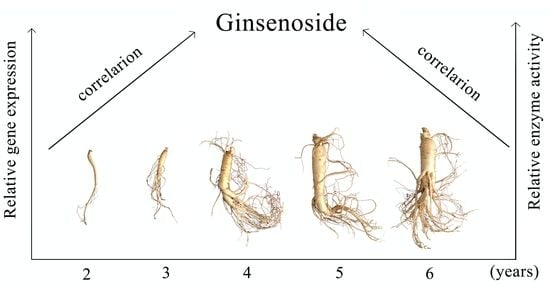
Study on the Correlation between Gene Expression and Enzyme Activity of Seven Key Enzymes and Ginsenoside Content in Ginseng in Over Time in Ji’an, China
Abstract
:1. Introduction
2. Results
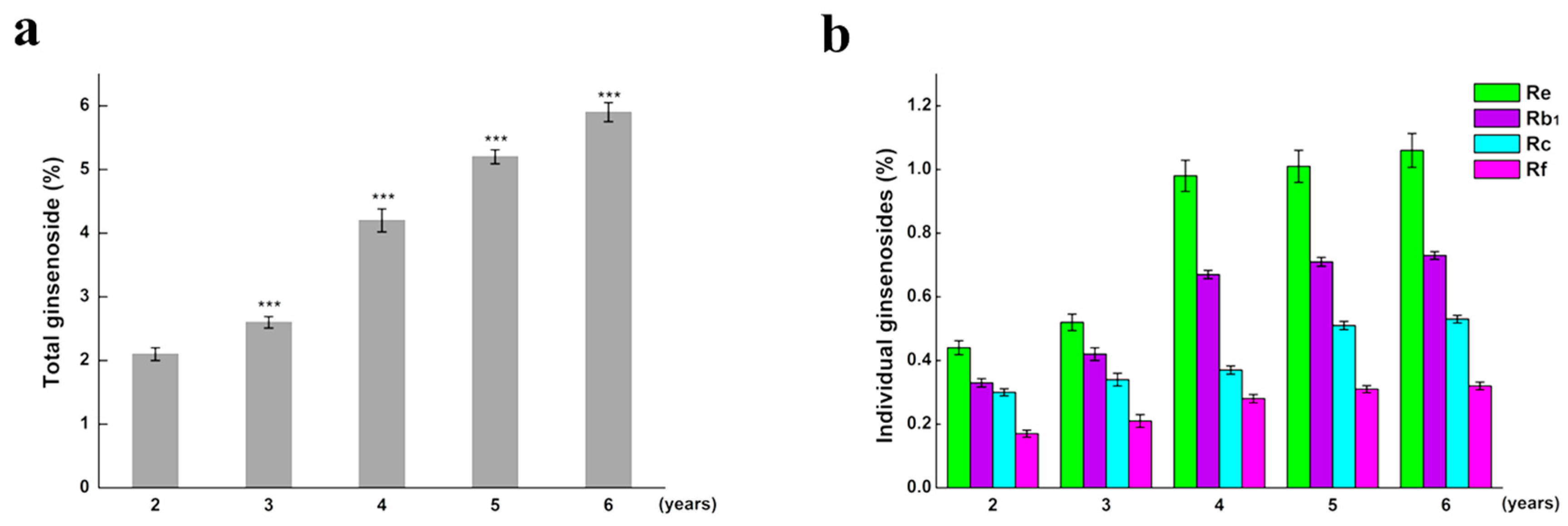
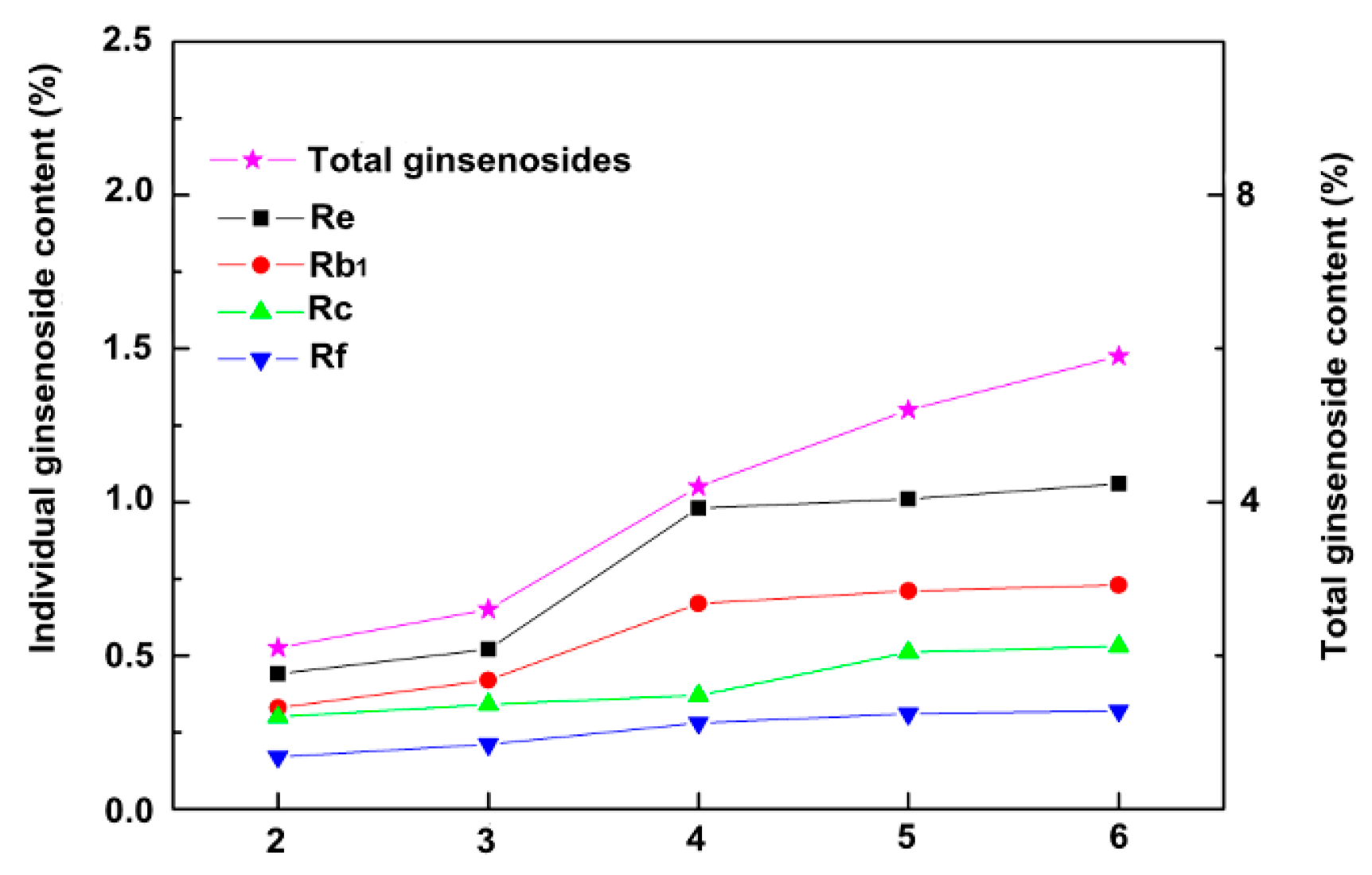
2.1. The Content of Total Saponin
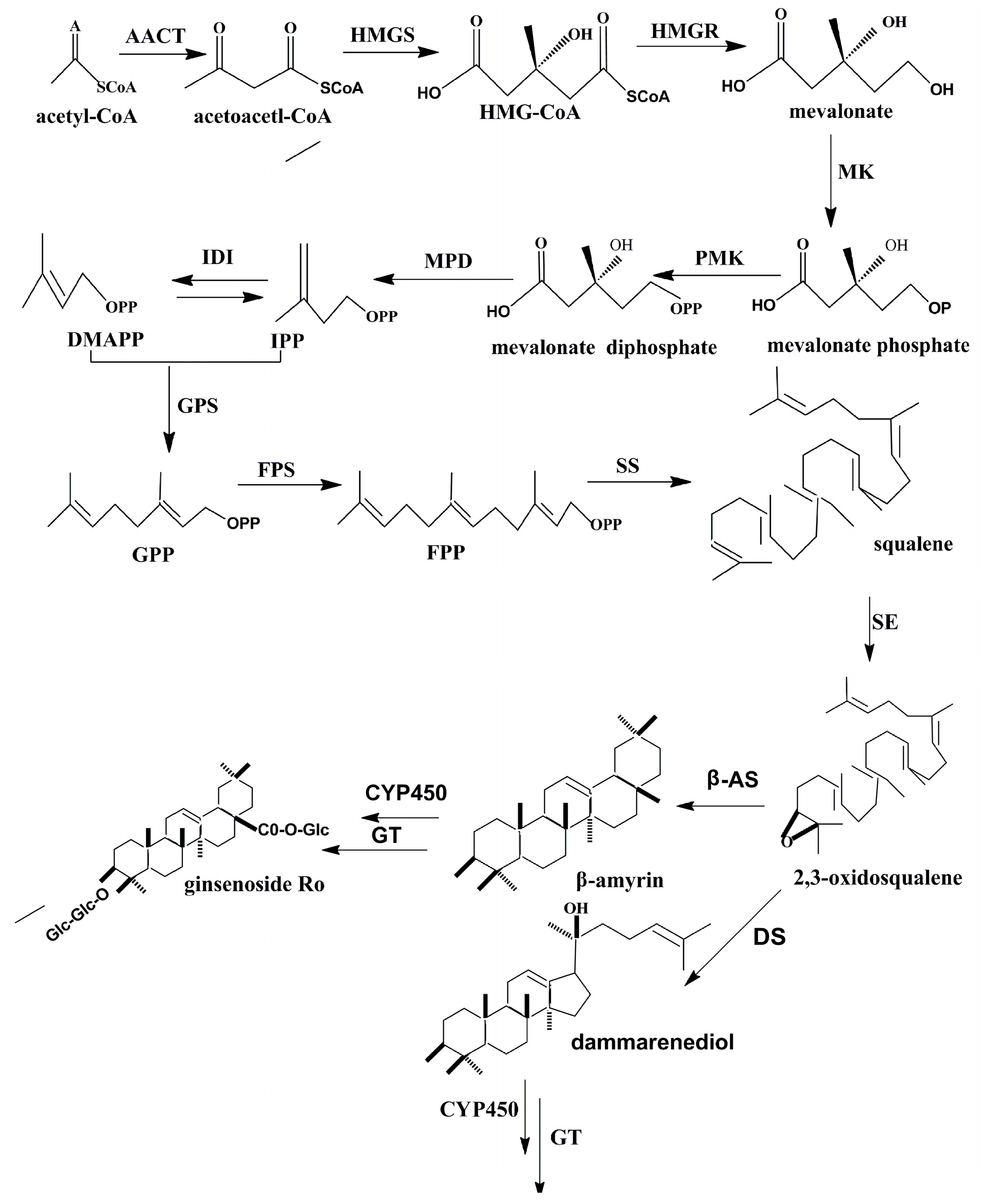
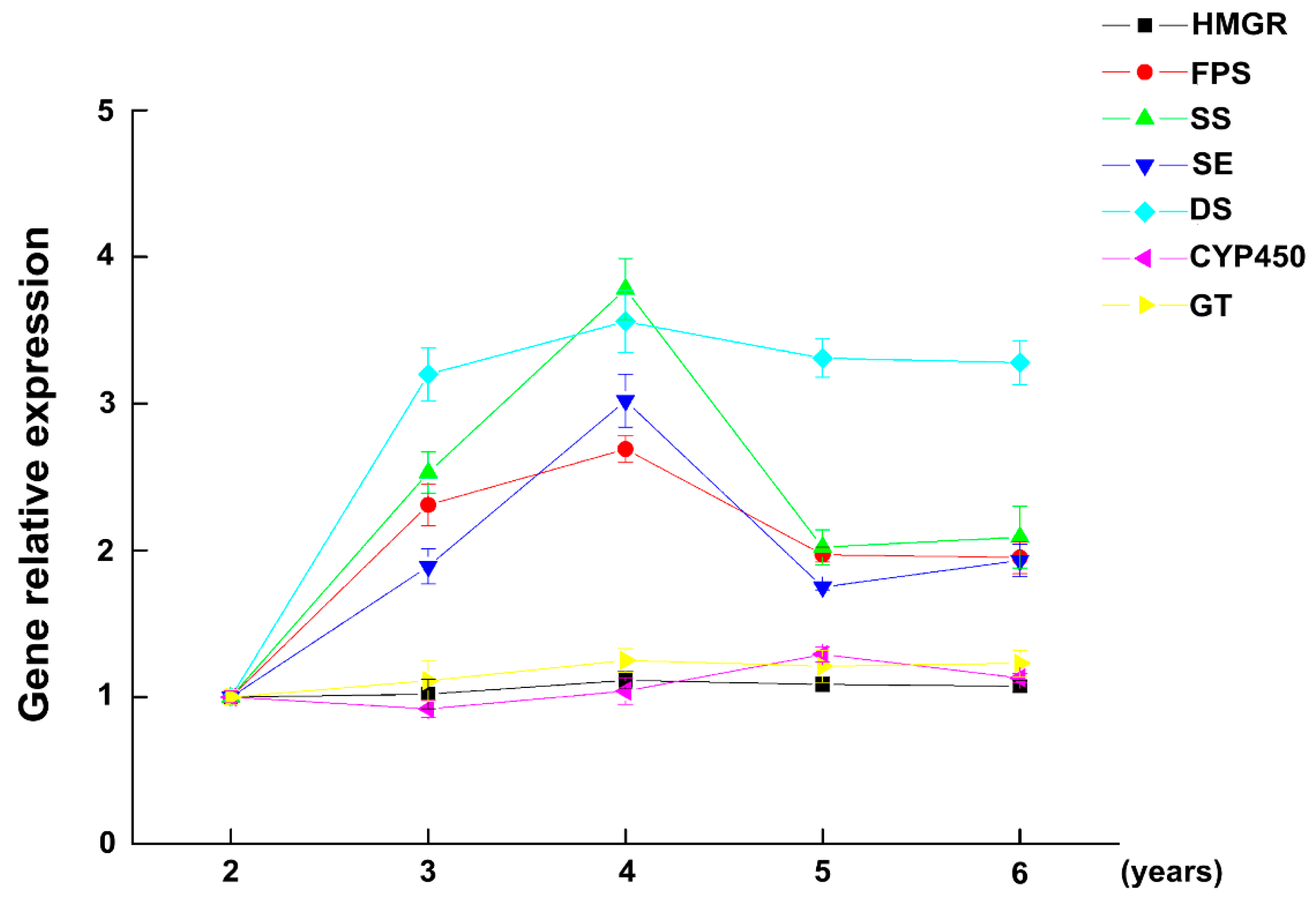
2.2. The Expression of Key Enzymes
2.3. The Activity of Key Enzymes
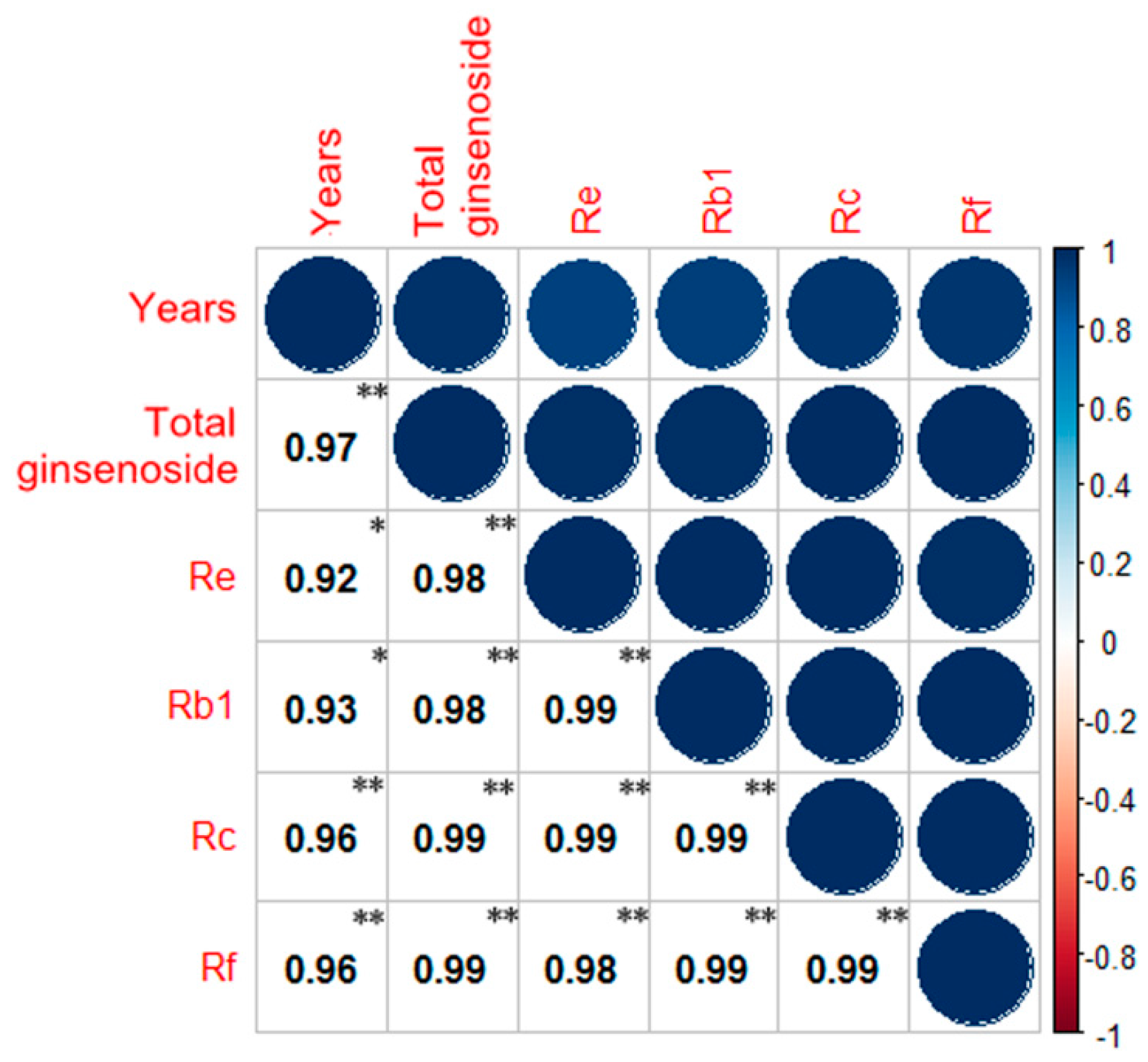
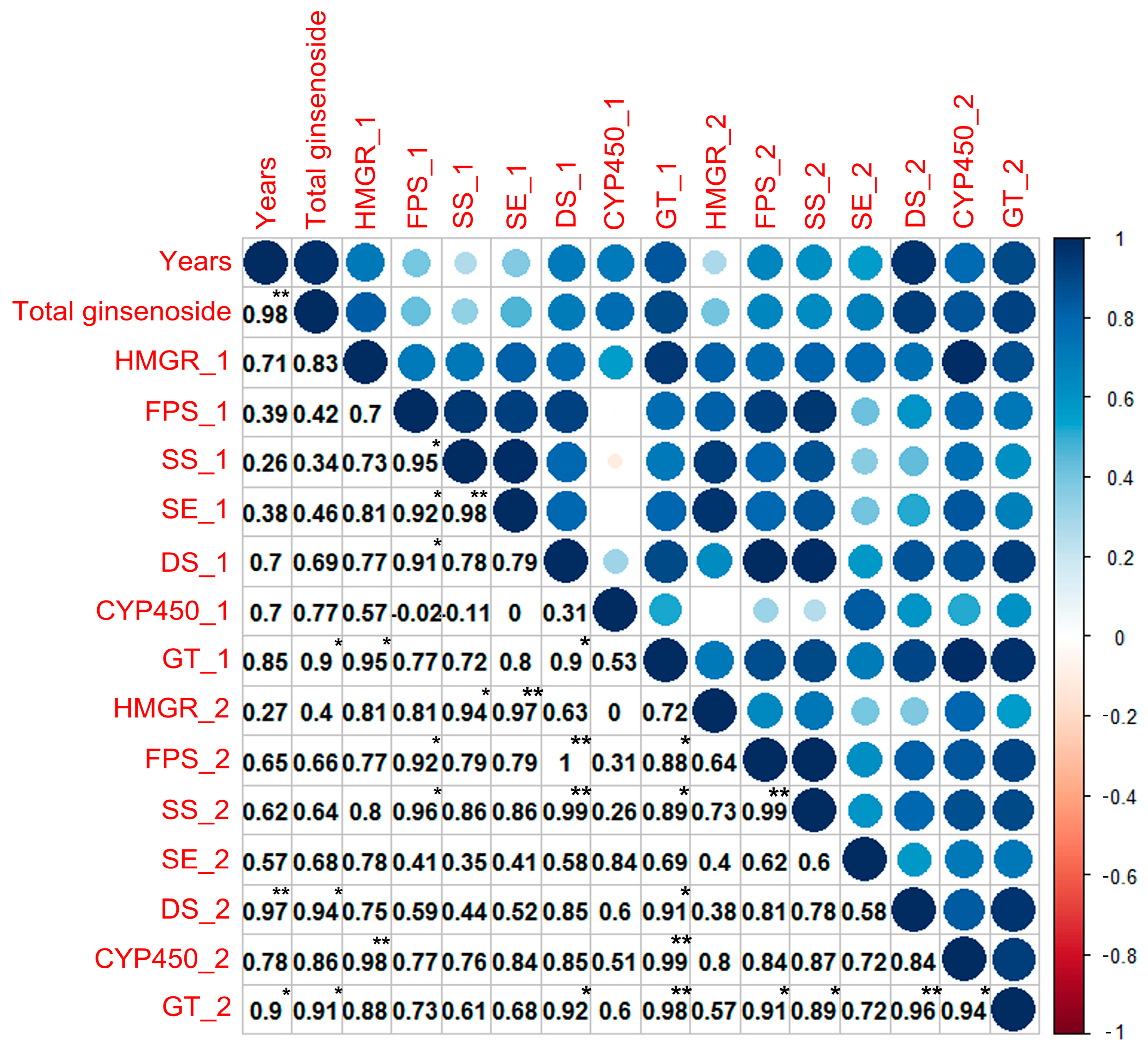
2.4. Correlation Analysis
3. Discussion
4. Methods
4.1. Plant Material
4.2. Extraction of Ginseng Total Saponins
4.3. Analysis of Ginsenosides by HPLC
4.4. Quantification of Transcript Levels
4.5. Enzyme Activity Assay
4.5.1. Protein Extraction
4.5.2. HMGR Activity Assay
4.5.3. FPS Activity Assay
4.5.4. SS Enzyme Activity
4.5.5. SE Enzyme Activity
4.5.6. DS Enzyme Activity
4.5.7. CYP450 Activity
4.5.8. GT Enzyme Activity
4.6. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Attele, A.S.; Wu, J.A.; Yuan, C.S. Ginseng pharmacology: Multiple constituents and multiple actions. Biochem. Pharmacol. 1999, 58, 1685–1693. [Google Scholar] [CrossRef]
- Kim, Y.J.; Jeon, J.N.; Jang, M.G.; Oh, J.Y.; Kwon, W.S.; Jung, S.K.; Yang, D.C. Ginsenoside profiles and related gene expression during foliation in Panax ginseng meyer. J. Ginseng Res. 2014, 38, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Chow, L.; Johnson, M.; Wells, A.; Dasgupta, A. Effect of the traditional Chinese Medicines Chan su, Lu-Shen-wan, Dan shen, and Asian ginseng on serum digoxin measurement by tina-quant (roche) and synchron Lx system (beckman) digoxin immunoassays. J. Clin. Lab. Anal. 2003, 17, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Nakanishi, K. Proof of the mysterious efficacy of ginseng: Basic and clinical trials: Clinical effects of medical ginseng, korean red ginseng: Specifically, its anti-stress action for prevention of disease. J. Pharmacol. Sci. 2004, 95, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.H.; Wu, C.F.; Duan, L.; Yang, J.Y. Protective effects of total saponins from stem and leaf of Panax ginseng against cyclophosphamide-induced genotoxicity and apoptosis in mouse bone marrow cells and peripheral lymphocyte cells. Food Chem. Toxicol. 2008, 46, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.T. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng c a meyer. Acta Pharmacol. Sin. 2008, 29, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.T.; Mehendale, S.R.; Li, X.M.; Quigg, R.; Wang, X.Y.; Wang, C.Z.; Wu, J.A.; Aung, H.H.; Rue, P.A.; Bell, G.I.; et al. Anti-diabetic effect of ginsenoside Re in ob/ob mice. Biochem. Biophys. Acta Mol. Basis Dis. 2005, 1740, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.J.; Wang, L.; Liu, L.; Liang, Y.L.; Sun, Y.; Wu, J.J. Both the mevalonate and the non-mevalonate pathways are involved in ginsenoside biosynthesis. Plant Cell Rep. 2014, 33, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Fazio, G.C.; Matsuda, S.P.T. On the origins of triterpenoid skeletal diversity. Phytochemistry 2004, 65, 261–291. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.Y.; Lu, Q.H.; Liu, S.P.; Chen, X.H.; Luo, J.Q.; Tan, L.J.; Hu, W.X. Screening and identification of novel genes involved in biosynthesis of ginsenoside in Panax ginseng plant. Acta Biochim. Biophys. Sin. 2003, 35, 554–560. [Google Scholar] [PubMed]
- Ahn, J.-J.; Akram, K.; Jo, D.; Kwon, J.-H. Investigation of different factors affecting the electron spin resomance-based characterization of γ-irradiated fresh, white, and red ginseng. J. Ginseng Res. 2012, 36, 308. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-R.; Whang, W.-K.; Shin, C.-G.; Lee, H.-S.; Han, S.-T.; Im, B.-O.; Ko, S.-K. Comparison of ginsenoside composition and contents in fresh ginseng roots cultivated in Korea, Japan, and China at various ages. Korean J. Food Sci. Technol. 2004, 36, 847–850. [Google Scholar]
- Wan, J.Y.; Fan, Y.; Yu, Q.T.; Ge, Y.Z.; Yan, C.P.; Alolga, R.N.; Li, P.; Ma, Z.H.; Qi, L.W. Integrated evaluation of malonyl ginsenosides, amino acids and polysaccharides in fresh and processed ginseng. J. Pharm. Biomed. Anal. 2015, 107, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.Z.; Huang, M.Y.; Niemira, B.A.; Cheng, L.Y. Shelf life extension of fresh ginseng roots using sanitiser washing, edible antimicrobial coating and modified atmosphere packaging. Int. J. Food Sci. Technol. 2016, 51, 2132–2139. [Google Scholar] [CrossRef]
- He, J.M.; Zhang, Y.Z.; Luo, J.P.; Zhang, W.J.; Mu, Q. Variation of ginsenosides in ginseng of different ages. Nat. Prod. Commun. 2016, 11, 739–740. [Google Scholar] [PubMed]
- Pan, H.Y.; Qu, Y.; Zhang, J.K.; Kang, T.G.; Dou, D.Q. Antioxidant activity of ginseng cultivated under mountainous forest with different growing years. J. Ginseng Res. 2013, 37, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Zhang, Z.H.; Zu, Y.G.; Tang, Z.H.; Efferth, T. Correlation of cultivation time of Panax ginseng with metabolic profiles of nine ginsenosides and mrna expression of genes encoding major biosynthetic enzymes. Acta. Physiol. Plant 2016, 38, 51. [Google Scholar] [CrossRef]
- Yeganehjoo, H.; DeBose-Boyd, R.; McFarlin, B.; Mo, H. Synergistic impact of d-δ-tocotrienol and geranylgeraniol on the growth and hmg coa reductase of human du145 prostate carcinoma cells. FASEB J. 2016, 30, 1176.27. [Google Scholar]
- Hu, W.Z.; Jiang, A.L.; Qi, H.P. Physiological behavior and quality of fresh ginseng stored in modified atmospheres generated by several package films. J. Food Sci. Technol. 2014, 51, 3862–3869. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.Z.; Tanaka, S.; Uchino, T.; Hamanaka, D.; Hori, Y. Effects of packaging film and storage temperature on the quality of fresh ginseng packaged in modified atmosphere. J. Fac. Agric. Kyushu Univ. 2004, 49, 139–147. [Google Scholar]
- Hu, W.Z.; Xu, P.; Uchino, T. Extending storage life of fresh ginseng by modified atmosphere packaging. J. Sci. Food Agric. 2005, 85, 2475–2481. [Google Scholar] [CrossRef]
- Macura, D.; McCannel, A.M.; Li, M.Z.C. Survival of clostridium botulinum in modified atmosphere packaged fresh whole North American ginseng roots. Food Res. Int. 2001, 34, 123–125. [Google Scholar] [CrossRef]
- Liang, J.B.; Jiang, C.; Peng, H.S.; Shi, Q.H.; Guo, X.; Yuan, Y.; Huang, L.Q. Analysis of the age of Panax ginseng based on telomere length and telomerase activity. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.S.; Yuan, Q.X.; Zhou, H.; Huang, L.Q. Nondestructive estimation of growth year in ginseng cultivars using the means of mathematical modeling on the basis of allometry. Microsc. Res. Tech. 2016, 79, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.K.; Ahn, M.S.; Park, J.S.; Liu, J.R.; In, D.S.; Min, B.W.; Kim, S.W. Discrimination of cultivation ages and cultivars of ginseng leaves using fourier transform infrared spectroscopy combined with multivariate analysis. J. Ginseng Res. 2014, 38, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.Q.; Wang, J.; Geng, L.H.; Wei, Z.B.; Tian, X.J. Determination of ginseng with different ages using a taste-sensing system. Sens. Mater. 2013, 25, 241–255. [Google Scholar]
- Cui, S.Q.; Wang, J.; Yang, L.C.; Wu, J.F.; Wang, X.L. Qualitative and quantitative analysis on aroma characteristics of ginseng at different ages using E-nose and GC-MS combined with chemometrics. J. Pharm. Biomed. Anal. 2015, 102, 64–77. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Wang, Y.T.; Li, J.; Zhang, H.Q.; Ding, L. Investigation of ginsenosides in different parts and ages of panax ginseng. Food Chem. 2007, 102, 664–668. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, J.Y.; Xiao, X.Y.; Lin, R.C.; Cheng, Y.Y. Simultaneous determination of ginsenosides in Panax ginseng with different growth ages using high-performance liquid chromatography-mass spectrometry. Phytochem. Anal. 2006, 17, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Wang, B.; Liu, Y.; Shi, M.; Wang, D.; Zhang, X.; Liu, T.; Huang, L.; Zhang, X. Producing aglycons of ginsenosides in bakers’ yeast. Sci. Rep. 2014, 4, 3698. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Liu, N.; Tian, Y.H.; Zhang, L.X. Molecular cloning, expression, purification, and functional characterization of dammarenediol synthase from Panax ginseng. Biomed. Res. Int. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Tansakul, P.; Shibuya, M.; Kushiro, T.; Ebizuka, Y. Dammarenediol-ii synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. FEBS. Lett. 2006, 580, 5143–5149. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Li, J.; Liu, S.; Wu, X.; Li, J.; Gao, W. Transcriptome profiling shows gene regulation patterns in ginsenoside pathway in response to methyl jasmonate in Panax quinquefolium adventitious root. Sci. Rep. 2016, 6, 37263. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Yamazaki, M.; Takahashi, H.; Nakamura, M.; Kojoma, M.; Suzuki, H.; Saito, K. RNA-seq transcriptome analysis of Panax japonicus, and its comparison with other Panax species to identify potential genes involved in the saponins biosynthesis. Front. Plant Sci. 2016, 7, 481. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Luo, H.; Li, Y.; Sun, Y.; Wu, Q.; Niu, Y.; Song, J.; Lv, A.; Zhu, Y.; Sun, C.; et al. 454 EST analysis detects genes putatively involved in ginsenoside biosynthesis in Panax ginseng. Plant Cell Rep. 2011, 30, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Dozier, J.K.; Distefano, M.D. An enzyme-coupled continuous fluorescence assay for farnesyl diphosphate synthases. Anal. Biochem. 2012, 421, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Rong, Q.; Chen, Y.; Yuan, Q.; Shen, Y.; Guo, J.; Yang, Y.; Zha, L.; Wu, H.; Huang, L.; et al. Molecular cloning and functional analysis of squalene synthase (ss) in Panax notoginseng. Int. J. Boil. Macromol. 2017, 95, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, M.; Eiben, S.; Asta, C.; Do, T.A.; Pleiss, J.; Urlacher, V.B. Cloning, expression and characterisation of CYP102A7, a self-sufficient P450 monooxygenase from Bacillus licheniformis. Appl. Microbiol. Biot. 2008, 79, 931–940. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, J.; Zhang, D.; Zhuang, J.; Huang, Y.; Mu, Y.; Lv, S. Study on the Correlation between Gene Expression and Enzyme Activity of Seven Key Enzymes and Ginsenoside Content in Ginseng in Over Time in Ji’an, China. Int. J. Mol. Sci. 2017, 18, 2682. https://doi.org/10.3390/ijms18122682
Yin J, Zhang D, Zhuang J, Huang Y, Mu Y, Lv S. Study on the Correlation between Gene Expression and Enzyme Activity of Seven Key Enzymes and Ginsenoside Content in Ginseng in Over Time in Ji’an, China. International Journal of Molecular Sciences. 2017; 18(12):2682. https://doi.org/10.3390/ijms18122682
Chicago/Turabian StyleYin, Juxin, Daihui Zhang, Jianjian Zhuang, Yi Huang, Ying Mu, and Shaowu Lv. 2017. "Study on the Correlation between Gene Expression and Enzyme Activity of Seven Key Enzymes and Ginsenoside Content in Ginseng in Over Time in Ji’an, China" International Journal of Molecular Sciences 18, no. 12: 2682. https://doi.org/10.3390/ijms18122682