf, Il6, Il1, Nfatc1, NFATc1, OCs number, resorbed area. MC3T3-E1: ↑ Tnfsf11, Il6 | [27] |
| Ti particles (ø 1–3 µm) | MC3T3-E1, hMSCs | Protein levels (p-p44; p-p42, pCREBβ, IL6, PGE2) Gene expression (Il6, Cox2) OCs number | ↑ Il6, Cox2, p-ERK, pCREBPb, IL6, PGE2, OCs number | [39] |
| 2.5 mg/mL Ti particles (ø < 4 µm) | Saos-2, OBs from pz | Cell viability Protein levels (IL6) Gene expression (Il6) | ↑ IL6, Il6 | [40] |
| 100 particles/cell TiO2 particles (ø 0.43 µm) | hMSC from OA pz | ALP activity Mineralization Cell viability Protein levels (IL6, IL8) Gene expression (TNFRSF11B, BCL10, COMP, FGFR2, IGF1, BMP6, SOX9, COLLs, CSF2, BAD, CD70, BCL2) | ↓ cell viability, ALP activity, mineralization, TNFRSF11B, BCL10, COMP, FGFR2, IGF1, BMP6, SOX9, COLLs; ↑ CSF2, BAD, CD70, BCL2 | [52] |
| 0.1 mg/mL Ti (ø 4.5 µm) or PMMA particles (ø 6 µm) | Mice BMM | Phagocytosis OCs number Gene expression (Nfatc1, Trap, Mmp9, CtsK, Calcr) Bone pit resorption assay Protein levels (TNFα, NFATc1) | ↑ OCs number, Nfatc1, Trap, Mmp9, Ctsk, Calcr, resorbed area, TNFα, NFATc1, phagocytosis | [49] |
| 1:100 (cells: particles) Ti or CE particles (ø 0.2 or 1.2 µm) | RAW264.7 | Gene expression (Pik3cb, Pik3r2, Tnf) Protein levels (TNFα, p-AKT) | ↑ Pik3cb, Tnf, TNFα, p-AKT | [34] |
| 5 mg/mL UHMWPE particles (ø 2.6 µm) | RAW264.7 alone or co-cultured with DC2.4 | OCs number Protein levels (TNFα, MCP-1, p65, p-p65) Gene expression (Mmp9, Calcr, Cstk) | Raw264.7: ↑ OCs number, TNFα, MCP-1, p-p65, p65, Mmp9, Ctsk, Calcr. Co-cultures: ↑ OCs number, MCP-1, p-p65, p65, Mmp9, Ctsk, Calcr | [35] |
| 0.5 or 0.75 mg/mL UHMWPE particles (ø < 9 µm) | RAW264.7 | DNA quantification Live/Dead Gene expression (Nfatc1, Tnfrsf11a, Tnfsf11, Tnfrsf11b) Pit resorption assay Protein levels (RANKL, OPG, TNFα, PGE2) NFκB assay | ↓ DNA quantification (at 3 days) ↑ Tnfrsf11a (at 7 days), Tnfsf11 (at 3 days), RANKL, TNFα, PGE2, NFκB activation ↑ Tnfsf11a (at 3 days), Nfatc1, RANKL, resorption activity in 0.75mg/mL PE than 0.5 mg/mL | [37] |
| 1:100 or 1:500 (cells: particles) UHMWPE particles (ø 1.74 µm) | MG-63 | Gene expression (TNFSF11, TNFRSF11B) Protein levels (RANKL, OPG) ALP activity | ↑ RANKL, TNFSF11 ↓ ALP activity. ↑ TNFSF11; ↑ TNFRSF11B, ALP activity in 1:500 than 1:100 | [43] |
| 1:100 or 1:500 (cells: particles) UHMWPE particles (ø 1.74 µm) | Primary hOBs from fractured pz | Gene expression (TNFRSF11B, TNFSF11) Protein levels (RANKL, OPG) ALP activity | ↑ TNFSF11, RANKL ↓ ALP activity ↑ TNFSF11 in 1:500 more than 1:100 | [48] |
| 1:100 or 1:500 (cells: particles) UHMWPE particles (ø 1.74 µm) | THP-1 | Gene expression (TNFRSF11A, TNF) Protein levels (RANK, TNFα, IL1β, IL6, CGRPR1) | ↑ CGRPR1, TNFα, IL1β, IL6, TNF ↑ IL1β, TNFα, TNF, IL6 in 1:500 more than 1:100 | [20] |
| 1 mg/mL UHMWPE, HA particles (ø 10 µm) | ROS 17/2.8 | Protein levels (TNFα, RANKL, OPG) Gene expression (Tnfrsf11b, Tnfsf11) | UHMWPE: ↑ TNFα ↓ OPG, Tnfrsf11b. HA: ↑ TNFα, OPG, Tnfrsf11b | [44] |
| 10 mg/mL HDP particles (ø 8.59 µm) | Primary FBs from the subcutaneous tissues of cranial capsules of mice, OBs from mice | Protein levels (IL6) Gene expression (Tnfsf11, Tnfrsf11b, Il6, Il1, Tnf, Cox2) | FBs: ↑ IL6, Il6 | [47] |
| 5 mg/mL PMMA particles (ø 0.33 µm) | RAW264.7 | TRAP function OCs number Gene expression (Ctsk, Trap, Tnfrsf11a, Cys1) | ↑ TRAP function, OCs number, Trap, Ctsk, Tnfrsf11a | [36] |
| 2 mg/mL PMMA particles (ø 0.1–10 µm) | Rat BMM | Gene expression (Il1, MhcII, Il10, Tgfb1, Ccr2) Protein levels (iNOS) | ↑ Il1, MhcII, Il10, Tgfb1, Ccr2, iNOS | [51] |
| 1 mg/mL PMMA particles (ø 6 mm) | MC3T3-E1 | Cell viability LDH ALP activity; Gene expression (Nfκb, Runx2, Sp7, Bglap) | ↑ LDH, Nfκb ↓ cell viability, ALP activity, Runx2, Sp7, Bglap | [41] |
| 0.3 or 2.5 mg/mL Co-Cr alloy particles (ø 5.7 µm); 62 or 500 µm Co (II) or Cr (III) (ø < 0.2 µm) | MC3T3-E1 | Cell viability ALP activity Gene expression (Ccl2, Tnf, Il6, Tnfsf11, Tnfrsf11b, Nfatc1, Runx2, Sp7, Lrp5) | Co-Cr alloy: ↓ cell viability, ALP activity, Runx2, Sp7 ↑ Nfatc1, Tnf, Il6, Ccl2 (dose-dependent manner). Co(II): ↓ cell viability, ALP activity, Tnfrsf11b, Lrp5, Runx2, Sp7 ↑ Tnfsf11, Tnf, Nfatc1 (dose-dependent manner). Co-Cr alloy: ↑ Ccl2, Tnf, Nfatc1 than Co(II) | [40] |
| 1 mg/mL Co-Ni-Cr-Mo-alloy or Co-Cr-Mo-alloy (ø < 5 µm) | MG-63, Saos-2 | Gene expression (CXCR4, TNF) Protein levels (CXCR4) | ↑ CXCR4, CXCR4, TNF. ↓ CXCR4, CXCR4, TNF in Co-Ni-Cr-Mo more than Co-Cr-Mo | [1] |
| 1:10, 1:100, 1:200, 1:500, 1:1000 (cells: particles) Co-Cr-Mo alloy from THA femoral head (ø 0.81 µm) | THP-1 | Cell viability Cytosol and nuclear protein levels (I-KB, NFκB, IL1β, TNFα, IL8) Gene expression (Il8) | ↑ IL1β (at 1:1000), IL8, Il8 (at ≥1:500), nuclear NFκB ↓ cytosolic I-KB, cytosolic NFκB, cell viability (at 1:1000) | [46] |
| Type of Wear Particles | Animal Number, Species and Model | Evaluations | Results: Osteolysis vs. Control | Ref. |
|---|---|---|---|---|
| 15 mg/mL Ti alloy particles (ø 0.1–20 µm) | 30 Female BALB/c mice (8–10 weeks old). Murine air-pouch model (also with injection of bone) | Infiltrated cells Pouch membrane thickness Bone erosion OCs number Protein levels (MMP9, TNFα, RANK, RANKL) Gene expression (Mmp9, Tnf, Tnfrsf11a, Tnfsf11) | ↑ Infiltrated cells, pouch membrane thickness, bone erosion, MMP9, TNFα, RANK, RANKL, OCs number, Mmp9, Tnf, Tnfrsf11a, Tnfsf11 | [57] |
| Ti-alloy pin (ø 0.8 mm, length 5 mm) | 36 BALB/c mice (10–12 weeks). Mouse pin-implantation model (before and after Ti-alloy implantation, 10 µL and 40 µL of Ti-challenged OBs and Ti particles were injected, respectively) | BMD Bone resorption Infiltrated cells Periprosthetic membrane thickness peak pulling force OCs number Gene expression (Sp7, Nfatc1, Runx2, Ccl2, Il1, Tnfsf11, Trap, Mmp2) | ↓ Pulling force, BMD ↑ bone resorption, infiltrated cells, periprosthetic membrane thickness, OCs number, Mmp2, Il1, Tnf, Tnfsf11, Trap. ↑ periprosthetic membrane thickness, OCs number, Mmp2, Il1, Tnf, Tnfsf11, Trap in Ti-challenged OBs more than Ti | [65] |
| 10 mg Ti particles (ø < 10 µm) | 16 female BALB/C mice (6 weeks old). Murine air-pouch model (also with injection of bone) | Gene expression (Tnfsf11, Tnfrsf11b, Vegfa, Traf6) Protein levels (RANKL, OPG, VEGF, TRAF6); Pouch membrane thickness Infiltrated cells Bone erosion Apoptosis assay CD68+ cells | ↑ Tnfsf11, Vegfa, Traf6, RANKL, VEGF, TRAF6, RANKL/OPG, CD68+ cells, apoptotic CD86+ cells, pouch membrane thickness, infiltrated cells, bone erosion | [29] |
| 10 mg/mL Ti particles (ø < 3.6 µm) | 20 female BALB/c mice (6–8 weeks old). Murine air-pouch model (also with injection of bone) | Protein levels (COX2, RANKL, PGE2, IL1β, TNFα) Gene expression (Tnfrsf11a, Tnfsf11, Cox2, Tnf, Il1) OCs number | ↑ Cox2, Tnfrsf11a, Tnfsf11, Tnf, Il1, TNFα, IL1β, PGE2, COX2, RANKL, OCs number | [16] |
| 10 mg/mL Ti particles (ø < 3.6 µm) | 20 female BALB/c mice (8–10 weeks old). Murine air-pouch model (also with injection of bone) | Gene expression (Tnfrsf11a, Tnfsf11, Cys1, Cnr2) Protein levels (CB2, RANKL) OCs number | ↑ Cnr2, Tnfrsf11a, Tnfsf11, Cys1, CB2, RANKL, OCs number | [58] |
| 50 mg/mL Ti particles (ø < 20 µm) | 30 female BALB/c mice (8–10 weeks old). Murine air-pouch model (also with injection of bone) | Gene expression (Tnfrsf11a, Tnfsf11) Protein levels (RANK, RANKL) Pouch membrane thickness, infiltrated cells Bone erosion OCs number | ↑ Tnfrsf11a, Tnfsf11, RANK, RANKL, bone erosion, pouch membrane thickness, infiltrated cells, OCs number | [59] |
| 50 mg/mL Ti particles (ø < 20 µm) | 30 female BALB/c mice (8–10 weeks old). Murine air-pouch model (also with injection of bone) | Pouch membrane thickness, infiltrated cells Bone erosion OCs number Protein levels (MMP9, TNFα, P38 MAPK, p-p38 MAPK) Gene expression (Mmp9, Tnf) | ↑ Mmp9, Tnf, MMP9, TNFα, p-p38/p-38 MAPK, pouch membrane thickness, bone erosion, infiltrated cells, OCs number | [60] |
| 10 mg/mL Ti particles (ø 4.50 µm) | 20 female BALB/c mice (6–8 weeks old). Murine air-pouch model (also with injection of bone) | Pouch membrane thickness Infiltrated cells Protein levels (IL1β, TNFα) Gene expression (Il1, Tnf) | ↑ Edema, pouch membrane thickness, infiltrated cells, IL1β, TNFα, Il1, Tnf | [33] |
| 10 mg/mL metallic wear particles from the stem of hip prosthesis mainly containing Ti, Co, Cr (ø < 6.67 µm) | 18 male BALB/c mice (8 weeks old). Murine air-pouch model | Pouch membrane thickness Infiltrated cells Gene expression (Tnf, Il1, Nfatc1) Protein levels (IL1β, TNFα, NFκB /p65) | ↑ Pouch membrane thickness, infiltrated cells, Tnf, Nfatc1, Il1, NFκB /p65, TNFα, IL1β | [56] |
| 30 mg/mL Ti particles (ø < 3.6 µm) | 14 C57BL/6J male mice (6–7 weeks old). Murine calvarial model | BV/TV, total porosity OCs number Bone resorption Fibrous tissue Gene expression (Tnfsf11) Protein levels (RANKL) | ↓ BMD, BV, BV/TV; ↑ OCs number, Tnfsf11, RANKL, bone resorption, fibrous tissue | [53] |
| 300 mg/mL Ti particles (ø 0.52 µm) | 14 C57BL/6J mice (6–7 weeks old). Murine calvarial model | OCs number BV/TV, Total porosity, bone resorption area Gene expression (Tnfsf11) | ↑ Bone resorption, OCs number, total porosity, Tnfsf11; ↓ BV/TV | [64] |
| 20 mg Ti particles (ø < 20 µm) | 54 Sprague Dawley rats (12 weeks old). Fluid flow (20 pressure cycles at 0.17 Hz twice a day) or particles induced models | OCs number Gene expression (Tnf, Il1, Cx3cl1, Nos2, Tnfrsf11b, Tnfrsf11a, Ctsk, Mmp9, Trap, Ptges, Ccl2) | Fluid flow: ↑ OCs number, Ptges, Nos2, Il6, Ctsk. Particles: ↑ OCs number, Ptges, Nos2, Tnf, Il1, Ccl2, Il6, Ctsk, Tnfrsf11a | [65] |
| 20 mg dried PE particles (ø 7.23 µm) | 6 C57BL/6 male mice (10 weeks old). Murine calvarial model (0.5 × 0.5 cm2 area of periosteum exposed) | BV/TV OCs number Protein levels (ALP, Osterix) Gene expression (Tnfrsf11a, Ctsk, Tnf, Il6, Il1, Cyc1, Rpl19) | ↑ Osteonecrosis, osteolytic lesions, empty lacunae, ALP, Osterix production, thickening of inflammatory membrane, infiltrated cells, Tnfrsf11a, Ctsk, Tnf, Il6, Il1 | [61] |
| 20 µg PE particles (ø 5.14 µm) | 24 C57BL/6J healthy female mice, 24 C57BL/6J OVX female mice (11 weeks old), 24 C57BL/6J OVX female mice + E. Murine calvarial model (0.5 × 0.5 cm2 area of periosteum exposed) | BV Tb.Th Bone erosion SSA OCs number Protein levels (IL1b, IL6, TNFa, RANKL) Gene expression (Tnfrsf11a, Tnfsf11, Tnfrsf11b) | In healthy and OVX mice ↑ bone erosion, fibrous and granulomatous scar tissue, SSA, OCs number, RANKL, Tnfrsf11a, Tnfsf11/Tnfrsf11b; ↓ BV, Tb.Th. | [18] |
| 10 mg PE particles (ø 7 µm) | 38 female Balb/c mice (6–8 weeks old). Murine calvarial model (1 × 1 cm2 area of periosteum exposed) | BV, BMD, Tb.Th.3D, BS/BV Protein levels (OPN) Gene expression (Nell1, Spp1, Tnfsf11, Runx2) Young’s modulus | ↓ BV, BMD, Tb.Th.3D, Young’s modulus ↑ BS/BV, bone resorption, Spp1, Tnfsf11 | [63] |
| 0.5, 2, 5 or 10 mg PE particles (ø 7 µm) | 61 NFκB/luc tg mice (7–8 weeks old). Murine calvarial model (1 × 1 cm2 area of periosteum exposed) | Total influx Gene expression (Nfκb (p100/p52), Tnf, Il1, Tnfsf11, Cox2) OCs number Bone erosion | ↑ Fibrous granulomatous tissues, bone resorption, OCs number, total influx, luciferase activity, Nfκb (p100/p52), Tnf, Il1, Tnsfs11, Cox2 (dose-dependent manner) | [62] |
| Type of Implant | Patients Groups | Evaluations | Results: Osteolysis vs. Control | Ref. |
|---|---|---|---|---|
| Not specified | (1) Synovial membrane from 15 pz with THR due to fracture of femoral neck (66.9 ± 6.23 years old) (2) Interfacial membrane from 15 pz with osteolysis of failed THR (66.5 ± 7 years old) | Number of FBs, histiocytes; Protein levels (MIF, CD68); Gene expression (Mif) | ↑ MIF, Mif, CD68+ cells, macrophages and FBs number | [67] |
| Not specified | Pseudosynovial membranes from: (1) pz with TKR due to OA (52–79 years old) (2) pz with THR due to OA (28–68 years old) (3) pz with osteolysis due to failed TKR (56–88 years old) (4) pz with osteolysis due to failed THR (31–86 years old) | Protein levels (RANKL, OPG, IL8, CCL3, DC-STAMP, SOCS3) Gene expression (Tnf, Il6, Chit1, Ccl18, Il8, Ccl3, Mmp9, Dcst2, Tnfrsf11b, Tnfsf11, Bmp4, Socs9, Nfκb, Hprt1) | ↑ Chit1, Il8, Mmp9; ↓ Tnf, Tnfrsf11b, Bmp4 in failed TKR for osteolysis ↓ Ccl3, Dcst2 in failed TKR more than in failed hip | [68] |
| Not specified | Synovial membrane from: (1) 3 healthy pz (12–41 years old) (2) 3 pz with TKR due to OA (71–82 years old) (3) 3 pz with osteolysis due to failed TKR (60–74 years old) | Protein levels (TWEAK, p-MAPK, MAPK) Gene expression (Tnfsf12, Mapk14) | ↑ inflammatory cells, hyperplasia, TWEAK, Tnfsf12, p-MAPK in failed TKR for osteolysis more than healthy pz or OA TKR | [19] |
| Cemented metal on conventional PE bearing couple | Peripheral whole blood from: (1) 356 pz with THR due to OA (65 ± 8 years old) (2) 275 pz with THR due to OA with osteolysis (59 ± 9 years old) | Genotyping (Md2, Msk1, Msk2, Myd88, Nod1, Nod2, P2y1, P2y6, P27, Sqstm1, Tlr1, Tlr2, Tlr4, Tlr5, Tlr6, Tlr9, Tram, Tirap, Trif, Dkk1, Kremen2, Lrp5, Lrp6, Sfrp1, Sost, Wnt3a, Tnfrsf11a, Tnfrsf11b, Tnfsf11) | SNPs associated with osteolysis susceptibility: 4 within RANK, 1 within KREMEN2, 1 within OPG, 1 within SFRP1, 1 within TIRAP; SNPs associated with time to implant failure: 2 within LRP6, 1 within LRP5, 1 within NOD2, 1 within SOST, 1 within SQSTH1, 1 within TIRAP, 1 within TRAM | [66] |
| PE particles | (1) Joint capsule and synovium of 12 pz with THR/TKR due to OA (2 male, 10 female; 60–81 years old) (2) Acetabular membrane, femoral membrane, joint capsule of 12 pz with osteolysis due to failed THR/TKR (4 male, 18 female; 60–81 years old) | % area resorbed Gene expression (Oscar, FcRg, Trem2, Dap12) | ↑ % resorbed area, OCs number, Oscar, FcRg, Trem2, Dap12 in THR/TKR due to OA more than failed THR/THR for osteolysis | [70] |
| Cemented stainless steel and cemented all-PE acetabular component | hPBMCs from: (1) 12 pz with THR/TKR due to OA (75 ± 6 years old) (2) 20 pz with osteolysis due to failed THR/TKR (75 ± 6 years old) | Gene expression (Il1a, Il1, Il1ra, Il6, Il10, Il18, Tnf) | ↑ Il1a, Il1, Il6, Il10, Il18, Tnf in THR/TKR due to OA more than failed THR/THR for osteolysis | [69] |
| Particle Types | Studies | Inflammation | Osteoclastogenesis | Osteoblastogenesis |
|---|---|---|---|---|
| Ti | In vitro studies | ↑ Tnfα, Il6, Il1, Nos2, Cox2, Nfκb, Mmp9 | ↑ CatK, Rank, Rankl, Trap, Nfatc1 | ↓ Tnfrsf11b |
| In vivo studies | ↑ Mmp9, Tnfα, Il1, Cox2, Nos2, Il6 | ↑ Nfatc1, Trap, Rank, Rankl | ↓ Tnfrsf11b | |
| PE | In vitro studies | ↑ Tnfα, Il6, Mmp9 | ↑ Cathepsin K, Rank, Rankl | ↓ Tnfrsf11b |
| In vivo studies | ↑ Tnfα, Il1, Il6, Cox2 | ↑ Rank, Rankl, Cathepsin K | ↓ Tnfrsf11b | |
| Clinical studies | ↑ Il1α, Il1, Il6, Tnfα, Il18 | ↑ Oscar, Trem2, Dap12, Fcrg | ||
| PMMA | In vitro studies | ↑ Il1, Nfκb | ↑ Cathepsin K, Rank, Trap | ↓ Runx2, Osx, Ocn |
| Co-Cr | In vitro studies | ↑ Tnf α, Il6 | ↑ Nfatc1, Rankl | ↓ Tnfrsf11b, Runx2, Osx |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veronesi, F.; Tschon, M.; Fini, M. Gene Expression in Osteolysis: Review on the Identification of Altered Molecular Pathways in Preclinical and Clinical Studies. Int. J. Mol. Sci. 2017, 18, 499. https://doi.org/10.3390/ijms18030499
Veronesi F, Tschon M, Fini M. Gene Expression in Osteolysis: Review on the Identification of Altered Molecular Pathways in Preclinical and Clinical Studies. International Journal of Molecular Sciences. 2017; 18(3):499. https://doi.org/10.3390/ijms18030499
Chicago/Turabian StyleVeronesi, Francesca, Matilde Tschon, and Milena Fini. 2017. "Gene Expression in Osteolysis: Review on the Identification of Altered Molecular Pathways in Preclinical and Clinical Studies" International Journal of Molecular Sciences 18, no. 3: 499. https://doi.org/10.3390/ijms18030499








