Every OGT Is Illuminated … by Fluorescent and Synchrotron Lights
Abstract
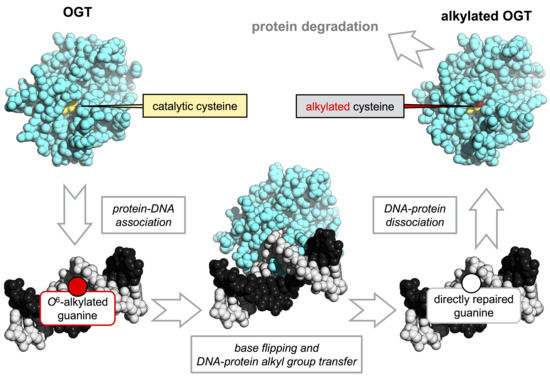
:1. Introduction
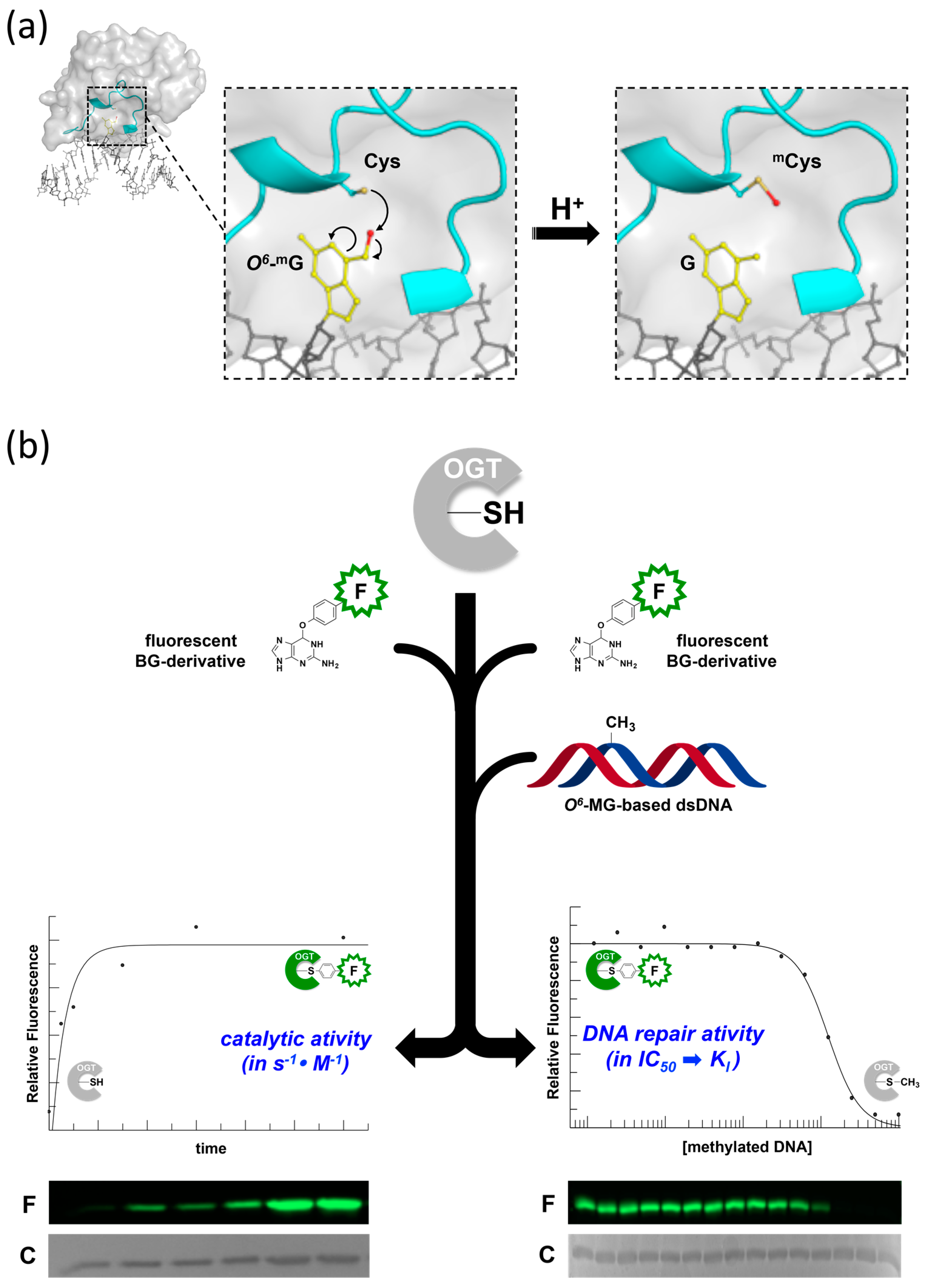
2. Development of Novel Alkyl-Transferase Assays
3. DNA Alkylation Damage and OGT-Mediated DNA Repair Response in M. tuberculosis
3.1. Biochemical and Structural Studies of M. tuberculosis OGT
3.2. The Active-Site Loop of MtOGT Is Intrinsically Flexible
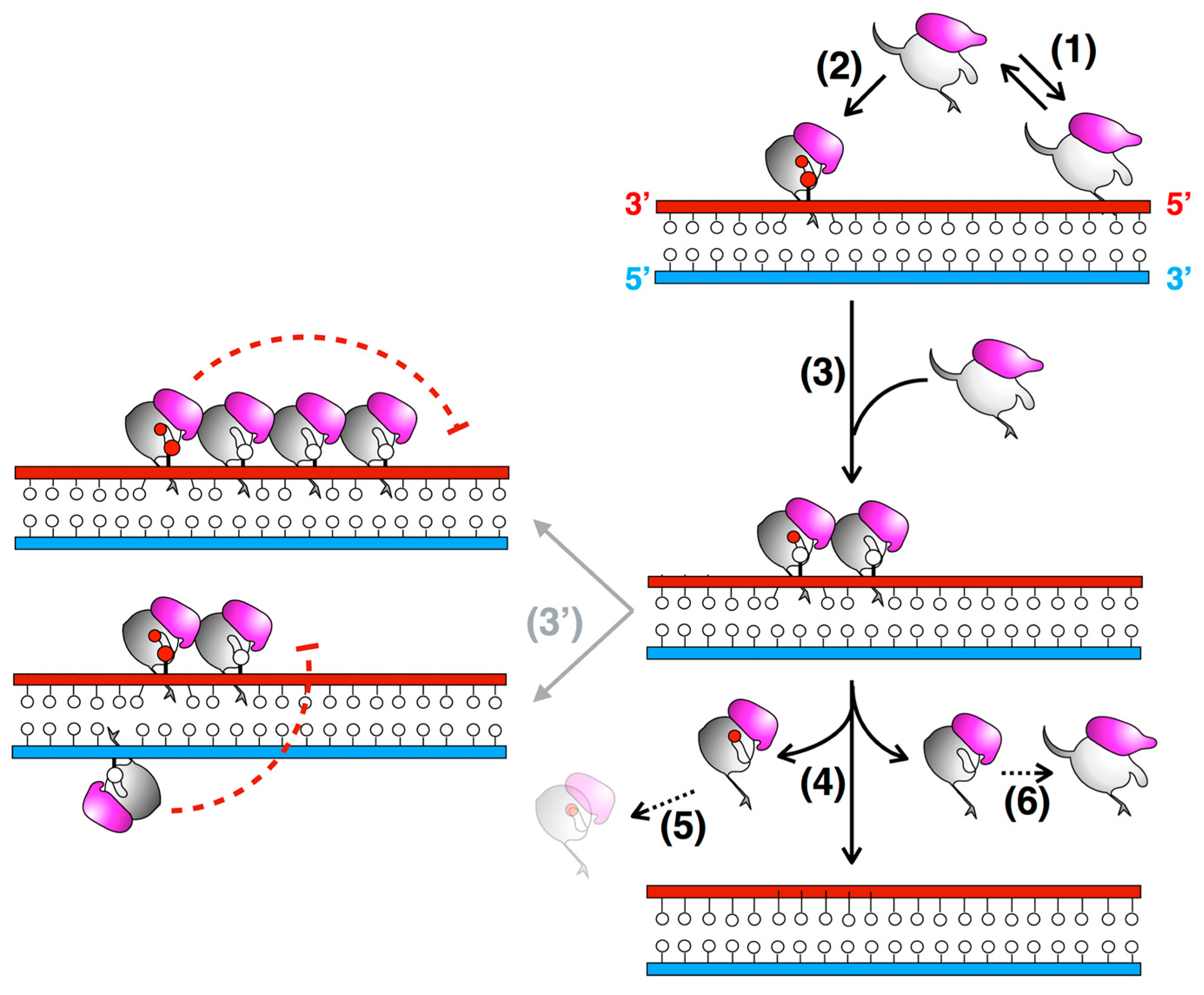
3.3. The N-Terminal Domain of MtOGT Attends DNA Cooperative Binding Process
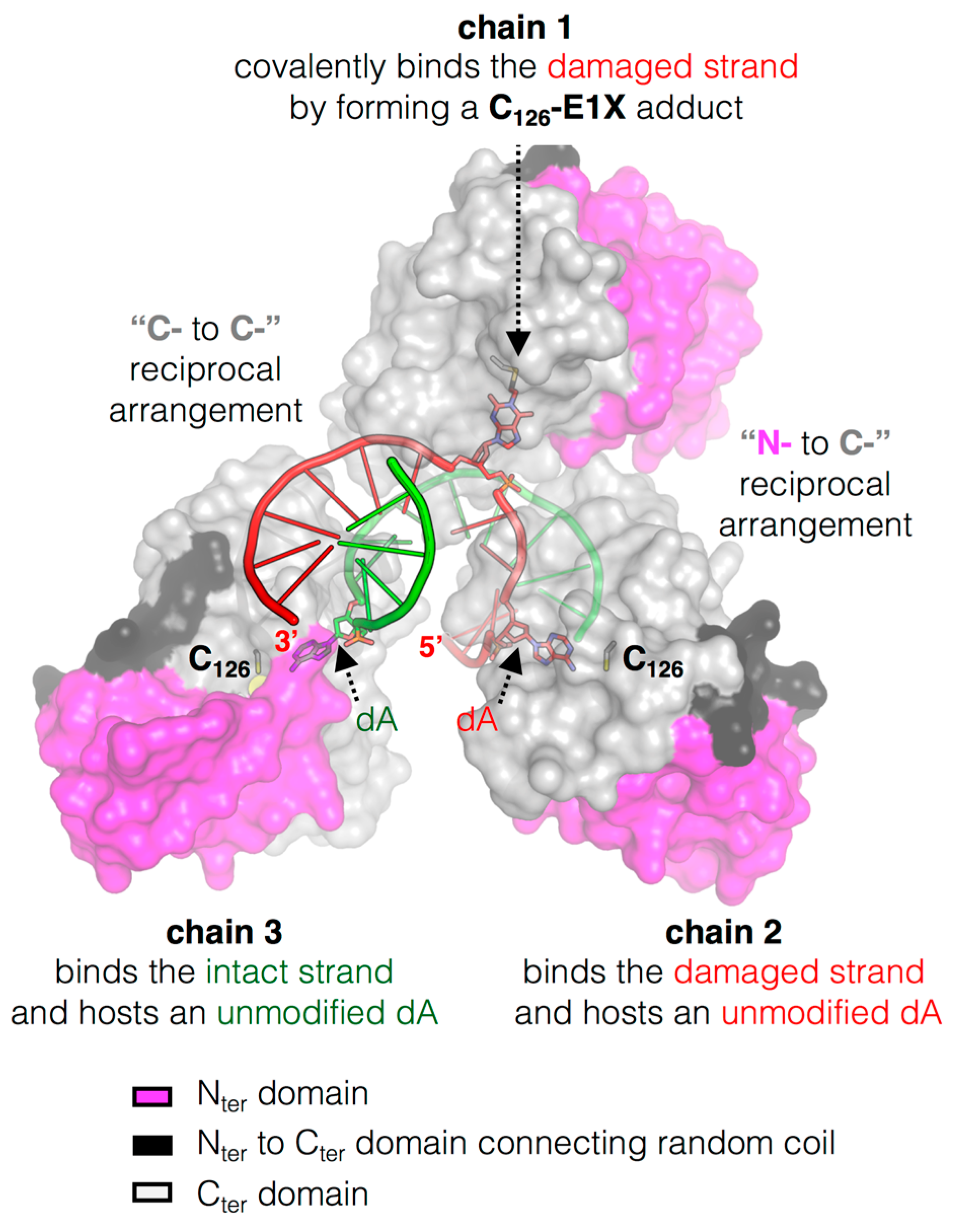
3.4. The Structure of MtOGT in Complex with Modified DNA Sheds Light on Cooperative Substrate Binding
4. The S. solfataricus DNA Alkyl-Transferase, SsOGT
4.1. Biochemical Characterization of SsOGT
4.2. SsOGT and the S. solfataricus Response to Alkylating Agents
4.3. Crystal Structure of Free and DNA-Bound SsOGT
4.4. Alkylation of the Active Site Induces Dramatic Conformational Changes and Destabilization of SsOGT
5. Development of OGT-Based Novel Protein-Tags
6. Conclusions and Perspective
Acknowledgments
Conflicts of Interest
References
- Pegg, A.E. Repair of O6-alkylguanine by alkyltransferases. Mutat. Res. 2000, 462, 83–100. [Google Scholar] [CrossRef]
- Drabløs, F.; Feyzi, E.; Aas, P.A.; Vaagbø, C.B.; Kavli, B.; Bratlie, M.S.; Peña-Diaz, J.; Otterlei, M.; Slupphaug, G.; Krokan, H.E. Alkylation damage in DNA and RNA-repair mechanisms and medical significance. DNA Repair 2004, 11, 1389–1407. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Multifaceted roles of alkyltransferase and related proteins in DNA repair, DNA damage, resistance to chemotherapy, and research tools. Chem. Res. Toxicol. 2011, 24, 618–639. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, J.L.; Pegg, A.E.; Tainer, J.A. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA-alkyltransferase and its implications for cancer chemotherapy. DNA Repair 2007, 6, 1100–1115. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H. Genome maintenance mechanisms for preventing cancer. Nature 2001, 411, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Mielecki, D.; Wrzesiński, M.; Grzesiuk, E. Inducible repair of alkylated DNA in microorganisms. Mutat. Res. Rev. Mutat. Res. 2015, 763, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Soll, J.M.; Sobol, R.W.; Mosammaparast, N. Regulation of DNA Alkylation Damage Repair: Lessons and Therapeutic Opportunities. Trends Biochem. Sci. 2017, 3, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Mishina, Y.; Duguid, E.M.; Chuan, H. Direct reversal of DNA alkylation damage. Chem. Rev. 2006, 106, 215–232. [Google Scholar] [CrossRef] [PubMed]
- Debiak, M.; Nikolova, T.; Kaina, B. Loss of ATM sensitizes against O6-methylguanine triggered apoptosis, SCEs and chromosomal aberrations. DNA Repair 2004, 3, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Gillingham, D.; Sauter, B. Genomic Studies Reveal New Aspects of the Biology of DNA Damaging Agents. Chembiochem 2017. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Kanugula, S.; Pegg, A.E. Function of domains of human O6-alkyl-guanine-DNA alkyltransferase. Biochemistry 2005, 44, 15396–15405. [Google Scholar] [CrossRef] [PubMed]
- Jena, N.R.; Shukla, P.K.; Jena, H.S.; Mishra, P.C.; Suhai, S. O6-methylguanine repair by O6-alkylguanine-DNA alkyltransferase. J. Phys. Chem. B 2009, 113, 16285–16290. [Google Scholar] [CrossRef] [PubMed]
- Xu-Welliver, M.; Pegg, A.E. Degradation of the alkylated form of the DNA repair protein, O6-alkylguanine-DNA alkyltransferase. Carcinogenesis 2002, 23, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Daniels, D.S.; Woo, T.T.; Luu, K.X.; Noll, D.M.; Clarke, N.D.; Pegg, A.E.; Tainer, J.A. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 2004, 11, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Mielecki, D.; Grzesiuk, E. Ada response—A strategy for repair of alkylated DNA in bacteria. FEMS Microbiol. Lett. 2014, 355, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sedgwick, B.; Lindahl, T. Recent progress on the Ada response for inducible repair of DNA alkylation damage. Oncogene 2002, 58, 8886–8894. [Google Scholar] [CrossRef] [PubMed]
- Takano, K.; Nakamura, T.; Sekiguchi, M. Roles of two types of O6-methylguanine-DNA methyltransferases in DNA repair. Mutat. Res. 1991, 254, 37–44. [Google Scholar] [CrossRef]
- Khan, O.; Middleton, M.R. The therapeutic potential of O6-alkylguanine DNA alkyltransferase inhibitors. Expert Opin. Investig. Drugs 2007, 10, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, K.; Zhu, Y.L.; Shyam, K.; Penketh, P.G.; Baumann, R.P.; Sartorelli, A.C. Quantitative relationship between guanine O6-alkyl lesions produced by OnriginTM and tumor resistance by O6-alkyl-guanine-DNA alkyltransferase. Biochem. Pharmacol. 2010, 80, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- Philip, S.; Swaminathan, S.G.; Kuznetsov, S.; Kanugula, K.; Biswas, S.; Chang, N.A.; Loktionova, D.C.; Haines, P.; Kaldis, A.E.; Pegg, S.K.; et al. Degradation of BRCA2 in alkyltransferase-mediated DNA repair and its clinical implications. Cancer Res. 2008, 68, 9973–9981. [Google Scholar] [CrossRef] [PubMed]
- Keppler, A.; Pick, H.; Arrivoli, C.; Vogel, H.; Johnsson, K. Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. USA 2004, 10, 9955–9959. [Google Scholar] [CrossRef] [PubMed]
- Keppler, A.; Gendreizig, S.; Gronemeyer, T.; Pick, H.; Vogel, H.; Johnsson, K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003, 21, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Gautier, A.; Juillerat, A.; Heinis, C.; Corrêa, I.R., Jr.; Kindermann, M.; Beaufils, F.; Johnsson, K. An engineered protein-tag for multi-protein labeling in living cells. Chem. Biol. 2008, 15, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, K.; Shyam, K.; Penketh, P.G.; Sartorelli, A.C. Development of an O6-alkyl-guanine-DNA alkyltransferase assay based on covalent transfer of the benzyl moiety from [benzene-3H]-O6-benzylguanine to the protein. Anal. Biochem. 2008, 83, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Tintoré, M.; Aviñó, A.; Ruiz, F.M.; Eritja, R.; Fábrega, C. Development of a novel fluorescence assay based on the use of the thrombin binding aptamer for the detection of O6-alkyl-guanine-DNA alkyltransferase activity. J. Nucleic Acids 2010, 2010, 632041. [Google Scholar] [CrossRef] [PubMed]
- Gronemeyer, T.; Chidley, C.; Juillerat, A.; Heinis, C.; Johnsson, K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for applications in protein labeling. Protein Eng. Des. Sel. 2006, 19, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Hinner, M.J.; Johnsson, K. How to obtain labeled proteins and what to do with them. Curr. Opin. Biotechnol. 2010, 21, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Perugino, G.; Vettone, A.; Illiano, G.; Valenti, A.; Ferrara, M.C.; Rossi, M.; Ciaramella, M. Activity and regulation of archaeal DNA alkyltransferase: Conserved protein involved in repair of DNA alkylation damage. J. Biol. Chem. 2012, 287, 4222–4231. [Google Scholar] [CrossRef] [PubMed]
- Miggiano, R.; Casazza, V.; Garavaglia, S.; Ciaramella, M.; Perugino, G.; Rizzi, M.; Rossi, F. Biochemical and structural studies of the Mycobacterium tuberculosis O6-methylguanine methyltransferase and mutated variants. J. Bacteriol. 2013, 195, 2728–2736. [Google Scholar] [CrossRef] [PubMed]
- Perugino, G.; Miggiano, R.; Serpe, M.; Vettone, A.; Valenti, A.; Lahiri, S.; Rossi, F.; Rossi, M.; Rizzi, M.; Ciaramella, M. Structure-function relationships governing activity and stability of a DNA alkylation damage repair thermostable protein. Nucleic Acids Res. 2015, 43, 8801–8816. [Google Scholar] [CrossRef] [PubMed]
- Gorna, A.E.; Bowater, R.P.; Dziadek, J. DNA repair systems and the pathogenesis of Mycobacterium tuberculosis: Varying activities at different stages of infection. Clin. Sci. 2010, 119, 187–202. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, E.C.; Walker, G.C.; Siede, W. DNA Repair and Mutagenesis; ASM Press: Washington, DC, USA, 1995. [Google Scholar]
- Durbach, S.I.; Springer, B.; Machowski, E.E.; North, R.J.; Papavinasasundaram, K.G.; Colston, M.J.; Bottger, E.C.; Mizrahi, V. DNA alkylation damage as a sensor of nitrosative stress in Mycobacterium tuberculosis. Infect. Immun. 2003, 71, 997–1000. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Aamodt, R.M.; Dalhus, B.; Balasingham, S.; Helle, I.; Andersen, P.; Tonjum, T.; Alseth, I.; Rognes, T.; Bjoras, M. The ada operon of Mycobacterium tuberculosis encodes two DNA-methyltransferases for inducible repair of DNA alkylation damage. DNA Repair 2011, 10, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Shrivastav, N.; Li, D.; Essigmann, J.M. Chemical biology of mutagenesis and DNA repair: Cellular responses to DNA alkylation. Carcinogenesis 2010, 3, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Sassetti, C.M.; Boyd, D.H.; Rubin, E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 2003, 48, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Boshoff, H.I.; Myers, T.G.; Copp, B.R.; McNeil, M.R.; Wilson, M.A.; Barry, C.E., III. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: Novel insights into drug mechanisms of action. J. Biol. Chem. 2004, 279, 40174–40184. [Google Scholar] [CrossRef] [PubMed]
- Schnappinger, D.; Ehrt, S.; Voskuil, M.I.; Liu, Y.; Mangan, J.A.; Monahan, I.M.; Dolganov, G.; Efron, B.; Butcher, P.D.; Nathan, C.; et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: Insights into the phagosomal environment. J. Exp. Med. 2003, 198, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.C.; Potter, P.M.; Cawkwell, L.; Georgiadis, P.; Patel, D.; Swann, P.F.; Margison, G.P. Purification of the E. coli ogt gene product to homogeneity and its rate of action on O6-methylguanine, O6-ethylguanine and O4-methylthymine in dodecadeoxyribonucleotides. Nucleic Acids Res. 1989, 17, 8475–8484. [Google Scholar] [CrossRef] [PubMed]
- Goodtzova, K.; Kanugula, S.; Edara, S.; Pauly, G.T.; Moschel, R.C.; Pegg, A.E. Repair of O6-benzylguanine by the Escherichia coli Ada and Ogt and the human O6-alkylguanine-DNA alkyltransferase. J. Biol. Chem. 1997, 272, 8332–8339. [Google Scholar] [CrossRef] [PubMed]
- Spratt, T.E.; Wu, J.D.; Levy, D.E.; Kanugula, S.; Pegg, A.E. Reaction and binding of oligodeoxynucleotides containing analogues of O6-methylguanine with wild-type and mutant human O6-alkylguanine-DNA alkyltransferase. Biochemistry 1999, 38, 6801–6806. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Inoue, T.; Nishioka, M.; Fujiwara, S.; Takagi, M.; Imanaka, T.; Kai, Y. Hyperthermostable protein structure maintained by intra and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase. J. Mol. Biol. 1999, 292, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Pelton, J.G.; Wemmer, D.E. Structural studies of MJ1529, an O6-methylguanine-DNA methyltransferase. Magn. Reson. Chem. 2006, 44, S71–S82. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.H.; Gulbis, J.M.; Dodson, E.J.; Demple, B.; Moody, P.C. Crystal structure of a suicidal DNA repair protein: The Ada O6-methylguanineDNA methyltransferase from E. coli. EMBO J. 1994, 13, 1495–1501. [Google Scholar] [PubMed]
- Miggiano, R.; Perugino, G.; Ciaramella, M.; Serpe, M.; Rejman, D.; Páv, O.; Pohl, R.; Garavaglia, S.; Lahiri, S.; Rizzi, M.; et al. Crystal structure of Mycobacterium tuberculosis O6-methylguanine-DNA methyltransferase protein clusters assembled on to damaged DNA. Biochem. J. 2016, 473, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Daniels, D.S.; Mol, C.D.; Arvai, A.S.; Kanugula, S.; Pegg, A.E.; Tainer, J.A. Active and alkylated human AGT structures: A novel zinc site, inhibitor and extrahelical base binding. EMBO J. 2000, 19, 1719–1730. [Google Scholar] [CrossRef] [PubMed]
- Wibley, J.E.; Pegg, A.E.; Moody, PC. Crystal structure of the human O6-alkylguanine-DNA alkyltransferase. Nucleic Acids Res. 2000, 28, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Duguid, E.M.; Rice, P.A.; He, C. The structure of the human AGT protein bound to DNA and its implications for damage detection. J. Mol. Biol. 2005, 350, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Olano, J.; Lopez, B.; Reyes, A.; Lemos, M.P.; Correa, N.; Del Portillo, P.; Barrera, L.; Robledo, J.; Ritacco, V.; Zambrano, M.M. Mutations in DNA repair genes are associated with the Haarlem lineage of Mycobacterium tuberculosis independently of their antibiotic resistance. Tuberculosis 2007, 87, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Rad, M.E.; Bifani, P.; Martin, C.; Kremer, K.; Samper, S.; Rauzier, J.; Kreiswirth, B.; Blazquez, J.; Jouan, M.; van Soolingen, D.; et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 2003, 9, 838. [Google Scholar]
- Adams, C.A.; Melikishvili, M.; Rodgers, D.W.; Rasimas, J.J.; Pegg, A.E.; Fried, M.G. Topologies of complexes containing O6-alkylguanine-DNA alkyltransferase and DNA. J. Mol. Biol. 2009, 389, 248–263. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.A.; Fried, M.G. Mutations that probe the cooperative assembly of O6-alkylguanine-DNA alkyltransferase complexes. Biochemistry 2011, 50, 1590–1598. [Google Scholar] [CrossRef] [PubMed]
- Melikishvili, M.; Fried, M.G. Quaternary interactions and supercoiling modulate the cooperative DNA binding of AGT. Nucleic Acids Res. 2017, 45, 7226–7236. [Google Scholar] [CrossRef] [PubMed]
- Tessmer, I.; Fried, M.G. Insight into the cooperative DNA binding of the O6-alkylguanine DNA alkyltransferase. DNA Repair 2014, 20, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Begley, T.J.; Samson, L.D. Reversing DNA damage with a directional bias. Nat. Struct. Mol. Biol. 2004, 11, 688–690. [Google Scholar] [CrossRef] [PubMed]
- Valenti, A.; Napoli, A.; Ferrara, M.C.; Nadal, M.; Rossi, M.; Ciaramella, M. Selective degradation of reverse gyrase and DNA fragmentation induced by alkylating agent in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2006, 34, 2098–2108. [Google Scholar] [CrossRef] [PubMed]
- Leclere, M.M.; Nishioka, M.; Yuasa, T.; Fujiwara, S.; Takagi, M.; Imanaka, T. The O6-methylguanine-DNA methyltransferase from the hyperthermophilic archaeon Pyrococcus sp. KOD1: A thermostable repair enzyme. Mol. Gen. Genet. 1998, 258, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Skorvaga, M.; Raven, N.D.; Margison, G.P. Thermostable archaeal O6-alkylguanine-DNA alkyltransferases. Proc. Natl. Acad. Sci. USA 1998, 95, 6711–6715. [Google Scholar] [CrossRef] [PubMed]
- Kanugula, S.; Pegg, A.E. Alkylation damage repair protein O6-alkyl-guanine-DNA alkyltransferase from the hyperthermophiles Aquifex aeolicus and Archaeoglobus fulgidus. Biochem. J. 2003, 375, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Vettone, A.; Perugino, G.; Rossi, M.; Valenti, A.; Ciaramella, M. Genome stability: Recent insights in the topoisomerase reverse gyrase and thermophilic DNA alkyltransferase. Extremophiles 2014, 18, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Morrone, C.; Miggiano, R.; Serpe, M.; Massarotti, A.; Valenti, A.; Del Monaco, G.; Rossi, M.; Rossi, F.; Rizzi, M.; Perugino, G.; et al. Interdomain interactions rearrangements control the reaction steps of a thermostable DNA alkyltransferase. Biochim. Biophys. Acta 2017, 1861, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Vettone, A.; Serpe, M.; Hidalgo, A.; Berenguer, J.; del Monaco, G.; Valenti, A.; Ciaramella, M.; Perugino, G. A novel thermostable protein-tag: Optimization of the Sulfolobus solfataricus DNA-alkyl-transferase by protein engineering. Extremophiles 2016, 20, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Visone, V.; Han, W.; Perugino, G.; Del Monaco, G.; She, Q.; Rossi, M.; Valenti, A.; Ciaramella, M. In vivo and in vitro protein imaging in thermophilic archaea by exploiting a novel protein tag. PLoS ONE 2017, 10, e0185791. [Google Scholar] [CrossRef] [PubMed]
- Valenti, A.; Perugino, G.; Nohmi, T.; Rossi, M.; Ciaramella, M. Inhibition of translesion DNA polymerase by archaeal reverse gyrase. Nucleic Acids Res. 2009, 37, 4287–4295. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.S.; Shemorry, A.; Varshavsky, A. Two proteolytic pathways regulate DNA repair by cotargeting the Mgt1 alkyl-guanine transferase. Proc. Natl. Acad. Sci. USA 2009, 106, 2142–2147. [Google Scholar] [CrossRef] [PubMed]
- Maupin-Furlow, J.A.; Humbard, M.A.; Kirkland, P.A.; Li, W.; Reuter, C.J.; Wright, A.J.; Zhou, G. Proteasomes from structure to function: Perspectives from archaea. Curr. Top. Dev. Biol. Rev. 2006, 75, 125–169. [Google Scholar]
- Mallick, P.; Boutz, D.R.; Eisenberg, D.; Yeates, T.O. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc. Natl. Acad. Sci. USA 2002, 99, 9679–9684. [Google Scholar] [CrossRef] [PubMed]
- Porcelli, M.; De Leo, E.; Del Vecchio, P.; Fuccio, F.; Cacciapuoti, G. Thermal unfolding of nucleoside hydrolases from the hyperthermophilic archaeon Sulfolobus solfataricus: Role of disulfide bonds. Protein Pept. Lett. 2012, 19, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Nishikori, S.; Shiraki, K.; Yokota, K.; Izumikawa, N.; Fujiwara, S.; Hashimoto, H.; Imanaka, T.; Takagi, M. Mutational effects on O6-methylguanine-DNA methyltransferase from hyperthermophile: Contribution of ion-pair network to protein thermostability. J. Biochem. 2004, 135, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Crone, T.M.; Goodtzova, K.; Pegg, A.E. Amino acid residues affecting the activity and stability of human O6-alkylguanine-DNA alkyltransferase. Mutat. Res. 1996, 63, 15–25. [Google Scholar] [CrossRef]
- Chalfie, M.; Tu, Y.; Euskirchen, G.; Ward, W.W.; Prasher, D.C. Green fluorescent protein as a marker for gene expression. Science 1994, 263, 802–805. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–554. [Google Scholar] [CrossRef] [PubMed]
- Perugino, G.; Valenti, A.; D’Amaro, A.; Rossi, M.; Ciaramella, M. Reverse gyrase and genome stability in hyperthermophilic organisms. Biochem. Soc. Trans. 2009, 37, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Jamroze, A.; Perugino, G.; Valenti, A.; Rashid, N.; Rossi, M.; Akhtar, M.; Ciaramella, M. The reverse gyrase from Pyrobaculum calidifontis, a novel extremely thermophilic DNA topoisomerase endowed with DNA unwinding and annealing activities. J. Biol. Chem. 2014, 289, 3231–3243. [Google Scholar] [CrossRef] [PubMed]
- Valenti, A.; Perugino, G.; Rossi, M.; Ciaramella, M. Positive supercoiling in thermophiles and mesophiles: Of the good and evil. Biochem. Soc. Trans. 2011, 39, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Valenti, A.; Perugino, G.; D’Amaro, A.; Cacace, A.; Napoli, A.; Rossi, M.; Ciaramella, M. Dissection of reverse gyrase activities: Insight into the evolution of a thermostable molecular machine. Nucleic Acids Res. 2008, 36, 4587–4597. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.; Nakagawa, N.; Kuramitsu, S.; Masui, R. An O6-methylguanine-DNA methyltransferase-like protein from Thermus thermophilus interacts with a nucleotide excision repair protein. J. Biochem. 2008, 144, 267–277. [Google Scholar] [CrossRef] [PubMed]


| Topt | 80 °C | |
|---|---|---|
| Activity | 25 °C | 25% |
| 37 °C | 45% | |
| 80 °C | 100% | |
| pHopt | 6.0 | |
| catalytic activity (M−1 s−1) | 25 °C | 0.28 ± 0.03 × 104 |
| 70 °C | 5.33 ± 1.49 × 104 | |
| thermal stability (t½, min) | 60 °C | 257.2 ± 10.3 |
| 70 °C | 165.1 ± 16.5 | |
| 80 °C | 18.7 ± 2.0 | |
| NaCl > 1.0 M | 100% activity | |
| EDTA > 10.0 mM | 100% activity | |
| Dithiotreithol (DTT) | not required | |
| O6-MG-DNA (Ki, µM) | close to 5′ end | 4.29 ± 0.39 |
| central | 0.83 ± 0.02 | |
| close to 3′ end | 56.6 ± 23.8 | |
| Type | Mutation | ∆Tm (°C) |
|---|---|---|
| - | SsOGT | a |
| disulphide bond | SsOGT C29A | −17.1 |
| catalytic cysteine | SsOGT C119m | −17.3 |
| SsOGT C119L | −35.4 | |
| SsOGT C119F | −35.1 | |
| Nter-Cter ion pair | SsOGT D27A | −8.0 |
| SsOGT D27K | −35.3 | |
| Nter-loop network | SsOGT E44L | +0.7 |
| SsOGT K48A | −8.0 | |
| SsOGT K48L | −9.6 | |
| DNA binding | SsOGT H5 | −5.0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miggiano, R.; Valenti, A.; Rossi, F.; Rizzi, M.; Perugino, G.; Ciaramella, M. Every OGT Is Illuminated … by Fluorescent and Synchrotron Lights. Int. J. Mol. Sci. 2017, 18, 2613. https://doi.org/10.3390/ijms18122613
Miggiano R, Valenti A, Rossi F, Rizzi M, Perugino G, Ciaramella M. Every OGT Is Illuminated … by Fluorescent and Synchrotron Lights. International Journal of Molecular Sciences. 2017; 18(12):2613. https://doi.org/10.3390/ijms18122613
Chicago/Turabian StyleMiggiano, Riccardo, Anna Valenti, Franca Rossi, Menico Rizzi, Giuseppe Perugino, and Maria Ciaramella. 2017. "Every OGT Is Illuminated … by Fluorescent and Synchrotron Lights" International Journal of Molecular Sciences 18, no. 12: 2613. https://doi.org/10.3390/ijms18122613