Comparative Analysis of Fruit Ripening-Related miRNAs and Their Targets in Blueberry Using Small RNA and Degradome Sequencing
Abstract
:1. Introduction
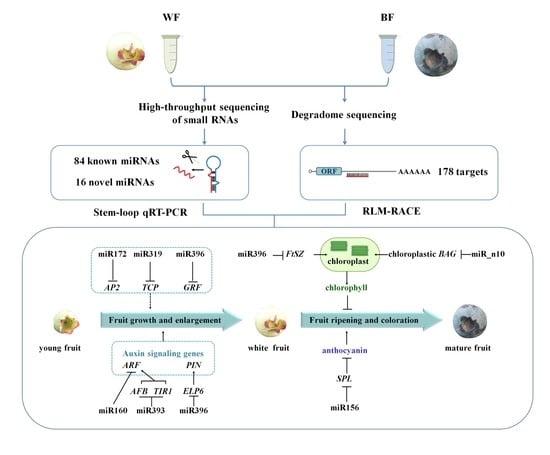
2. Results
2.1. Global Analysis of Sequences from Small RNA and Degradome Libraries
2.2. Identification of Fruit Ripening-Responsive miRNAs in Blueberry
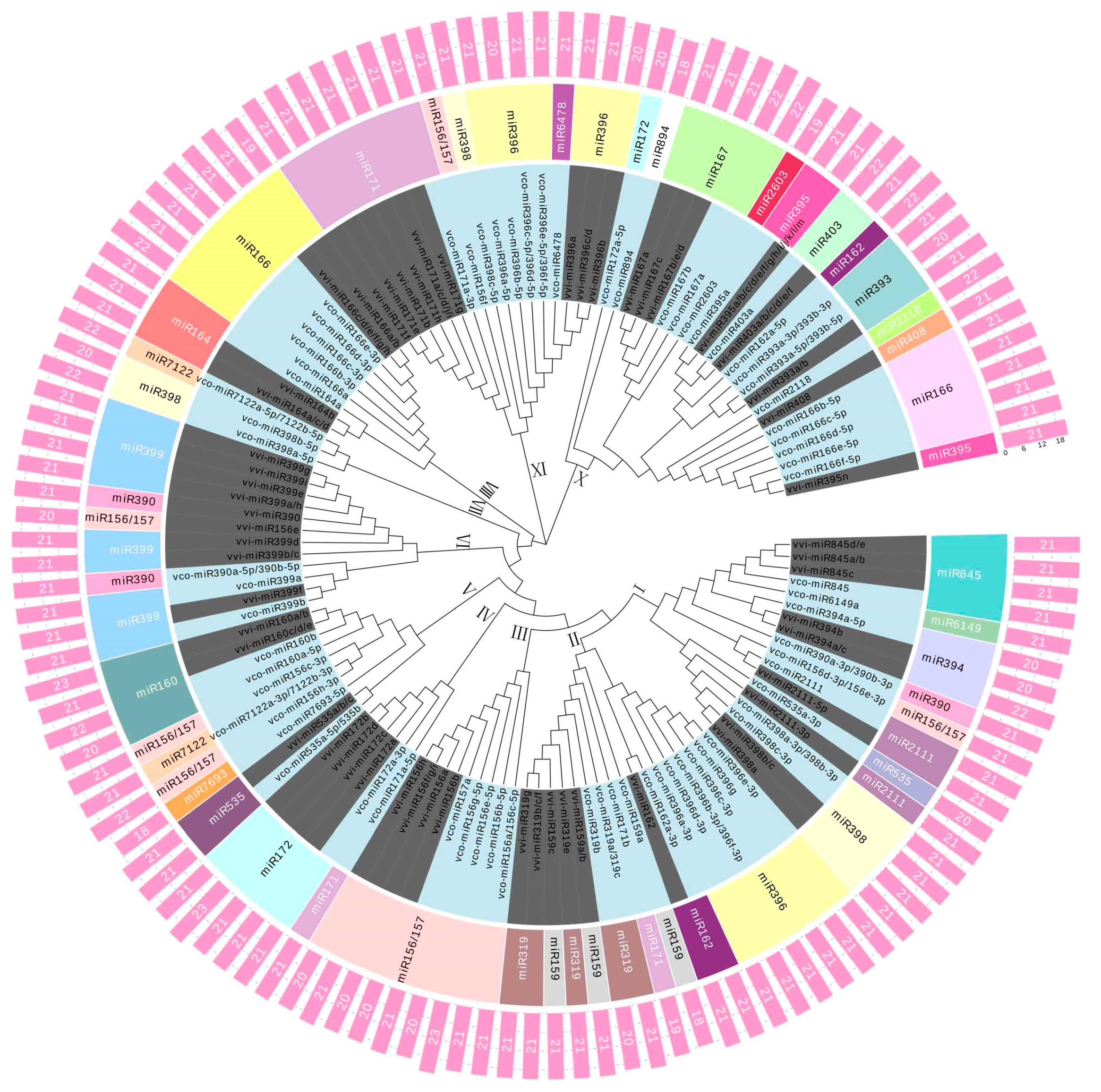
2.3. Phylogenetic Analysis of the Known and Novel miRNAs in Blueberry Fruit
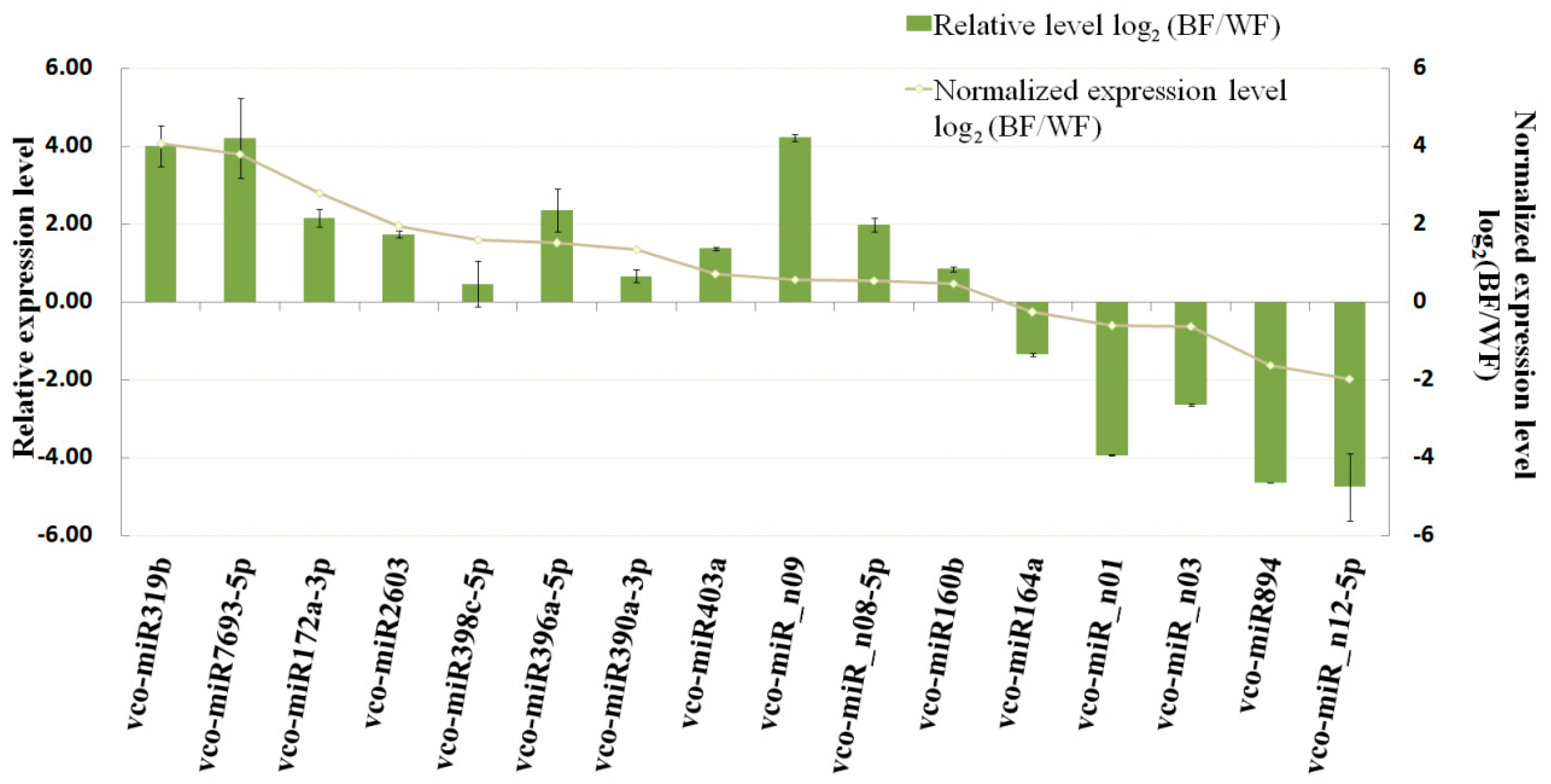
2.4. Accumulation Patterns of Blueberry miRNAs during Fruit Ripening
2.5. Identification of miRNA Targets during Fruit Ripening by Degradome Sequencing
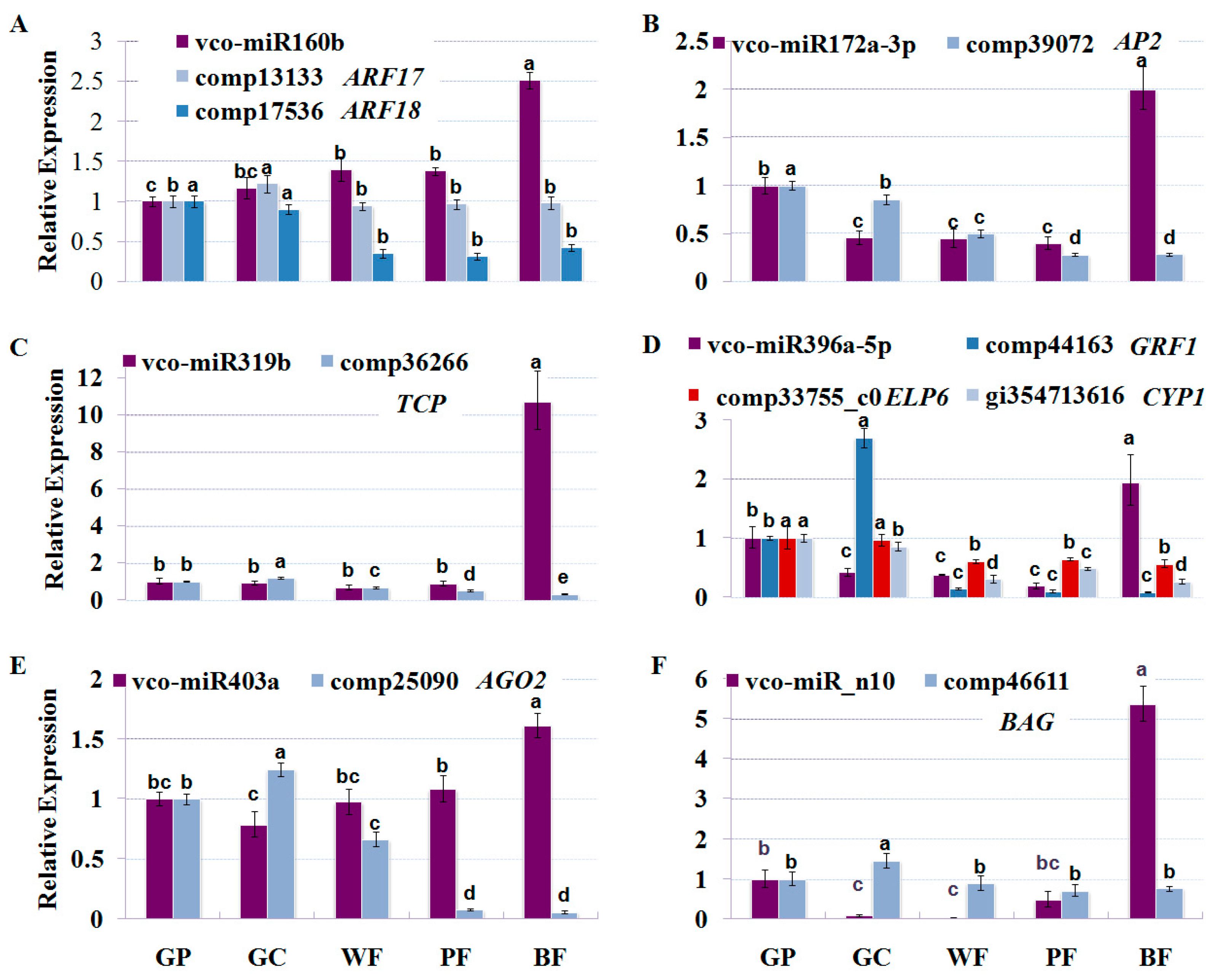
2.6. Dynamic Expression Patterns of Fruit-Related miRNAs and Their Targets during Fruit Development and Maturation
3. Discussion
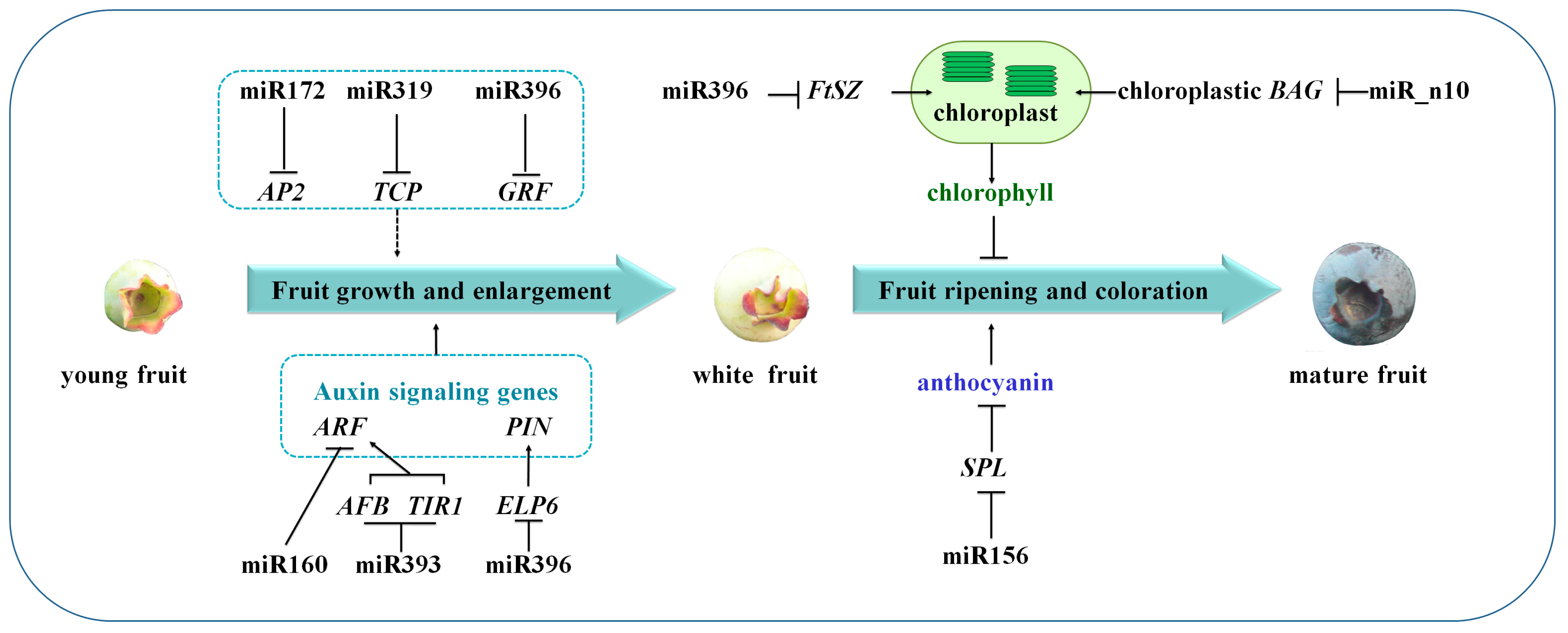
3.1. miRNAs and Their Targets during Blueberry Fruit Maturation
3.2. miRNA-Mediated Regulation Network during Blueberry Fruit Maturation
4. Materials and Methods
4.1. Plant Materials and RNA Extraction
4.2. Construction and Sequencing of sRNA and Degradome Libraries
4.3. Analysis of Small RNA Sequencing Data
4.4. Phylogenetic Analysis of Known miRNAs and Their Precursors in Blueberry
4.5. Differential Expression Analysis of miRNAs Between WF and BF Libraries
4.6. Analysis of Degradome Sequencing Data
4.7. qRT-PCR Validation of miRNAs and Their Potential Targets from Degradome Data
4.8. RNA Ligase-Mediated 5′ RACE
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wu, X.L.; Beecher, G.R.; Holden, J.M.; Haytowitz, D.B.; Gebhardt, S.E.; Prior, R.L. Concentrations of anthocyanins in common foods in the United States and estimation of normal consumption. J. Agric. Food Chem. 2006, 54, 4069–4075. [Google Scholar] [CrossRef] [PubMed]
- Routray, W.; Orsat, V. Blueberries and Their Anthocyanins: Factors Affecting Biosynthesis and Properties. Compr. Rev. Food Sci. Food Saf. 2011, 10, 303–320. [Google Scholar] [CrossRef]
- Rowland, L.J.; Alkharouf, N.; Darwish, O.; Ogden, E.L.; Polashock, J.J.; Bassil, N.V.; Main, D. Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol. 2012, 12, 46. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Estrada, A.D.; Blakley, I.; Reid, R.; Patel, K.; Meyer, M.D.; Andersen, S.U.; Brown, A.F.; Lila, M.A.; Loraine, A.E. RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. Gigascience 2015, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Zhang, H.H.; Liu, Z.S.; Cui, X.Y.; Zhang, T.; Li, Y.F.; Zhang, L.Y. Comparative transcriptome sequencing and de novo analysis of Vaccinium corymbosum during fruit and color development. BMC Plant Biol. 2016, 16, 223. [Google Scholar] [CrossRef] [PubMed]
- Zifkin, M.; Jin, A.; Ozga, J.A.; Zaharia, L.I.; Schernthaner, J.P.; Gesell, A.; Abrams, S.R.; Kennedy, J.A.; Constabel, C.P. Gene Expression and Metabolite Profiling of Developing Highbush Blueberry Fruit Indicates Transcriptional Regulation of Flavonoid Metabolism and Activation of Abscisic Acid Metabolism. Plant Physiol. 2012, 158, 200–224. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.M. Small RNAs and Their Roles in Plant Development. Annu. Rev. Cell Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Brousse, C.; Liu, Q.; Beauclair, L.; Deremetz, A.; Axtell, M.J.; Bouche, N. A non-canonical plant microRNA target site. Nucleic Acids Res. 2014, 42, 5270–5279. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Lelandais-Briere, C.; Sorin, C.; Declerck, M.; Benslimane, A.; Crespi, M.; Hartmann, C. Small RNA Diversity in Plants and its Impact in Development. Curr. Genom. 2010, 11, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.R.; Bian, S.M.; Tang, M.J.; Lu, Q.; Li, S.B.; Liu, X.G.; Tian, G.; Nguyen, V.; Tsang, E.W.T.; Wang, A.M.; et al. MicroRNA-Mediated Repression of the Seed Maturation Program during Vegetative Development in Arabidopsis. PLoS Genet. 2012, 8, e1003091. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.F.F.E.; Silva, E.M.; Azevedo, M.D.; Guivin, M.A.C.; Ramiro, D.A.; Figueiredo, C.R.; Carrer, H.; Peres, L.E.P.; Nogueira, F.T.S. MicroRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 2014, 78, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Shen, J.; Liu, H.; Li, F.; Ding, N.; Gao, C.; Pattanaik, S.; Patra, B.; Li, R.; Yuan, L. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 2015, 242, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Csukasi, F.; Donaire, L.; Casanal, A.; Martinez-Priego, L.; Botella, M.A.; Medina-Escobar, N.; Llave, C.; Valpuesta, V. Two strawberry miR159 family members display developmental-specific expression patterns in the fruit receptacle and cooperatively regulate Fa-GAMYB. New Phytol. 2012, 195, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Mou, W.; Luo, Z.; Li, L.; Limwachiranon, J.; Mao, L.; Ying, T. Developmental and stress regulation on expression of a novel miRNA, Fan-miR73, and its target ABI5 in strawberry. Sci. Rep. 2016, 6, 28385. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Yan, F.; Meng, R.; Jiang, X.B.; Yang, H.J.; Gao, Z.Y.; Dong, Y.H.; Yang, Y.Z.; Zhao, Z.Y. Identification of MicroRNAs and Their Targets Associated with Fruit-Bagging and Subsequent Sunlight Re-exposure in the “Granny Smith” Apple Exocarp Using High-Throughput Sequencing. Front. Plant Sci. 2016, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Han, J.; Liu, C.H.; Kibet, K.N.; Kayesh, E.; Shangguan, L.F.; Li, X.Y.; Fang, J.G. Identification of microRNAs from Amur grape (vitis amurensis Rupr.) by deep sequencing and analysis of microRNA variations with bioinformatics. BMC Genom. 2012, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Leng, X.P.; Zhang, Y.Y.; Kayesh, E.; Zhang, Y.P.; Sun, X.; Fang, J.G. Transcriptome-wide analysis of dynamic variations in regulation modes of grapevine microRNAs on their target genes during grapevine development. Plant Mol. Biol. 2014, 84, 269–285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Zhang, B.B.; Ma, R.J.; Yu, M.L.; Guo, S.L.; Guo, L.; Korir, N.K. Identification of Known and Novel microRNAs and Their Targets in Peach (Prunus persica) Fruit by High-Throughput Sequencing. PLoS ONE 2016, 11, e0159253. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xia, R.; Zhao, B.Y.; An, Y.Q.; Dardick, C.D.; Callahan, A.M.; Liu, Z.R. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 2012, 12, 149. [Google Scholar] [CrossRef] [PubMed]
- Saminathan, T.; Bodunrin, A.; Singh, N.V.; Devarajan, R.; Nimmakayala, P.; Jeff, M.; Aradhya, M.; Reddy, U.K. Genome-wide identification of microRNAs in pomegranate (Punica granatum L.) by high-throughput sequencing. BMC Plant Biol. 2016, 16, 122. [Google Scholar] [CrossRef] [PubMed]
- Xin, C.Q.; Liu, W.F.; Lin, Q.; Zhang, X.W.; Cui, P.; Li, F.S.; Zhang, G.Y.; Pan, L.L.; Al-Amer, A.; Mei, H.L.; et al. Profiling microRNA expression during multi-staged date palm (Phoenix dactylifera L.) fruit development. Genomics 2015, 105, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.X.; Zheng, S.S.; Feng, G.Z.; Yi, H.L. Comparative Analysis of miRNAs and Their Target Transcripts between a Spontaneous Late-Ripening Sweet Orange Mutant and Its Wild-Type Using Small RNA and Degradome Sequencing. Front. Plant Sci. 2016, 7, 1416. [Google Scholar] [CrossRef] [PubMed]
- Costich, D.E.; Ortiz, R.; Meagher, T.R.; Bruederle, L.P.; Vorsa, N. Determination of ploidy level and nuclear DNA content in blueberry by flow cytometry. Theor. Appl. Genet. 1993, 86, 1001–1006. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hou, Y.; Zhang, L.; Zhang, W.; Quan, C.; Cui, Y.; Bian, S. Computational identification of conserved microRNAs and their targets from expression sequence tags of blueberry (Vaccinium corybosum). Plant Signal. Behav. 2014, 9, e29462. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.B.; Yin, L.L.; Ying, Q.C.; Song, H.M.; Xue, D.W.; Lai, T.F.; Xu, M.J.; Shen, B.; Wang, H.Z.; Shi, X.Q. High-Throughput Sequencing and Degradome Analysis Identify miRNAs and Their Targets Involved in Fruit Senescence of Fragaria ananassa. PLoS ONE 2013, 8, e70959. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.H.; Pan, X.P.; Cox, S.B.; Cobb, G.P.; Anderson, T.A. Evidence that miRNAs are different from other RNAs. Cell. Mol. Life Sci. 2006, 63, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Din, M.; Barozai, M.Y.K. Profiling microRNAs and their targets in an important fleshy fruit: Tomato (Solanum lycopersicum). Gene 2014, 535, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Jagadeeswaran, G.; Nimmakayala, P.; Zheng, Y.; Gowdu, K.; Reddy, U.K.; Sunkar, R. Characterization of the small RNA component of leaves and fruits from four different cucurbit species. BMC Genom. 2012, 13, 329. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, B.H. MicroRNAs in Control of Plant Development. J. Cell. Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W. Conservation and divergence in plant microRNAs. Plant Mol. Biol. 2012, 80, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Eldem, V.; Okay, S.; Unver, T. Plant microRNAs: New players in functional genomics. Turk. J. Agric. For. 2013, 37, 1–21. [Google Scholar]
- Omidbakhshfard, M.A.; Proost, S.; Fujikura, U.; Mueller-Roeber, B. Growth-Regulating Factors (GRFs): A Small Transcription Factor Family with Important Functions in Plant Biology. Mol. Plant 2015, 8, 998–1010. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, C.; Keyzers, R.A.; Boss, P.K.; Davies, C. Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 2010, 61, 3615–3625. [Google Scholar] [CrossRef] [PubMed]
- Fortes, A.M.; Teixeira, R.T.; Agudelo-Romero, P. Complex Interplay of Hormonal Signals during Grape Berry Ripening. Molecules 2015, 20, 9326–9343. [Google Scholar] [CrossRef] [PubMed]
- Li, S.B.; Xie, Z.Z.; Hu, C.G.; Zhang, J.Z. A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 2016, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Grones, P.; Friml, J. Auxin transporters and binding proteins at a glance. J. Cell Sci. 2015, 128, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Leitner, J.; Retzer, K.; Malenica, N.; Bartkeviciute, R.; Lucyshyn, D.; Jager, G.; Korbei, B.; Bystrom, A.; Luschnig, C. Meta-regulation of Arabidopsis Auxin Responses Depends on tRNA Maturation. Cell Rep. 2015, 11, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Ziliotto, F.; Corso, M.; Rizzini, F.M.; Rasori, A.; Botton, A.; Bonghi, C. Grape berry ripening delay induced by a pre-veraison NAA treatment is paralleled by a shift in the expression pattern of auxin- and ethylene-related genes. BMC Plant Biol. 2012, 12, 185. [Google Scholar] [CrossRef] [PubMed]
- Gubler, F.; Chandler, P.M.; White, R.G.; Llewellyn, D.J.; Jacobsen, J.V. Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 2002, 129, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Chiba, Y.; Shimizu, T.; Miyakawa, S.; Kanno, Y.; Koshiba, T.; Kamiya, Y.; Seo, M. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 2015, 128, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Karidas, P.; Challa, K.R.; Nath, U. The tarani mutation alters surface curvature in Arabidopsis leaves by perturbing the patterns of surface expansion and cell division. J. Exp. Bot. 2015, 66, 2107–2122. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Guo, L.X.; Jin, L.F.; Liu, Y.Z.; Liu, T.; Fan, Y.H.; Peng, S.A. Identification and transcript profiles of citrus growth-regulating factor genes involved in the regulation of leaf and fruit development. Mol. Biol. Rep. 2016, 43, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.L.; Xu, J.; Cornille, A.; Tomes, S.; Karunairetnam, S.; Luo, Z.; Bassett, H.; Whitworth, C.; Rees-George, J.; Ranatunga, C.; et al. A microRNA allele that emerged prior to apple domestication may underlie fruit size evolution. Plant J. 2015, 84, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Oh, K.; Ivanchenko, M.G.; White, T.J.; Lomax, T.L. The diageotropica gene of tomato encodes a cyclophilin: A novel player in auxin signaling. Planta 2006, 224, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Balbi, V.; Lomax, T.L. Regulation of early tomato fruit development by the diageotropica gene. Plant Physiol. 2003, 131, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.Y.; Vrebalov, J.; Alba, R.; Lee, J.; McQuinn, R.; Chung, J.D.; Klein, P.; Giovannoni, J. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010, 64, 936–947. [Google Scholar] [CrossRef] [PubMed]
- Callis, J. The ubiquitination machinery of the ubiquitin system. Arabidopsis Book 2014, 12, e0174. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Smalle, J.; Lee, S.; Yan, N.; Emborg, T.J.; Vierstra, R.D. Ubiquitin C-terminal hydrolases 1 and 2 affect shoot architecture in Arabidopsis. Plant J. 2007, 51, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.H.; Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009, 10, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.J.W.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstadt, S.; Carr, J.P.; Baulcombe, D.C. An Antiviral Defense Role of AGO2 in Plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef] [PubMed]
- Fatyol, K.; Ludman, M.; Burgyan, J. Functional dissection of a plant Argonaute. Nucleic Acids Res. 2016, 44, 1384–1397. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, M.; Sugita, M. RNA Editing and Its Molecular Mechanism in Plant Organelles. Genes 2016, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Vicente, A.R.; Ortugno, C.; Rosli, H.; Powell, A.L.T.; Greve, L.C.; Labavitch, J.M. Temporal sequence of cell wall disassembly events in developing fruits. 2. Analysis of blueberry (Vaccinium Species). J. Agric. Food Chem. 2007, 55, 4125–4130. [Google Scholar] [CrossRef] [PubMed]
- Yip, H.K.; Floyd, S.K.; Sakakibara, K.; Bowman, J.L. Class III HD-Zip activity coordinates leaf development in Physcomitrella patens. Dev. Biol. 2016, 419, 184–197. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.S.; Wickneswari, R. Characterization of microRNAs Expressed during Secondary Wall Biosynthesis in Acacia mangium. PLoS ONE 2012, 7, e49662. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Doesseger, B.; Guyot, R.; Diet, A.; Parsons, R.L.; Clark, M.A.; Simmons, M.P.; Bedinger, P.; Goff, S.A.; Ringli, C.; et al. Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 2003, 131, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Vargas, L.; Cesarino, I.; Vanholme, R.; Voorend, W.; Saleme, M.D.S.; Morreel, K.; Boerjan, W. Improving total saccharification yield of Arabidopsis plants by vessel-specific complementation of caffeoyl shikimate esterase (cse) mutants. Biotechnol. Biofuels 2016, 9, 139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative Regulation of Anthocyanin Biosynthesis in Arabidopsis by a miR156-Targeted SPL Transcription Factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [PubMed]
- Galpaz, N.; Wang, Q.; Menda, N.; Zamir, D.; Hirschberg, J. Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 2008, 53, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Kabbage, M.; Dickman, M.B. The BAG proteins: A ubiquitous family of chaperone regulators. Cell Mol. Life Sci. 2008, 65, 1390–1402. [Google Scholar] [CrossRef] [PubMed]
- Li, R.Q.; Yu, C.; Li, Y.R.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.K.; Wang, Y.B.; Liu, Z.X.; Cheng, H.; Xue, Y. HemI: A Toolkit for Illustrating Heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- Addo-Quaye, C.; Miller, W.; Axtell, M.J. CleaveLand: A pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009, 25, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Xia, Z.H.; Chen, L.; Shi, M.J.; Pu, J.J.; Guo, J.R.; Fan, Z.F. Rapid and Efficient Isolation of High-Quality Small RNAs from Recalcitrant Plant Species Rich in Polyphenols and Polysaccharides. PLoS ONE 2014, 9, e95687. [Google Scholar] [CrossRef] [PubMed]




| ID | miRNA Sequences (5′-3′) | LT 1 | TP 2 | Accession 3 | Str 4 | Start | End | HOMO 5/NM 6 | MFEI 7 | FPKM 8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BF | WF | ||||||||||
| vco-miR156a | CUGACAGAAGAGAGUGAGCAC | 21 | 5′ | JOTO01194981 | − | 104 | 229 | gma-miR156a | 1 | 9.57 | 5.41 |
| vco-miR156b-5p | UUGACAGAAGAGAGUGAGCAC | 21 | 5′ | JOTO01158632 | + | 1177 | 1373 | mtr-miR156b-5p | 0.9 | 2.50 | 2.40 |
| vco-miR156c-3p | GCUCACUUUUCUUCUGUCAUU | 21 | 3′ | JOTO01174941 | − | 2100 | 2189 | mdm-miR156w-p3 | 1.3 | 9.16 | 0.00 |
| vco-miR156c-5p | CUGACAGAAGAGAGUGAGCAC | 21 | 5′ | JOTO01174941 | − | 2100 | 2189 | gma-miR156a | 1.3 | 9.57 | 5.41 |
| vco-miR156d-3p | GCUCUCUAUGCUUCUGUCAUC | 21 | 3′ | JOTO01177646 | + | 179 | 337 | stu-miR156d- | 0.8 | 58.27 | 36.64 |
| vco-miR156e-3p | GCUCUCUAUGCUUCUGUCAUC | 21 | 3′ | JOTO01177646 | − | 1 | 159 | mtr-miR156h-3p | 0.8 | 58.27 | 36.64 |
| vco-miR156e-5p | UUGACAGAAGAUAGAGAGCACUG | 23 | 5′ | JOTO01177646 | − | 1 | 159 | mtr-miR156e | 0.8 | 1.66 | 6.01 |
| vco-miR156f | GCUCUCUAGUCUUCUGUCAUC | 21 | 3′ | JOTO01152293 | + | 62 | 166 | gma-miR156e-p3 | 1.1 | 10.82 | 0.00 |
| vco-miR156g-5p | UGACAGAAGAGAGUGAGCAC | 20 | 5′ | JOTO01184662 | + | 3109 | 3203 | mtr-miR156b-5p | 1 | 32.46 | 4.81 |
| vco-miR156h-3p | GCUCACUUCUCUCUCUGUCAUC | 22 | 3′ | JOTO01167006 | + | 3494 | 3585 | mtr-miR156i-3p | 1.2 | 23.31 | 3.60 |
| vco-miR157a | UUGACAGAAGAUAGAGAGCAC | 21 | 5′ | JOTO01152293 | + | 65 | 163 | gra-miR157a | 1.1 | 11.65 | 39.64 |
| vco-miR159a | GCGGCGACGGGGGCGGUU | 18 | 3′ | JOTO01109152 | − | 4086 | 4230 | zma-miR159e-p3 | 0.6 | 3.75 | 0.00 |
| vco-miR160a-5p | UGCCUGGCUCCCUGUAUGCA | 20 | 5′ | JOTO01191503 | + | 4712 | 4793 | ath-miR160a-5p | 1.1 | 7.49 | 0.00 |
| vco-miR160b | UGCCUGGCUCCCUGAAUGCCAU | 22 | 5′ | JOTO01197084 | − | 3443 | 3530 | mes-miR160e | 1.1 | 1.66 | 1.20 |
| vco-miR162a-3p | UCGAUAAACCUCUGCAUCCAG | 21 | 3′ | JOTO01180996 | + | 1229 | 1340 | mtr-miR162 | 0.8 | 414.55 | 432.47 |
| vco-miR162a-5p | UGGACGCAGCGGUUCAUCGAUC | 22 | 5′ | JOTO01180996 | + | 1229 | 1340 | ath-miR162a-5p | 0.8 | 0.83 | 1.20 |
| vco-miR164a | AUGGAGAAGCAGGGCACGUGU | 21 | 5′ | JOTO01183560 | − | 3074 | 3198 | mtr-miR164a | 1 | 4.99 | 6.01 |
| vco-miR166a | UCGGACCAGGCUUCAUUCCCCU | 22 | 3′ | JOTO01182330 | − | 3021 | 3176 | mtr-miR166a | 0.9 | 89.49 | 30.03 |
| vco-miR166b-3p | UCGGACCAGGCUUCAUUCCCC | 21 | 3′ | JOTO01175424 | − | 3267 | 3364 | ptc-miR166a | 1 | 1377.66 | 980.27 |
| vco-miR166b-5p | GGAAUGCUGUCUGGUUCGAGG | 21 | 5′ | JOTO01175424 | − | 3267 | 3364 | ptc-miR166g-p5 | 1 | 77.42 | 1.20 |
| vco-miR166c-3p | UGUCGGACCAGGCUUCAUUCC | 21 | 3′ | JOTO01191740 | − | 2020 | 2122 | mtr-miR166a | 1.2 | 10.82 | 11.41 |
| vco-miR166c-5p | AAUGUUGUCUGGUUCGAGAUC | 21 | 5′ | JOTO01191740 | − | 2020 | 2122 | mdm-miR166c-p5 | 1.2 | 1.66 | 2.40 |
| vco-miR166d-3p | UCGGACCAGGCUUCAUUCCUC | 21 | 3′ | JOTO01177553 | + | 1331 | 1462 | mtr-miR166c | 0.9 | 469.90 | 853.53 |
| vco-miR166d-5p | GGGAUGUUGUCUGGCUCGAUG | 21 | 5′ | JOTO01177553 | + | 1331 | 1462 | sly-miR166c-5p | 0.9 | 318.82 | 461.30 |
| vco-miR166e-3p | UCUCGGACCAGGCUUCAUUCU | 21 | 3′ | JOTO01175424 | − | 3207 | 3307 | cme-miR166i-p3 | 1 | 159.83 | 72.08 |
| vco-miR166e-5p | GGAAUGCUGUCUGGCUCGAGG | 21 | 5′ | JOTO01175424 | − | 3207 | 3307 | mtr-miR166g-5p | 1 | 0.00 | 2.40 |
| vco-miR166f-5p | GGAAUGUUGUCUGGCUCGAGG | 21 | 5′ | JOTO01193807 | + | 4411 | 4505 | mtr-miR166g-5p | 1 | 469.49 | 353.19 |
| vco-miR167a | UGAAGCUGCCAGCAUGAUCUAA | 22 | 5′ | JOTO01143215 | − | 81 | 168 | mdm-miR167b | 0.9 | 0.83 | 2.40 |
| vco-miR167b | UGAAGCUGCCAGCAUGAUCUGA | 22 | 5′ | JOTO01169118 | − | 703 | 820 | mtr-miR167b-5p | 1 | 21.64 | 21.62 |
| vco-miR171a-3p | UUGAGCCGCGCCAAUAUCACU | 21 | 3′ | JOTO01145355 | − | 1932 | 2031 | vvi-miR171f | 1.1 | 39.12 | 0.00 |
| vco-miR171a-5p | CGAUGUUGGUGAGGUUCAAUC | 21 | 5′ | JOTO01145355 | − | 1932 | 2031 | mtr-miR171e-5p | 1.1 | 7.49 | 0.00 |
| vco-miR171b | AAGGGUGUUGUUCGAUUAA | 19 | 5′ | JOTO01109152 | − | 2162 | 2277 | ath-miR171b-p5 | 0.6 | 6.66 | 0.00 |
| vco-miR172a-3p | AGAAUCUUGAUGAUGCUGCAU | 21 | 3′ | JOTO01171463 | + | 458 | 595 | mtr-miR172b | 1.1 | 8.32 | 1.20 |
| vco-miR172a-5p | GCGGCAUUAUCAAGAUUCAC | 20 | 5′ | JOTO01171463 | + | 458 | 595 | gra-miR172a | 1.1 | 64.10 | 7.21 |
| vco-miR319a | UGGACUGAAGGGAGCUCCCUU | 21 | 5′ | JOTO01138414 | − | 1309 | 1470 | mtr-miR319a-3p | 0.7 | 365.02 | 49.85 |
| vco-miR319b | UUGGACUGAAGGGAGCUCCC | 20 | 3′ | JOTO01184324 | + | 5377 | 5438 | mtr-miR319a-3p | 0.7 | 262.63 | 15.62 |
| vco-miR319c | UGGACUGAAGGGAGCUCCCUU | 21 | 3′ | JOTO01169050 | + | 2402 | 2535 | mtr-miR319a-3p | 0.6 | 365.02 | 49.85 |
| vco-miR390a-3p | CGCUAUCCAUCCUGAGUUUCA | 21 | 3′ | JOTO01159657 | − | 2355 | 2501 | gma-miR390a-3p | 1 | 4.58 | 1.80 |
| vco-miR390a-5p | AAGCUCAGGAGGGAUAGCGCC | 21 | 5′ | JOTO01159657 | − | 2355 | 2501 | mtr-miR390 | 1 | 1.25 | 1.20 |
| vco-miR390b-3p | CGCUAUCCAUCCUGAGUUUCA | 21 | 3′ | JOTO01180795 | + | 4526 | 4630 | gma-miR390 | 1 | 4.58 | 1.80 |
| vco-miR390b-5p | AAGCUCAGGAGGGAUAGCGCC | 21 | 5′ | JOTO01180795 | + | 4526 | 4630 | mtr-miR390 | 1 | 1.25 | 1.20 |
| vco-miR393a-3p | AUCAUGCGAUCCCUUAGGAUU | 21 | 3′ | JOTO01199451 | + | 28 | 171 | ptc-miR393a-3p | 1 | 2.91 | 0.60 |
| vco-miR393a-5p | UCCAAAGGGAUCGCAUUGAU | 20 | 5′ | JOTO01199451 | + | 28 | 171 | mtr-miR393a | 1 | 2.50 | 1.20 |
| vco-miR393b-3p | AUCAUGCGAUCCCUUAGGAUU | 21 | 3′ | JOTO01199451 | + | 2549 | 2697 | ptc-miR393a-3p | 1 | 2.91 | 0.60 |
| vco-miR393b-5p | UCCAAAGGGAUCGCAUUGAU | 20 | 5′ | JOTO01199451 | + | 2549 | 2697 | mtr-miR393a | 1 | 2.50 | 1.20 |
| vco-miR394a-5p | UUGGCAUUCUGUCCACCUCC | 20 | 5′ | JOTO01174460 | + | 2780 | 2935 | gma-miR394a-5p | 0.8 | 87.40 | 0.00 |
| vco-miR395a | CUGAAGUGUUUGGGGGAACUC | 21 | 3′ | JOTO01195534 | − | 9719 | 9799 | vvi-miR395a | 1 | 70.76 | 82.89 |
| vco-miR396a-3p | GUUCAAGAAAUCUGUGGGAGA | 21 | 3′ | JOTO01179423 | + | 3728 | 3868 | mtr-miR396a-3p | 1.3 | 4.16 | 6.01 |
| vco-miR396a-5p | UUCCACAGCUUUCUUGAACUU | 21 | 5′ | JOTO01179423 | + | 3728 | 3868 | mtr-miR396a-5p | 1.3 | 432.86 | 151.97 |
| vco-miR396b-3p | GUUCAAUAAAGCUGUGGGAAG | 21 | 3′ | JOTO01198645 | + | 118 | 264 | mtr-miR396b-3p | 1.1 | 60.77 | 34.24 |
| vco-miR396b-5p | UUCCACAGCUUUCUUGAACU | 20 | 5′ | JOTO01198645 | + | 118 | 264 | mtr-miR396b-5p | 1.1 | 69.51 | 22.82 |
| vco-miR396c-3p | GUUCGAGAAAGCUGUGGGAAG | 21 | 3′ | JOTO01196729 | − | 5681 | 5840 | mtr-miR396b-3p | 0.9 | 3.33 | 2.40 |
| vco-miR396c-5p | UUCCACAGCUUUCUUGAACUC | 21 | 5′ | JOTO01196729 | − | 5681 | 5840 | mtr-miR396a-5p | 0.9 | 165.24 | 34.24 |
| vco-miR396d-3p | GUUCAAUUAAGCUGUGGGAAG | 21 | 3′ | JOTO01198645 | − | 421 | 567 | mtr-miR396b-3p | 1.1 | 26.64 | 9.61 |
| vco-miR396d-5p | UUCCACAGCUUUCUUGAACUC | 21 | 5′ | JOTO01198645 | − | 421 | 567 | mtr-miR396b-5p | 1.1 | 165.24 | 34.24 |
| vco-miR396e-3p | GUUCACUAAAGCUGUGGAAAG | 21 | 3′ | JOTO01155840 | + | 2054 | 2198 | mtr-miR396b-3p | 1 | 27.47 | 21.62 |
| vco-miR396e-5p | UUCCACAGCUUUCUUGAACUG | 21 | 5′ | JOTO01155840 | + | 2054 | 2198 | mtr-miR396b-5p | 1 | 124.45 | 16.22 |
| vco-miR396f-3p | GUUCAAUAAAGCUGUGGGAAG | 21 | 3′ | JOTO01198645 | + | 8127 | 8264 | mtr-miR396b-3p | 1.1 | 60.77 | 34.24 |
| vco-miR396f-5p | UUCCACAGCUUUCUUGAACUG | 21 | 5′ | JOTO01198645 | + | 8127 | 8264 | mtr-miR396b-5p | 1.1 | 124.45 | 16.22 |
| vco-miR396g | GCUCAAGAAAGCUGUGGGAAG | 21 | 5′ | JOTO01192216 | + | 28 | 102 | mtr-miR396b-3p | 0.6 | 3.33 | 14.42 |
| vco-miR398a-3p | UGUGUUCUCAGGUCACCCCUU | 21 | 3′ | JOTO01181367 | − | 2816 | 2942 | mtr-miR398a-3p | 0.8 | 4.16 | 0.00 |
| vco-miR398a-5p | GGAGUGACACUUAGAACACACG | 22 | 5′ | JOTO01181367 | − | 2816 | 2942 | mtr-miR398a-5p | 0.8 | 0.42 | 1.20 |
| vco-miR398b-3p | UGUGUUCUCAGGUCACCCCUU | 21 | 3′ | JOTO01178370 | − | 4047 | 4163 | mtr-miR398a-3p | 1.1 | 4.16 | 0.00 |
| vco-miR398b-5p | GGAGUGACACUGAGAACACU | 20 | 5′ | JOTO01178370 | − | 4047 | 4163 | mtr-miR398a-5p | 1.1 | 11.24 | 0.00 |
| vco-miR398c-3p | UGUGUUCUCAGGUCGCCCCUG | 21 | 3′ | JOTO01177986 | − | 4722 | 4858 | mtr-miR398b | 1 | 17.48 | 12.01 |
| vco-miR398c-5p | GGAGCGACCUGAGAUCACAUG | 21 | 5′ | JOTO01177986 | − | 4722 | 4858 | ptc-miR398c-5p | 1 | 69.09 | 22.82 |
| vco-miR399a | UGCCAAAGGAGAAUUGCCCUG | 21 | 3′ | JOTO01171567 | − | 2276 | 2397 | mtr-miR399c | 1 | 4.99 | 7.21 |
| vco-miR399b | UGCCAAAGGAGAGUUGCCCUG | 21 | 3′ | JOTO01187770 | − | 6181 | 6281 | mtr-miR399l | 1 | 1.66 | 10.81 |
| vco-miR403a | UUAGAUUCACGCACAAACUCUG | 22 | 3′ | JOTO01197485 | − | 10,488 | 10,629 | gma-miR403a | 1.1 | 89.90 | 34.84 |
| vco-miR535a-3p | GUGCUCCCUAUCGUCGUCAU | 20 | 3′ | JOTO01190921 | + | 974 | 1058 | mes-miR535a-p3 | 1 | 276.36 | 241.46 |
| vco-miR535a-5p | UGACGACGAGAGAGAGCACGC | 21 | 5′ | JOTO01190921 | + | 974 | 1058 | mdm-miR535d | 1 | 17.06 | 66.07 |
| vco-miR535b | UGACGACGAGAGAGAGCACGC | 21 | 5′ | JOTO01171383 | + | 217 | 311 | mdm-miR535d | 1.1 | 17.06 | 66.07 |
| vco-miR845 | UGCUCUGAUACCAAUUGUUGA | 21 | 5′ | JOTO01134176 | − | 526 | 624 | bdi-miR845 | 0.9 | 3.33 | 4.81 |
| vco-miR894 | UUCACGUCGGGUUCACCA | 18 | 5′ | JOTO01175668 | + | 4734 | 4832 | ppt-miR894 | 0.5 | 340.46 | 1055.95 |
| vco-miR2111 | ACCGGGUAAUCUGCAUCCUGA | 21 | 5′ | JOTO01195655 | − | 6181 | 6282 | tcc-miR2111-p5 | 1.1 | 0.83 | 1.20 |
| vco-miR2118 | UUACCGACUCCACCCAUACCUA | 22 | 3′ | JOTO01101378 | − | 99 | 199 | mtr-miR2118 | 1.3 | 26.64 | 20.42 |
| vco-miR2603 | GGUCCCUGCCCUUUGUACA | 19 | 5′ | JOTO01109152 | − | 4086 | 4230 | mtr-miR2603 | 0.6 | 4.58 | 1.20 |
| vco-miR6149a | CUUCAUACGCACCUGAAUCGG | 21 | 5′ | JOTO01189719 | + | 7347 | 7491 | nta-miR6149a | 1 | 14.15 | 6.01 |
| vco-miR6478 | UCGACCUUAGCUCAGUUGGUA | 21 | 5′ | JOTO01186167 | − | 6643 | 6834 | ptc-miR6478 | 0.6 | 201.03 | 330.36 |
| vco-miR7122a-3p | ACCGUGUUUCUCUGUAUAAAU | 21 | 3′ | JOTO01165247 | + | 1957 | 2063 | mdm-miR7122a-p3 | 1.1 | 748.35 | 595.85 |
| vco-miR7122a-5p | UUAUACAGAAAAAUCACGGUCG | 22 | 5′ | JOTO01165247 | + | 1957 | 2063 | mdm-miR7122a | 1.1 | 2.08 | 0.00 |
| vco-miR7122b-3p | ACCGUGUUUCUCUGUAUAAAU | 21 | 3′ | JOTO01194243 | − | 8897 | 9055 | mdm-miR7122a-p3 | 1 | 748.35 | 595.85 |
| vco-miR7122b-5p | UUAUACAGAAAAAUCACGGUCG | 22 | 5′ | JOTO01194243 | − | 8897 | 9055 | mdm-miR7122a | 1 | 2.08 | 0.00 |
| vco-miR7693-5p | GCAUCGAUGAAGAGCGUA | 18 | 5′ | JOTO01109152 | − | 3655 | 3758 | osa-miR7693-p5 | 0.5 | 8.32 | 0.60 |
| vco-miR_n01 | UCCGAUCUGAAUUCAAGCGAA | 21 | 3′ | JOTO01190684 | + | 1726 | 1822 | 1 | 1.4 | 6.66 | 10.21 |
| vco-miR_n02 | UCCGAUCUGAAUUCAAGCGAA | 21 | 3′ | JOTO01190684 | − | 1689 | 1787 | 0 | 1.4 | 6.66 | 10.21 |
| vco-miR_n03 | GUGUGGCUGGGAGUGGCUCCU | 21 | 3′ | JOTO0118763 | − | 468 | 619 | 0 | 1.6 | 11.65 | 18.02 |
| vco-miR_n04 | GUGUGGCUGGGAGUGGCUCCC | 21 | 3′ | JOTO0118763 | + | 541 | 703 | 1 | 1.5 | 11.65 | 29.43 |
| vco-miR_n05 | ACACAUCUGUGUCUGAAUCUGUGC | 24 | 3′ | JOTO01183568 | + | 4231 | 4401 | 1 | 1.3 | 633.47 | 1071.57 |
| vco-miR_n06 | CCGAUAUGAGUCUAACAUGCC | 21 | 3′ | JOTO01181717 | − | 2207 | 2389 | 1 | 1.7 | 21.23 | 10.21 |
| vco-miR_n07 | CCCGUUGGAUGCAUUUUCU | 19 | 3′ | JOTO01180185 | + | 2426 | 2591 | 1 | 1.5 | 69.92 | 16.82 |
| vco-miR_n08-5p | UGAGUUGUAAGCAAAGUCCGC | 21 | 5′ | JOTO01179705 | − | 4366 | 4501 | 2 | 1.2 | 26.22 | 18.02 |
| vco-miR_n08-3p | GGCGGACUUUGCUUAGAACCC | 21 | 3′ | JOTO01179705 | − | 4366 | 4501 | 2 | 1.2 | 198.95 | 201.82 |
| vco-miR_n09 | UGGACGAUCGGGAUGUGCUAGUGU | 24 | 3′ | JOTO01178111 | − | 3319 | 3476 | 0 | 1.4 | 19.56 | 13.21 |
| vco-miR_n10 | GUGCUUUCUAUCGUCGUCAU | 20 | 3′ | JOTO01171383 | + | 217 | 311 | 2 | 1.1 | 350.45 | 97.31 |
| vco-miR_n11 | CCCGUUGGAUGCAUUUUCUGA | 21 | 3′ | JOTO01167166 | − | 2168 | 2315 | 1 | 1.1 | 116.12 | 21.02 |
| vco-miR_n12-5p | CCUCCGACUCUAUAGACGAAG | 21 | 5′ | JOTO01135109 | − | 1462 | 1683 | 1 | 1.5 | 13.32 | 52.86 |
| vco-miR_n12-3p | UCGUAUAUAGAGUCGGAGGCU | 21 | 3′ | JOTO01135109 | − | 1462 | 1683 | 1 | 1.5 | 11.65 | 58.86 |
| vco-miR_n13 | GUUGAUCUCGGAUCGGACGG | 20 | 5′ | JOTO01100636 | + | 836 | 981 | 0 | 1.3 | 10.41 | 11.41 |
| vco-miR_n14 | CGTCCGAUCCAAGAUCAACGGUCU | 24 | 3′ | JOTO01100636 | − | 731 | 878 | 1 | 1.3 | 19.15 | 4.81 |
| miRNAs | BF (norm) | WF (norm) | Fold Change | log2(Fold Change) 1 | p-Value (Fisher Test) | Sig-Lable 2 | Up/Down |
|---|---|---|---|---|---|---|---|
| vco-miR156c-3p | 9.16 | 0.00 | inf | inf | 1.29 × 10−5 | ** | up |
| vco-miR156f | 10.82 | 0.00 | inf | inf | 1.06 × 10−6 | ** | up |
| vco-miR159a | 3.75 | 0.00 | inf | inf | 6.72 × 10−3 | ** | up |
| vco-miR160a-5p | 7.49 | 0.00 | inf | inf | 1.58×10-4 | ** | up |
| vco-miR171a-3p | 39.12 | 0.00 | inf | inf | 6.54 × 10−22 | ** | up |
| vco-miR171a-5p | 7.49 | 0.00 | inf | inf | 1.58 × 10−4 | ** | up |
| vco-miR171b | 6.66 | 0.00 | inf | inf | 1.58 × 10−4 | ** | up |
| vco-miR394a-5p | 87.40 | 0.00 | inf | inf | 5.47 × 10−48 | ** | up |
| vco-miR398a-3p | 4.16 | 0.00 | inf | inf | 6.72 × 10−3 | ** | up |
| vco-miR398b-3p | 4.16 | 0.00 | inf | inf | 6.72 × 10−3 | ** | up |
| vco-miR398b-5p | 11.24 | 0.00 | inf | inf | 1.06 × 10−6 | ** | up |
| vco-miR166b-5p | 77.42 | 1.20 | 64.44 | 6.01 | 8.31 × 10−41 | ** | up |
| vco-miR319b | 262.63 | 15.62 | 16.82 | 4.07 | 2.33 × 10−120 | ** | up |
| vco-miR7693-5p | 8.32 | 0.60 | 13.86 | 3.79 | 3.03 × 10−4 | ** | up |
| vco-miR172a-5p | 64.10 | 7.21 | 8.89 | 3.15 | 2.25 × 10−27 | ** | up |
| vco-miR396e-5p | 124.45 | 16.22 | 7.67 | 2.94 | 8.80 × 10−50 | ** | up |
| vco-miR396f-5p | 124.45 | 16.22 | 7.67 | 2.94 | 8.80 × 10−50 | ** | up |
| vco-miR319a | 365.02 | 49.85 | 7.32 | 2.87 | 2.54 × 10−141 | ** | up |
| vco-miR319c | 365.02 | 49.85 | 7.32 | 2.87 | 2.54 × 10−141 | ** | up |
| vco-miR172a-3p | 8.32 | 1.20 | 6.93 | 2.79 | 3.03 × 10−4 | ** | up |
| vco-miR156h-3p | 23.31 | 3.60 | 6.47 | 2.69 | 1.57 × 10−9 | ** | up |
| vco-miR396c-5p | 165.24 | 34.24 | 4.83 | 2.27 | 6.37 × 10−57 | ** | up |
| vco-miR396d-5p | 165.24 | 34.24 | 4.83 | 2.27 | 6.37 × 10−57 | ** | up |
| vco-miR2603 | 4.58 | 1.20 | 3.81 | 1.93 | 8.79 × 10−3 | ** | up |
| vco-miR396b-5p | 69.51 | 22.82 | 3.05 | 1.61 | 1.73 × 10−20 | ** | up |
| vco-miR398c-5p | 69.09 | 22.82 | 3.03 | 1.60 | 4.57 × 10−20 | ** | up |
| vco-miR166a | 89.49 | 30.03 | 2.98 | 1.58 | 2.66 × 10−25 | ** | up |
| vco-miR396a-5p | 432.86 | 151.97 | 2.85 | 1.51 | 3.86 × 10−114 | ** | up |
| vco-miR396d-3p | 26.64 | 9.61 | 2.77 | 1.47 | 3.00 × 10−8 | ** | up |
| vco-miR403a | 89.90 | 34.84 | 2.58 | 1.37 | 1.39 × 10−23 | ** | up |
| vco-miR390a-3p | 4.58 | 1.80 | 2.54 | 1.35 | 2.35 × 10−2 | * | up |
| vco-miR390b-3p | 4.58 | 1.80 | 2.54 | 1.35 | 2.35 × 10−2 | * | up |
| vco-miR6149a | 14.15 | 6.01 | 2.36 | 1.24 | 1.51 × 10−4 | ** | up |
| vco-miR166e-3p | 159.83 | 72.08 | 2.22 | 1.15 | 5.95 × 10−37 | ** | up |
| vco-miR_n11 | 116.12 | 21.02 | 5.52 | 2.47 | 2.51 × 10−42 | ** | up |
| vco-miR_n07 | 69.92 | 16.82 | 4.16 | 2.06 | 1.67 × 10−23 | ** | up |
| vco-miR_n14 | 19.15 | 4.81 | 3.98 | 1.99 | 4.17 × 10−7 | ** | up |
| vco-miR_n10 | 350.45 | 97.31 | 3.60 | 1.85 | 6.93 × 10−105 | ** | up |
| vco-miR_n06 | 21.23 | 10.21 | 2.08 | 1.06 | 7.25 × 10−6 | ** | up |
| vco-miR894 | 340.46 | 1055.95 | 0.32 | −1.63 | 3.70 × 10−4 | ** | down |
| vco-miR_n12-3p | 11.65 | 58.86 | 0.20 | −2.34 | 3.46 × 10−2 | * | down |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Y.; Zhai, L.; Li, X.; Xue, Y.; Wang, J.; Yang, P.; Cao, C.; Li, H.; Cui, Y.; Bian, S. Comparative Analysis of Fruit Ripening-Related miRNAs and Their Targets in Blueberry Using Small RNA and Degradome Sequencing. Int. J. Mol. Sci. 2017, 18, 2767. https://doi.org/10.3390/ijms18122767
Hou Y, Zhai L, Li X, Xue Y, Wang J, Yang P, Cao C, Li H, Cui Y, Bian S. Comparative Analysis of Fruit Ripening-Related miRNAs and Their Targets in Blueberry Using Small RNA and Degradome Sequencing. International Journal of Molecular Sciences. 2017; 18(12):2767. https://doi.org/10.3390/ijms18122767
Chicago/Turabian StyleHou, Yanming, Lulu Zhai, Xuyan Li, Yu Xue, Jingjing Wang, Pengjie Yang, Chunmei Cao, Hongxue Li, Yuhai Cui, and Shaomin Bian. 2017. "Comparative Analysis of Fruit Ripening-Related miRNAs and Their Targets in Blueberry Using Small RNA and Degradome Sequencing" International Journal of Molecular Sciences 18, no. 12: 2767. https://doi.org/10.3390/ijms18122767