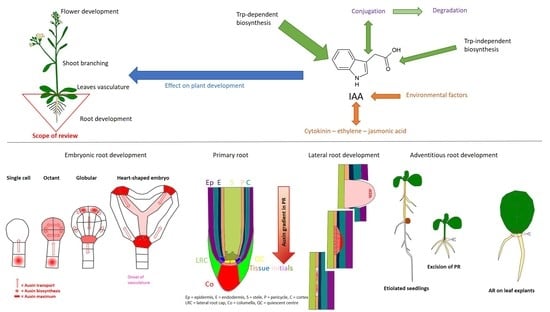
Control of Endogenous Auxin Levels in Plant Root Development
Abstract
:1. Introduction
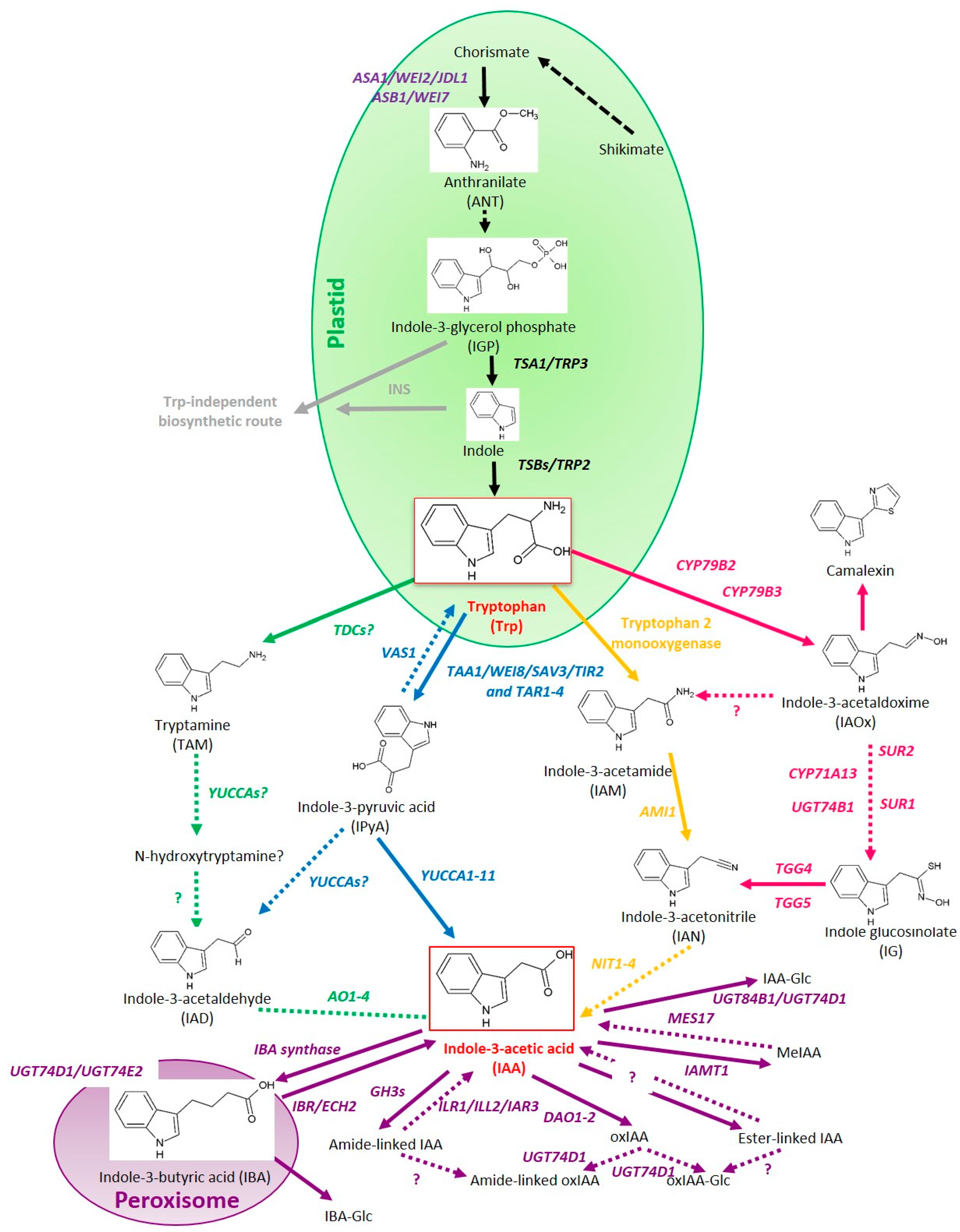
2. Auxin Biosynthesis
2.1. The Indole-3-Pyruvic Acid (IPyA) Pathway
2.2. The Indole-3-Acetamide (IAM) Pathway
2.3. The Tryptamine (TAM) Pathway
2.4. The Indole-3-Acetaldoxime (IAOx) Pathway
2.5. Tryptophan-Independent Auxin Biosynthesis Pathways
3. Auxin Biosynthesis Regulates Root Embryogenesis
4. Local Auxin Accumulation Regulates Root Development and Branching
5. Importance of Auxin Biosynthesis for Adventitious Root Development
6. Regulation of Auxin Homeostasis during Root Development by Other Hormones
7. Auxin Biosynthesis as an Integrator of Environmental Factors and Root Development
8. Meta- and Catabolic Processes Controlling Auxin Levels
9. Conclusions
Author Contributions
Conflicts of Interest
Abbreviations
| 2,4-d | 2,4-dichlorophenoxy acetic acid |
| 4-Cl-IAA | 4-chloroindole-3-acetic acid |
| ABP1 | auxin binding protein 1 |
| AFB | AUXIN SIGNALING F-BOX (AFB) family of auxin receptors |
| AO | aldehyde oxidases |
| AR(s) | adventitious root(s) |
| ARF | auxin response factor |
| Aux/IAA | AUXIN/INDOLE-3-ACETIC ACID proteins |
| IAA | indole-3-acetic acid |
| IAD | indole-3-acetaldehyde |
| IAM | indole-3-acetamide |
| IAN | indole-3-acetonitrile |
| IAOx | indole-3-acetaldoxime |
| IBA | indole-3-butyric acid |
| IG | indole glucosinolates |
| IGS | indole-3-glycerol phosphate synthase |
| IPyA | indole-3-pyruvic acid |
| LR(s) | lateral root(s) |
| NAA | 1-naphthaleneacetic acid |
| PAA | phenyl acetic acid |
| PIN | PIN-formed family of auxin transporters |
| PHYB | PHYTOCHROME B |
| PR | primary root |
| QC | quiescent center |
| RAM | root apical meristem |
| SAM | shoot apical meristem |
| SCF | ubiquitin protein ligase complex, with subunits: Skp1, Cullin and F-box |
| TAA1/TAR1-4 | TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1/TRYPTOPHAN AMINOTRANSFERASE RELATED 1-4 |
| TAM | Tryptamine |
| TIR1/2 | TRANSPORT INHIBITOR RESPONSE ½ |
| Trp | Tryptophan |
| TSA1 | Trp synthase a |
| TSB | Trp synthase b |
| YUC1-11 | yucca genes encoding flavin-containing monooxygenases |
References
- Palmer, C.M.; Bush, S.M.; Maloof, J.N. Phenotypic and Developmental Plasticity in Plants; Encyclopedia of Life Science, John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2012. [Google Scholar]
- Gaillochet, C.; Lohmann, J.U. The never-ending story: From pluripotency to plant developmental plasticity. Development 2015, 142, 2237–2249. [Google Scholar] [CrossRef] [PubMed]
- Stahl, Y.; Simon, R. Plant primary meristems: Shared functions and regulatory mechanisms. Curr. Opin. Plant Biol. 2010, 13, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Wendrich, J.R.; Möller, B.K.; Li, S.; Saiga, S.; Sozzani, R.; Benfey, P.N.; De Rybel, B.; Weijers, D. Framework for gradual progression of cell ontogeny in the Arabidopsis root meristem. Proc. Natl. Acad. Sci. USA 2017, 111, E8922–E8929. [Google Scholar] [CrossRef] [PubMed]
- Kenrick, P.; Strullu-Derrien, C. The origin and early evolution of roots. Plant Physiol. 2014, 166, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Overvoorde, P.; Fukaki, H.; Beeckman, T. Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2010, 2, a001537. [Google Scholar] [CrossRef] [PubMed]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; De Smet, I. Root system architecture: Insights from Arabidopsis and cereal crops. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2012, 367, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Scheres, B.; Wolkenfelt, H.; Willemsen, V.; Terlouw, M.; Lawson, E.; Dean, C.; Weisbeek, P. Embryonic origin of the Arabidopsis primary root and root-meristem initials. Development 1994, 120, 2475–2487. [Google Scholar]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef] [PubMed]
- Verstraeten, I.; Schotte, S.; Geelen, D. Hypocotyl adventitious root organogenesis differs from lateral root development. Front. Plant Sci. 2014, 5, 495. [Google Scholar] [PubMed]
- Coudert, P.; van Anh Le, T.; Gantet, P. Rice: A model plant to decipher the hidden origin of adventitious roots. In Plant Roots: The Hidden Half; Beeckman, T., Ed.; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Palme, K.; Hesse, T.; Moore, I.; Campos, N.; Feldwisch, J.; Garbers, C.; Hesse, F.; Schell, J. Hormonal modulation of plant growth: The role of auxin perception. Mech. Dev. 1991, 33, 97–106. [Google Scholar] [CrossRef]
- Benkova, E.; Hejatko, J. Hormone interactions at the root apical meristem. Plant Mol. Biol. 2009, 69, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Peer, W.A.; Blakeslee, J.J.; Yang, H.; Murphy, A.S. Seven things we think we know about auxin transport. Mol. Plant 2011, 4, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Zazimalová, E.; Murphy, A.S.; Yang, H.; Hoyerová, K.; Hošek, P. Auxin transporters—Why so many? Cold Spring Harb. Perspect. Biol. 2010, 2, a001552. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Muller, J. Indole-3-butyric acid in plant growth and development. Plant Growth Regul. 2000, 32, 219–230. [Google Scholar] [CrossRef]
- Mellor, N.; Band, L.R.; Pencik, A.; Novak, O.; Rashed, A.; Holman, T.; Wilson, M.H.; Voss, U.; Bishopp, A.; King, J.R.; et al. Dynamic regulation of auxin oxidase and conjugating enzymes AtDAO1 and GH3 modulates auxin homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 11022–11027. [Google Scholar] [CrossRef] [PubMed]
- Rosquete, M.R.; Barbez, E.; Kleine-Vehn, J. Cellular auxin homeostasis: Gatekeeping is housekeeping. Mol. Plant 2012, 5, 772–786. [Google Scholar] [CrossRef] [PubMed]
- Rozov, S.; Zagorskaya, A.; Deineko, E.; Shumny, V. Auxins: Biosynthesis, metabolism, and transport. Biol. Bull. Rev. 2013, 3, 286–295. [Google Scholar] [CrossRef]
- Kramer, E.M.; Ackelsberg, E.M. Auxin metabolism rates and implications for plant development. Front. Plant Sci. 2015, 6, 150. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.D.W.; Cho, H.T. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front. Plant Sci. 2013, 4, 448. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jurgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Greenham, K.; Prigge, M.J.; Jensen, P.J.; Estelle, M. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. 2009, 151, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.D. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis: The Arabidopsis Book. BioOne 2014, 12, e0173. [Google Scholar] [CrossRef] [PubMed]
- Band, L.R.; Wells, D.M.; Fozard, J.A.; Ghetiu, T.; French, A.P.; Pound, M.P.; Wilson, M.H.; Yu, L.; Li, W.D.; Hijazi, H.I.; et al. Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 2014, 26, 862–875. [Google Scholar] [CrossRef] [PubMed]
- Benkova, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertova, D.; Jurgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Petrasek, J.; Friml, J. Auxin transport routes in plant development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Skupa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klima, P.; Carna, M.; Rolcik, J.; De Rycke, R.; Moreno, I.; et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Swarup, K.; Benkova, E.; Swarup, R.; Casimiro, I.; Peret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bargmann, B.O.; Estelle, M. Auxin perception: In the IAA of the beholder. Physiol. Plant 2014, 151, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Farcot, E.; Lavedrine, C.; Vernoux, T. A modular analysis of the auxin signalling network. PLoS ONE 2015, 10, e0122231. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.C.; Zhao, Y.D. Auxin perception and downstream events. Curr. Opin. Plant Biol. 2016, 33, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin signaling. Plant Physiol. 2017, 175. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos Maraschin, F.; Memelink, J.; Offringa, R. Auxin-induced, SCFTIR1-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 2009, 59, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Greenham, K.; Zhang, Y.; Santner, A.; Castillejo, C.; Mutka, A.M.; O’Malley, R.C.; Ecker, J.R.; Kunkel, B.N.; Estelle, M. The Arabidopsis auxin receptor F-Box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 2016, 6, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Paponov, I.A.; Paponov, M.; Teale, W.; Menges, M.; Chakrabortee, S.; Murray, J.A.H.; Palme, K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 2008, 1, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Vernoux, T.; Brunoud, G.; Farcot, E.; Morin, V.; Van den Daele, H.; Legrand, J.; Oliva, M.; Das, P.; Larrieu, A.; Wells, D.; et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011, 7, 508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calderon Villalobos, L.I.A.; Lee, S.; De Oliveira, C.; Ivetac, A.; Brandt, W.; Armitage, L.; Sheard, L.B.; Tan, X.; Parry, G.; Mao, H.; et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 2012, 8, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Jones, A.R. Endoplasmic reticulum: The rising compartment in auxin biology. Plant Physiol. 2010, 154, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Petrášek, J. Why plants need more than one type of auxin. Plant Sci. 2011, 180, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Feng, M.; Kim, J.Y. Revisiting apoplastic auxin signaling mediated by AUXIN BINDING PROTEIN 1. Mol. Cell 2015, 38, 829–835. [Google Scholar]
- Gao, Y.; Zhang, Y.; Zhang, D.; Dai, X.; Estelle, M.; Zhao, Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc. Natl. Acad. Sci. USA 2015, 112, 2275–2280. [Google Scholar] [CrossRef] [PubMed]
- Dahlke, R.I.; Fraas, S.; Ullrich, K.K.; Heinemann, K.; Romeiks, M.; Rickmeyer, T.; Klebe, G.; Palme, K.; Luthen, H.; Steffens, B. Protoplast swelling and hypocotyl growth depend on different auxin signaling pathways. Plant Physiol. 2017, 175, 982–994. [Google Scholar] [CrossRef] [PubMed]
- Cook, S.D.; Ross, J.J. The auxins, IAA and PAA, are synthesized by similar steps catalyzed by different enzymes. Plant Signal. Behav. 2016, 11, e1250993. [Google Scholar] [CrossRef] [PubMed]
- Enders, T.A.; Strader, L.C. Auxin activity: Past, present, and future. Am. J. Bot. 2015, 102, 180–196. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, S.; Mashiguchi, K.; Tanaka, K.; Hishiyama, S.; Sakai, T.; Hanada, K.; Kinoshita-Tsujimura, K.; Yu, H.; Dai, X.H.; Takebayashi, Y.; et al. Distinct characteristics of indole-3-acetic acid and phenylacetic acid, two common auxins in plants. Plant Cell Physiol. 2015, 56, 1641–1654. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.C.; Wheeler, D.L.; Christensen, S.E.; Berens, J.C.; Cohen, J.D.; Rampey, R.A.; Bartel, B. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 2011, 23, 984–999. [Google Scholar] [CrossRef] [PubMed]
- Zolman, B.K.; Nyberg, M.; Bartel, B. IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 2007, 64, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Zolman, B.R.; Martinez, N.; Millius, A.; Adham, A.R.; Bartel, B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 2008, 180, 237–251. [Google Scholar] [CrossRef] [PubMed]
- De Rybel, B.; Audenaert, D.; Xuan, W.; Overvoorde, P.; Strader, L.C.; Kepinski, S.; Hoye, R.; Brisbois, R.; Parizot, B.; Vanneste, S.; et al. A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat. Chem. Biol. 2012, 8, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Audenaert, D.; Parizot, B.; Möller, B.K.; Njo, M.F.; De Rybel, B.; De Rop, G.; Van Isterdael, G.; Mähönen, A.P.; Vanneste, S.; et al. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr. Biol. 2015, 25, 1381–1388. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.C.; Nemhauser, J.L. Auxin 2012: A rich mea ho’oulu. Development 2013, 140, 1153–1157. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.; Leyser, O. The auxin question: A philosophical overview. In Auxin and Its Role in Plant Development; Zazimalova, E., Petrasek, J., Benkova, E., Eds.; Springer: Wien, Austria, 2014; pp. 3–19. [Google Scholar]
- Petersson, S.V.; Johansson, A.I.; Kowalczyk, M.; Makoveychuk, A.; Wang, J.Y.; Moritz, T.; Grebe, M.; Benfey, P.N.; Sandberg, G.; Ljung, K. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 2009, 21, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Niu, T.; Yu, Q.; Quan, T.; Ding, Z. Auxin gradient is crucial for the maintenance of root distal stem cell identity in Arabidopsis. Plant Signal. Behav. 2013, 8, e26429. [Google Scholar] [CrossRef] [PubMed]
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Ferrer, J.-L.; Ljung, K.; Pojer, F.; Hong, F.; Long, J.A.; Li, L.; Moreno, J.E.; Bowman, M.E.; Ivans, L.J. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 2008, 133, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Bhalerao, R.P.; Sandberg, G. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 2001, 28, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Hull, A.K.; Celenza, J.; Yamada, M.; Estelle, M.; Normanly, J.; Sandberg, G. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 2005, 17, 1090–1104. [Google Scholar] [CrossRef] [PubMed]
- Bhalerao, R.P.; Eklof, J.; Ljung, K.; Marchant, A.; Bennett, M.; Sandberg, G. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 2002, 29, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Renton, M.; Hanan, J.; Ferguson, B.J.; Beveridge, C.A. Models of long-distance transport: How is carrier-dependent auxin transport regulated in the stem? New Phytol. 2012, 194, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Kosuge, T.; Heskett, M.; Wilson, E. Microbial synthesis and degradation of indole-3-acetic acid I. The conversion of L-tryptophan to indole-3-acetamide by an enzyme system from Pseudomonas savastanoi. J. Biol. Chem. 1966, 241, 3738–3744. [Google Scholar] [PubMed]
- Klee, H.J.; Horsch, R.B.; Hinchee, M.A.; Hein, M.B.; Hoffmann, N.L. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes Dev. 1987, 1, 86–96. [Google Scholar] [CrossRef]
- Badenoch-Jones, J.; Summons, R.E.; Djordjevic, M.A.; Shine, J.; Letham, D.S.; Rolfe, B.G. Mass spectrometric quantification of indole-3-acetic acid in Rhizobium culture supernatants: Relation to root hair curling and nodule initiation. Appl. Environ. Microbiol. 1982, 44, 275–280. [Google Scholar] [PubMed]
- Badenoch-Jones, J.; Summons, R.E.; Entsch, B.; Rolfe, B.G.; Parker, C.W.; Letham, D.S. Mass spectrometric identification of indole compounds produced by Rhizobium strains. Biol. Mass Spectrom. 1982, 9, 429–437. [Google Scholar] [CrossRef]
- Koga, J.; Adachi, T.; Hidaka, H. Purification and characterization of indolepyruvate decarboxylase. A novel enzyme for indole-3-acetic acid biosynthesis in Enterobacter cloacae. J. Biol. Chem. 1992, 267, 15823–15828. [Google Scholar] [PubMed]
- Wright, A.D.; Sampson, M.B.; Neuffer, M.G.; Michalczuk, L.; Slovin, J.P.; Cohen, J.D. Indole-3-acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 1991, 254, 998–1000. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J.; Cohen, J.D.; Fink, G.R. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc. Natl. Acad. Sci. USA 1993, 90, 10355–10359. [Google Scholar] [CrossRef] [PubMed]
- Radwanski, E.R.; Last, R.L. Tryptophan biosynthesis and metabolism: Biochemical and molecular genetics. Plant Cell 1995, 7, 921–934. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Shao, X.; Li, J. Indole-3-glycerol phosphate, a branchpoint of indole-3-acetic acid biosynthesis from the tryptophan biosynthetic pathway in Arabidopsis thaliana. Plant J. 2000, 24, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Tzin, V.; Galili, G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 2010, 3, 956–972. [Google Scholar] [CrossRef] [PubMed]
- Last, R.L.; Fink, G.R. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 1988, 240, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Estelle, M.A.; Somerville, C. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Genet. Genom. 1987, 206, 200–206. [Google Scholar] [CrossRef]
- Cooney, T.P.; Nonhebel, H.M. The measurement and mass-spectral identification of indole-3-pyruvate from tomato shoots. Biochem. Biophys. Res. Commun. 1989, 162, 761–766. [Google Scholar] [CrossRef]
- Tivendale, N.D.; Ross, J.J.; Cohen, J.D. The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 2014, 19, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhao, Y. Auxin biosynthesis and catabolism. In Auxin and Its Role in Plant Development; Zazimalova, E., Petrasek, J., Benkova, E., Eds.; Springer: Wien, Austria, 2014; pp. 21–38. [Google Scholar]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.R.; Guo, H.W.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef] [PubMed]
- Won, C.; Shen, X.L.; Mashiguchi, K.; Zheng, Z.Y.; Dai, X.H.; Cheng, Y.F.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y.D. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis: A simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Tivendale, N.D.; Davidson, S.E.; Davies, N.W.; Smith, J.A.; Dalmais, M.; Bendahmane, A.I.; Quittenden, L.J.; Sutton, L.; Bala, R.K.; Le Signor, C.; et al. Biosynthesis of the halogenated auxin, 4-chloroindole-3-acetic acid. Plant Physiol. 2012, 159, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.D.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.H.; Mashiguchi, K.; Chen, Q.G.; Kasahara, H.; Kamiya, Y.; Ojha, S.; DuBois, J.; Ballou, D.; Zhao, Y.D. The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 2013, 288, 1448–1457. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [PubMed]
- Gallavotti, A.; Barazesh, S.; Malcomber, S.; Hall, D.; Jackson, D.; Schmidt, R.J.; McSteen, P. Sparse inflorescence 1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc. Natl. Acad. Sci. USA 2008, 105, 15196–15201. [Google Scholar] [CrossRef] [PubMed]
- Phillips, K.A.; Skirpan, A.L.; Liu, X.; Christensen, A.; Slewinski, T.L.; Hudson, C.; Barazesh, S.; Cohen, J.D.; Malcomber, S.; McSteen, P. Vanishing tassel 2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 2011, 23, 550–566. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.F.; Dai, X.H.; Zhao, Y.D. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef] [PubMed]
- Mano, Y.; Nemoto, K. The pathway of auxin biosynthesis in plants. J. Exp. Bot. 2012, 63, 2853–2872. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Akaba, S.; Oritani, T.; Delarue, M.; Bellini, C.; Caboche, M.; Koshiba, T. Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol. 1998, 116, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Novak, O.; Henykova, E.; Sairanen, I.; Kowalczyk, M.; Pospisil, T.; Ljung, K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012, 72, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, S.; Müller, A.; Piotrowski, M.; Weiler, E.W. Occurrence and formation of indole-3-acetamide in Arabidopsis thaliana. Planta 2002, 216, 155–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugawara, S.; Hishiyama, S.; Jikumaru, Y.; Hanada, A.; Nishimura, T.; Koshiba, T.; Zhao, Y.; Kamiya, Y.; Kasahara, H. Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5430–5435. [Google Scholar] [CrossRef] [PubMed]
- Pollmann, S.; Neu, D.; Weiler, E.W. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry 2003, 62, 293–300. [Google Scholar] [CrossRef]
- Nemoto, K.; Hara, M.; Suzuki, M.; Seki, H.; Muranaka, T.; Mano, Y. The NtAMI1 gene functions in cell division of tobacco BY-2 cells in the presence of indole-3-acetamide. FEBS Lett. 2009, 583, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Parra, B.; Frerigmann, H.; Alonso, M.-M.P.; Loba, V.C.; Jost, R.; Hentrich, M.; Pollmann, S. Characterization of four bifunctional plant IAM/PAM-amidohydrolases capable of contributing to auxin biosynthesis. Plants 2014, 3, 324–347. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, T.; Hoffmann, M.; Hentrich, M.; Pollmann, S. Indole-3-acetamide-dependent auxin biosynthesis: A widely distributed way of indole-3-acetic acid production? Eur. J. Cell Biol. 2010, 89, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Stirk, W.A.; Novák, O.; Hradecká, V.; Pĕnčík, A.; Rolčík, J.; Strnad, M.; Van Staden, J. Endogenous cytokinins, auxins and abscisic acid in Ulva fasciata (Chlorophyta) and Dictyota humifusa (Phaeophyta): Towards understanding their biosynthesis and homoeostasis. Eur. J. Phycol. 2009, 44, 231–240. [Google Scholar] [CrossRef]
- Abu-Zaitoon, Y.; Aladaileh, S.; Tawaha, A.R.A. Contribution of the IAM pathway to IAA pool in developing rice grains. Braz. Arch. Biol. Technol. 2016, 59, e16150677. [Google Scholar] [CrossRef]
- Mikkelsen, M.D.; Hansen, C.H.; Wittstock, U.; Halkier, B.A. Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 2000, 275, 33712–33717. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.D.; Hull, A.K.; Gupta, N.R.; Goss, K.A.; Alonso, J.; Ecker, J.R.; Normanly, J.; Chory, J.; Celenza, J.L. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 2002, 16, 3100–3112. [Google Scholar] [CrossRef] [PubMed]
- Tivendale, N.D.; Davies, N.W.; Molesworth, P.P.; Davidson, S.E.; Smith, J.A.; Lowe, E.K.; Reid, J.B.; Ross, J.J. Reassessing the role of N-hydroxytryptamine in auxin biosynthesis. Plant Physiol. 2010, 154, 1957–1965. [Google Scholar] [CrossRef] [PubMed]
- Bender, J.; Celenza, J.L. Indolic glucosinolates at the crossroads of tryptophan metabolism. Phytochem. Rev. 2009, 8, 25–37. [Google Scholar] [CrossRef]
- Bak, S.; Tax, F.E.; Feldmann, K.A.; Galbraith, D.W.; Feyereisen, R. CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 2001, 13, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Douglas Grubb, C.; Zipp, B.J.; Ludwig-Müller, J.; Masuno, M.N.; Molinski, T.F.; Abel, S. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 2004, 40, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Naur, P.; Halkier, B.A. Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004, 37, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Nafisi, M.; Goregaoker, S.; Botanga, C.J.; Glawischnig, E.; Olsen, C.E.; Halkier, B.A.; Glazebrook, J. Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 2007, 19, 2039–2052. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.L.; Wang, M.; Han, B.Y.; Tan, D.G.; Sun, X.P.; Zhang, J.M. Arabidopsis myrosinase genes AtTGG4 and AtTGG5 are root-tip specific and contribute to auxin biosynthesis and root growth regulation. Int. J. Mol. Sci. 2016, 17, 892. [Google Scholar] [CrossRef] [PubMed]
- Bartel, B.; Fink, G.R. Differential regulation of an auxin-producing Nitrilase gene family in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1994, 91, 6649–6653. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J.; Grisafi, P.; Fink, G.R.; Bartel, B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 1997, 9, 1781–1790. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, B.; Ouyang, J.; Li, J.; Wang, Y. Arabidopsis indole synthase, a homolog of tryptophan synthase alpha, is an enzyme involved in the trp-independent indole-containing metabolite biosynthesis. J. Integr. Plant Biol. 2008, 50, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Eilbert, E.; Bolbol, A.; Lukowitz, W. Going mainstream: How is the body axis of plants first initiated in the embryo? Dev. Biol. 2016, 419, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Mironova, V.; Teale, W.; Shahriari, M.; Dawson, J.; Palme, K. The systems biology of auxin in developing embryos. Trends Plant Sci. 2017, 22, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Grones, P.; Stepanova, A.N.; Robles, L.M.; Lokerse, A.S.; Alonso, J.M.; Weijers, D.; Friml, J. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 2013, 23, 2506–2512. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, S.; Saiga, S.; Weijers, D. Auxin regulation of embryonic root formation. Plant Cell Physiol. 2013, 54, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Sauer, M.; Meurette, O.; Friml, J.; Ljung, K.; Sandberg, G.; Hooykaas, P.; Offringa, R. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell 2005, 17, 2517–2526. [Google Scholar] [CrossRef] [PubMed]
- Geldner, N.; Hamann, T.; Jürgens, G. Is there a role for auxin in early embryogenesis? Plant Growth Regul. 2000, 32, 187–191. [Google Scholar] [CrossRef]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jurgens, G. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Benjamins, R.; Scheres, B. Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 2008, 59, 443–465. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Chen, J.; Yang, Z. Auxin regulation of cell polarity in plants. Curr. Opin. Plant Biol. 2015, 28, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Wabnik, K.; Robert, H.S.; Smith, R.S.; Friml, J. Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr. Biol. 2013, 23, 2513–2518. [Google Scholar] [CrossRef] [PubMed]
- Picciarelli, P.; Ceccarelli, N.; Paolicchi, F.; Calistri, G. Endogenous auxins and embryogenesis in Phaseolus coccineus. Aust. J. Plant Physiol. 2001, 28, 73–78. [Google Scholar] [CrossRef]
- Smit, M.E.; Weijers, D. The role of auxin signaling in early embryo pattern formation. Curr. Opin. Plant Biol. 2015, 28, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Möller, B.; Weijers, D. Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. 2009, 1, a001545. [Google Scholar] [CrossRef] [PubMed]
- Takatsuka, H.; Umeda, M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Chu, J.; Yu, T.; Xu, Q.; Sun, X.; Yuan, J.; Xiong, G.; Wang, G.; Wang, Y.; Li, J. Tryptophan-independent auxin biosynthesis contributes to early embryogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 4821–4826. [Google Scholar] [CrossRef] [PubMed]
- Nibau, C.; Gibbs, D.J.; Coates, J.C. Branching out in new directions: The control of root architecture by lateral root formation. New Phytol. 2008, 179, 595–614. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Gutjahr, C.; Li, C.J.; Hochholdinger, F. Genetic control of lateral root formation in cereals. Trends Plant Sci. 2016, 21, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, S.; Beis, D.; Wolkenfelt, H.; Murfett, J.; Guilfoyle, T.; Malamy, J.; Benfey, P.; Leyser, O.; Bechtold, N.; Weisbeek, P. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 1999, 99, 463–472. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Benkova, E.; Blilou, I.; Wisniewska, J.; Hamann, T.; Ljung, K.; Woody, S.; Sandberg, G.; Scheres, B.; Jurgens, G.; et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef]
- Grieneisen, V.A.; Xu, J.; Maree, A.F.M.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, M.; Liang, N.; Zheng, Y.; Yu, Q.; Wu, S. Symplastic communication spatially directs local auxin biosynthesis to maintain root stem cell niche in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 4005–4010. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.G.; Dai, X.H.; De-Paoli, H.; Cheng, Y.F.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y.D. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Pinon, V.; Prasad, K.; Grigg, S.P.; Sanchez-Perez, G.F.; Scheres, B. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Ito, M.; Sumikura, T.; Nakayama, A.; Nishimura, T.; Kitano, H.; Yamaguchi, I.; Koshiba, T.; Hibara, K.I.; Nagato, Y.; et al. The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. 2014, 78, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kamiya, N.; Morinaka, Y.; Matsuoka, M.; Sazuka, T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 2007, 143, 1362–1371. [Google Scholar] [CrossRef] [PubMed]
- Sazuka, T.; Kamiya, N.; Nishimura, T.; Ohmae, K.; Sato, Y.; Imamura, K.; Nagato, Y.; Koshiba, T.; Nagamura, Y.; Ashikari, M.; et al. A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 2009, 60, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xie, W.F.; Zhang, L.; Valpuesta, V.; Ye, Z.W.; Gao, Q.H.; Duan, K. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J. Integr. Plant Biol. 2014, 56, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Xiong, L. Localized auxin biosynthesis and postembryonic root development in Arabidopsis. Plant Signal. Behav. 2009, 4, 752–754. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, T.; Janowitz, T.; Sanchez-Parra, B.; Alonso, M.M.P.; Trompetter, I.; Piotrowski, M.; Pollmann, S. Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of auxin biosynthesis in seedlings. Front. Plant Sci. 2017, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Beeckman, T.; Burssens, S.; Inze, D. The peri-cell-cycle in Arabidopsis. J. Exp. Bot. 2001, 52, 403–411. [Google Scholar] [PubMed]
- De Smet, I.; Tetsumura, T.; De Rybel, B.; Frei dit Frey, N.; Laplaze, L.; Casimiro, I.; Swarup, R.; Naudts, M.; Vanneste, S.; Audenaert, D.; et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 2007, 134, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M. Lateral root initiation is a probabilistic event whose frequency is set by fluctuating levels of auxin response. J. Exp. Bot. 2013, 64, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; Van Norman, J.M.; Moreno, A.; Zhang, J.Y.; Ahnert, S.E.; Benfey, P.N. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 2010, 329, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Van Norman, J.M.; Xuan, W.; Beeckman, T.; Benfey, P.N. To branch or not to branch: The role of pre-patterning in lateral root formation. Development 2013, 140, 4301–4310. [Google Scholar] [CrossRef] [PubMed]
- Xuan, W.; Band, L.R.; Kumpf, R.P.; Van Damme, D.; Parizot, B.; De Rop, G.; Opdenacker, D.; Möller, B.K.; Skorzinski, N.; Njo, M.F.; et al. Cyclic programmed cell death stimulates hormone signaling and root development in Arabidopsis. Science 2016, 351, 384–387. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 2005, 28, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Benkova, E.; Bielach, A. Lateral root organogenesis—From cell to organ. Curr. Opin. Plant Biol. 2010, 13, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Dastidar, M.G.; Jouannet, V.; Maizel, A. Root branching: Mechanisms, robustness, and plasticity. WIREs Dev. Biol. 2012, 1, 329–343. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Teeples, M.; Lanctot, A.; Nemhauser, J.L. As above, so below: Auxin’s role in lateral organ development. Dev. Biol. 2016, 419, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Dubrovsky, J.G.; Sauer, M.; Napsucialy-Mendivil, S.; Ivanchenko, M.G.; Friml, J.; Shishkova, S.; Celenza, J.; Benková, E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 2008, 105, 8790–8794. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Cervera, M.-T.; Delarue, M.; Beeckman, T.; Dewitte, W.; Bellini, C.; Caboche, M.; Van Onckelen, H.; Van Montagu, M.; Inzé, D. Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 1995, 7, 1405–1419. [Google Scholar] [CrossRef] [PubMed]
- Delarue, M.; Prinsen, E.; Va, H.; Caboche, M.; Bellini, C. Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 1998, 14, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, S.; Friml, J. Auxin: A trigger for change in plant development. Cell 2009, 136, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.Z.; Wei, J.; Xu, J.; Sun, M.X. Inducible knock-down of GNOM during root formation reveals tissue-specific response to auxin transport and its modulation of local auxin biosynthesis. J. Exp. Bot. 2014, 65, 1165–1179. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Sun, B.; Xu, L.; Liu, W. Wound signaling: The missing link in plant regeneration. Plant Signal. Behav. 2016, 11, e1238548. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, N. Getting to the root of regeneration: Adventitious rooting and callus formation. Plant Cell 2014, 26, 845. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Pacurar, D.I.; Perrone, I.; Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant. 2014, 151, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Da Rocha Correa, L.; Troleis, J.; Mastroberti, A.; Mariath, J.; Fett-Neto, A. Distinct modes of adventitious rooting in Arabidopsis thaliana. Plant Biol. 2012, 14, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Pop, T.I.; Pamfil, D.; Bellini, C. Auxin control in the formation of adventitious roots. Not. Bot. Horti Agrobot. Cluj-Napoca 2011, 39, 307–316. [Google Scholar]
- Ahkami, A.H.; Lischewski, S.; Haensch, K.T.; Porfirova, S.; Hofmann, J.; Rolletschek, H.; Melzer, M.; Franken, P.; Hause, B.; Druege, U.; et al. Molecular physiology of adventitious root formation in Petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytol. 2009, 181, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Druege, U.; Franken, P.; Lischewski, S.; Ahkami, A.H.; Zerche, S.; Hause, B.; Hajirezaei, M.R. Transcriptomic analysis reveals ethylene as stimulator and auxin as regulator of adventitious root formation in petunia cuttings. Front. Plant Sci. 2014, 5, 494. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Miao, J.; Yumoto, E.; Yokota, T.; Asahina, M.; Watahiki, M. YUCCA9-mediated auxin biosynthesis and polar auxin transport synergistically regulate regeneration of root systems following root cutting. Plant Cell Physiol. 2017, 58, 1710–1723. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.Q.; Tong, J.H.; Xiao, L.T.; Ruan, Y.; Liu, J.C.; Zeng, M.H.; Huang, H.; Wang, J.W.; Xu, L. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J. Exp. Bot. 2016, 67, 4273–4284. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.L.; Cheng, Z.J.; Zhang, X.S. Endogenous auxin biosynthesis and de novo root organogenesis. J. Exp. Bot. 2016, 67, 4011–4013. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.B.; Chen, L.Q.; Liu, J.C.; Zhang, X.N.; Yang, Z.N.; Liu, W.; Xu, L. TAA family contributes to auxin production during de novo regeneration of adventitious roots from Arabidopsis leaf explants. Sci. Bull. 2016, 61, 1728–1731. [Google Scholar] [CrossRef]
- Della Rovere, F.; Fattorini, L.; D’Angeli, S.; Veloccia, A.; Falasca, G.; Altamura, M.M. Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann. Bot. 2013, 112, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Della Rovere, F.; Fattorini, L.; Ronzan, M.; Falasca, G.; Altamura, M.M. The quiescent center and the stem cell niche in the adventitious roots of Arabidopsis thaliana. Plant Signal. Behav. 2016, 11, e1176660. [Google Scholar] [CrossRef] [PubMed]
- Blakesley, D. Auxin metabolism and adventitious root initiation in Biology of adventitious root formation. Eds. Davis, T.D.; Haissig, B.E. Springer, Boston MA. Basic Life Sci. 1994, 62, 143. [Google Scholar]
- Zhang, W.; Swarup, R.; Bennett, M.; Schaller, G.E.; Kieber, J.J. Cytokinin induces cell division in the quiescent center of the Arabidopsis root apical meristem. Curr. Biol. 2013, 23, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.J.; Estelle, M. Cytokinin and auxin intersection in root meristems. Genome Biol. 2009, 10, 210. [Google Scholar] [CrossRef]
- Marhavy, P.; Duclercq, J.; Weller, B.; Feraru, E.; Bielach, A.; Offringa, R.; Friml, J.; Schwechheimer, C.; Murphy, A.; Benkova, E. Cytokinin controls polarity of PIN1-dependent auxin transport during lateral root organogenesis. Curr. Biol. 2014, 24, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Simaskova, M.; O’Brien, J.A.; Khan, M.; Van Noorden, G.; Otvos, K.; Vieten, A.; De Clercq, I.; Van Haperen, J.M.; Cuesta, C.; Hoyerova, K.; et al. Cytokinin response factors regulate PIN-FORMED auxin transporters. Nat. Commun. 2015, 6, 8717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.Y.; Zhang, C.G.; Wu, L.; Zhang, C.G.; Chai, J.; Wang, M.; Jha, A.; Jia, P.F.; Cui, S.J.; Yang, M. Functional characterization of the CKRC1/TAA1 gene and dissection of hormonal actions in the Arabidopsis root. Plant J. 2011, 66, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.; Gunnerås, S.A.; Petersson, S.V.; Tarkowski, P.; Graham, N.; May, S.; Dolezal, K.; Sandberg, G.; Ljung, K. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 2010, 22, 2956–2969. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.I.; Noh, E.W.; Kim, H.J.; Park, W.J. Differential regulation of cytokinin oxidase genes and cytokinin-induced auxin biosynthesis by cellular cytokinin level in transgenic poplars. Plant Cell Rep. 2014, 33, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.; Ljung, K. Auxin and cytokinin regulate each other’s levels via a metabolic feedback loop. Plant Signal. Behav. 2011, 6, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Di, D.W.; Wu, L.; Luo, P.; Zhang, L.; Zhang, T.Z.; Sun, X.; Wei, S.D.; An, C.W.; Guo, G.Q. Analysis the role of arabidopsis CKRC6/ASA1 in auxin and cytokinin biosynthesis. J. Plant Biol. 2016, 59, 162–171. [Google Scholar] [CrossRef]
- Moubayidin, L.; Di Mambro, R.; Sozzani, R.; Pacifici, E.; Salvi, E.; Terpstra, I.; Bao, D.; Van Dijken, A.; Ioio, R.D.; Perilli, S. Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev. Cell 2013, 26, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Ivanchenko, M.G.; Muday, G.K.; Dubrovsky, J.G. Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J. 2008, 55, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, K.; Ljung, K.; Vanneste, S.; Podhorska, R.; Beeckman, T.; Friml, J.; Benkova, E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepanova, A.N.; Hoyt, J.M.; Hamilton, A.A.; Alonso, J.M. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 2005, 17, 2230–2242. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Perry, P.; Hagenbeek, D.; Van Der Straeten, D.; Beemster, G.T.S.; Sandberg, G.; Bhalerao, R.; Ljung, K.; Bennett, M.J. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 2007, 19, 2186–2196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, J.L.; Miao, Z.Q.; Wang, Z.; Yu, L.H.; Cai, X.T.; Xiang, C.B. Arabidopsis ERF1 mediates crosstalk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet. 2016, 12, e1005760. [Google Scholar]
- He, W.R.; Brumos, J.; Li, H.J.; Ji, Y.S.; Ke, M.; Gong, X.Q.; Zeng, Q.L.; Li, W.Y.; Zhang, X.Y.; An, F.Y.; et al. A small-molecule screen identifies L-Kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 2011, 23, 3944–3960. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Zhang, Z.; Wang, J.; Chen, X.; Wei, P.; Huang, R. The activation of OsEIL1 on YUC8 transcription and auxin biosynthesis is required for ethylene-inhibited root elongation in rice early seedling development. PLoS Genet. 2017, 13, e1006955. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.Y.; Chao, Y.Y.; Kao, C.H. Methyl jasmonate-induced lateral root formation in rice: The role of heme oxygenase and calcium. J. Plant Physiol. 2013, 170, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Raya-Gonzalez, J.; Pelagio-Flores, R.; Lopez-Bucio, J. The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana. J. Plant Physiol. 2012, 169, 1348–1358. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Q.; Xu, Y.X.; Ye, S.Q.; Jiang, H.L.; Chen, Q.; Liu, F.; Zhou, W.K.; Chen, R.; Li, X.G.; Tietz, O.; et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 2009, 21, 1495–1511. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.T.; Xu, P.; Zhao, P.X.; Liu, R.; Yu, L.H.; Xiang, C.B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014, 5, 5833. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Galván-Ampudia, C.S.; Demarsy, E.; Langowski, L.; Kleine-Vehn, J.; Fan, Y.; Morita, M.T.; Tasaka, M.; Fankhauser, C.; Offringa, R. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat. Cell Biol. 2011, 13, 447–452. [Google Scholar] [CrossRef] [PubMed]
- Sassi, M.; Lu, Y.; Zhang, Y.; Wang, J.; Dhonukshe, P.; Blilou, I.; Dai, M.; Li, J.; Gong, X.; Jaillais, Y. COP1 mediates the coordination of root and shoot growth by light through modulation of PIN1-and PIN2-dependent auxin transport in Arabidopsis. Development 2012, 139, 3402–3412. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.-L.; Okamoto, H.; Wang, M.; Ang, L.-H.; Matsui, M.; Goodman, H.; Deng, X.W. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 2000, 14, 1958–1970. [Google Scholar] [PubMed]
- Nakazawa, M.; Yabe, N.; Ichikawa, T.; Yamamoto, Y.Y.; Yoshizumi, T.; Hasunuma, K.; Matsui, M. DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 2001, 25, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Sorin, C.; Bussell, J.D.; Camus, I.; Ljung, K.; Kowalczyk, M.; Geiss, G.; McKhann, H.; Garcion, C.; Vaucheret, H.; Sandberg, G.; et al. Auxin and light control of adventitious rooting in Arabidopsis require ARGONAUTE1. Plant Cell 2005, 17, 1343–1359. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.-I.; Mochizuki, N.; Nagatani, A. Expression of the AtGH3a gene, an Arabidopsis homologue of the soybean GH3 gene, is regulated by phytochrome B. Plant Cell Physiol. 2002, 43, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Hoecker, U.; Toledo-Ortiz, G.; Bender, J.; Quail, P.H. The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta 2004, 219, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Yokawa, K.; Nakano, S.; Yoshida, Y.; Fabrissin, I.; Okamoto, T.; Baluska, F.; Koshiba, T. Root cap-dependent gravitropic U-turn of maize root requires light-induced auxin biosynthesis via the YUC pathway in the root apex. J. Exp. Bot. 2016, 67, 4581–4591. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Guo, Y.; Novák, O.; Chen, W.; Ljung, K.; Noel, J.P.; Chory, J. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat. Plants 2016, 2, 16025. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ljung, K.; Breton, G.; Schmitz, R.J.; Pruneda-Paz, J.; Cowing-Zitron, C.; Cole, B.J.; Ivans, L.J.; Pedmale, U.V.; Jung, H.S.; et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012, 26, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Sairanen, I.; Novák, O.; Pěnčík, A.; Ikeda, Y.; Jones, B.; Sandberg, G.; Ljung, K. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 2012, 24, 4907–4916. [Google Scholar] [CrossRef] [PubMed]
- Mishra, B.S.; Singh, M.; Aggrawal, P.; Laxmi, A. Glucose and auxin signaling interaction in controlling Arabidopsis thaliana seedlings root growth and development. PLoS ONE 2009, 4, e4502. [Google Scholar] [CrossRef] [PubMed]
- Lilley, J.L.S.; Gee, C.W.; Sairanen, I.; Ljung, K.; Nemhauser, J.L. An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol. 2012, 160, 2261–2270. [Google Scholar] [CrossRef] [PubMed]
- Giehl, R.F.; von Wiren, N. Root nutrient foraging. Plant Physiol. 2014, 166, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Gruber, B.D.; Giehl, R.F.H.; Friedel, S.; von Wirén, N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013, 163, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Hodge, A. The plastic plant: Root responses to heterogeneous supplies of nutrients. New Phytol. 2004, 162, 9–24. [Google Scholar] [CrossRef]
- Ristova, D.; Busch, W. Natural variation of root traits: From development to nutrient uptake. Plant Physiol. 2014, 166, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Nacry, P.; Pervent, M.; Girin, T.; Tillard, P.; Lepetit, M.; Gojon, A. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006, 140, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Forde, B.G. Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 2000, 51, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Jennings, A.; Barlow, P.W.; Forde, B.G. Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 1999, 96, 6529–6534. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Chen, F.; Liu, J.; Zhang, F.; Mi, G. Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J. Plant Physiol. 2008, 165, 942–951. [Google Scholar] [CrossRef] [PubMed]
- Walch-Liu, P.; Ivanov, I.I.; Filleur, S.; Gan, Y.; Remans, T.; Forde, B.G. Nitrogen regulation of root branching. Ann. Bot. 2005, 97, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.Y.; Li, J.J.; Qu, B.Y.; He, X.; Zhao, X.Q.; Li, B.; Fu, X.D.; Tong, Y.P. Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 2014, 78, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Shao, A.; Ma, W.; Zhao, X.; Hu, M.; He, X.; Teng, W.; Li, H.; Tong, Y. The auxin biosynthetic TRYPTOPHAN AMINOTRANSFERASE RELATED TaTAR2.1-3A increases grain yield of wheat. Plant Physiol. 2017, 174, 2274–2288. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.H.; Miao, Z.Q.; Qi, G.F.; Wu, J.; Cai, X.T.; Mao, J.L.; Xiang, C.B. MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 2014, 7, 1653–1669. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.F.; Chai, R.S.; Jin, G.L.; Wang, H.; Tang, C.X.; Zhang, Y.S. Responses of root architecture development to low phosphorus availability: A review. Ann. Bot. 2013, 112, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Lee, J.; Gong, Q.; Ma, S.; Jin, J.B.; Yoo, C.Y.; Miura, T.; Sato, A.; Bohnert, H.J.; Hasegawa, P.M. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiol. 2011, 155, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Qu, B.; He, X.; Wang, J.; Zhao, Y.; Teng, W.; Shao, A.; Zhao, X.; Ma, W.; Wang, J.; Li, B.; et al. A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 2015, 167, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Kutz, A.; Muller, A.; Hennig, P.; Kaiser, W.M.; Piotrowski, M.; Weiler, E.W. A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 2002, 30, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Blancaflor, E.B.; Masson, P.H. Plant gravitropism. Unraveling the ups and downs of a complex process. Plant Physiol. 2003, 133, 1677–1690. [Google Scholar] [CrossRef] [PubMed]
- Band, L.R.; Wells, D.M.; Larrieu, A.; Sun, J.Y.; Middleton, A.M.; French, A.P.; Brunoud, G.; Sato, E.M.; Wilson, M.H.; Peret, B.; et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 4668–4673. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Wang, B.; Zhu, J. Auxin transport during root gravitropism: Transporters and techniques. Plant Biol. 2014, 16 (Suppl. S1), 50–57. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.Y.; Kim, W.Y.; Kang, S.B.; Kim, J.I.; Baek, D.; Jung, I.J.; Kim, M.R.; Li, N.; Kim, H.J.; Nakajima, M.; et al. A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin biosynthesis. Nat. Commun. 2015, 6, 8041. [Google Scholar] [CrossRef] [PubMed]
- Ke, Q.; Wang, Z.; Ji, C.Y.; Jeong, J.C.; Lee, H.S.; Li, H.; Xu, B.; Deng, X.; Kwak, S.S. Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 2015, 94, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Baek, D.; Park, H.C.; Chun, H.J.; Oh, D.H.; Lee, M.K.; Cha, J.Y.; Kim, W.Y.; Kim, M.C.; Chung, W.S.; et al. Overexpression of Arabidopsis YUCCA6 in potato results in high auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant 2013, 6, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Jung, J.-H.; Han, D.Y.; Seo, P.J.; Park, W.J.; Park, C.M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 2012, 235, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chen, L.; Ye, T.; Liu, X.; Ding, K.; Chan, Z. Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 2014, 82, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, N.; Wang, H.; Kasahara, H.; Liu, J.; MacPherson, C.; Machida, Y.; Kamiya, Y.; Hannah, M.A.; Chua, N.H. IAA-Ala Resistant 3, an evolutionarily conserved target of miR167, mediates Arabidopsis root architecture changes during high osmotic stress. Plant Cell 2012, 24, 3590–3602. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.E.; Dinneny, J.R. The divining root: Moisture-driven responses of roots at the micro- and macro-scale. J. Exp. Bot. 2015, 66, 2145–2154. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Aggarwal, P.; Robbins, N.E.; Sturrock, C.J.; Thompson, M.C.; Tan, H.Q.; Tham, C.; Duan, L.N.; Rodriguez, P.L.; Vernoux, T.; et al. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc. Natl. Acad. Sci. USA 2014, 111, 9319–9324. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Tripathi, D.K.; Singh, S.; Sharma, S.; Dubey, N.K.; Chauhan, D.K.; Vaculik, M. Toxicity of aluminium on various levels of plant cells and organism: A review. Environ. Exp. Bot. 2017, 137, 177–193. [Google Scholar] [CrossRef]
- Sun, L.; Tian, J.; Zhang, H.; Liao, H. Phytohormone regulation of root growth triggered by P deficiency or Al toxicity. J. Exp. Bot. 2016, 67, 3655–3664. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Tian, Q.Y.; Chen, J.; Zhang, W.H. Aluminium-induced inhibition of root elongation in Arabidopsis is mediated by ethylene and auxin. J. Exp. Bot. 2010, 61, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.Y.; Ren, X.Y.; Huang, B.R.; Wang, G.; Zhou, P.; An, Y. Aluminium-induced reduction of plant growth in alfalfa (Medicago sativa) is mediated by interrupting auxin transport and accumulation in roots. Sci. Rep. 2016, 6, 30079. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.B.; Geng, X.Y.; He, C.M.; Zhang, F.; Wang, R.; Horst, W.J.; Ding, Z.J. TAA1-regulated local auxin biosynthesis in the root apex transition zone mediates the Aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell 2014, 26, 2889–2904. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.B.; Liu, G.; Liu, J.; Zhang, B.; Meng, W.; Müller, B.; Hayashi, K.I.; Zhang, X.; Zhao, Z.; De Smet, I.; et al. Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. EMBO Rep. 2017, 18, 1213–1230. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.C.; Gao, S.; Tian, H.Y.; Wu, W.W.; Robert, H.S.; Ding, Z.J. Local transcriptional control of YUCCA regulates auxin promoted root growth inhibition in response to Aluminium stress in Arabidopsis. PLoS Genet. 2016, 12, e1006360. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.F.; Lei, G.J.; Wang, Z.W.; Shi, Y.Z.; Braam, J.; Li, G.X.; Zheng, S.J. Coordination between apoplastic and symplastic detoxification confers plant aluminum resistance. Plant Physiol. 2013, 162, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Hou, N.; Schlicht, M.; Wan, Y.; Mancuso, S.; Baluska, F. Aluminium toxicity targets PIN2 in Arabidopsis root apices: Effects on PIN2 endocytosis, vesicular recycling, and polar auxin transport. Chin. Sci. Bull. 2008, 53, 2480. [Google Scholar] [CrossRef]
- Wu, D.; Shen, H.; Yokawa, K.; Baluška, F. Alleviation of aluminium-induced cell rigidity by overexpression of OsPIN2 in rice roots. J. Exp. Bot. 2014, 65, 5305–5315. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wang, W.; Zhou, H.; Wang, R.; Zhang, P.; Wang, H.; Pan, X.; Xu, J. Manganese toxicity inhibited root growth by disrupting auxin biosynthesis and transport in Arabidopsis. Front. Plant Sci. 2017, 8, 272. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, Y.; Pan, Z.; Xie, S.; Peng, S.A. Boron deficiency alters root growth and development and interacts with auxin metabolism by influencing the expression of auxin synthesis and transport genes. Biotechnol. Biotechnol. Equip. 2016, 30, 661–668. [Google Scholar] [CrossRef]
- Sukumar, P.; Legue, V.; Vayssieres, A.; Martin, F.; Tuskan, G.A.; Kalluri, U.C. Involvement of auxin pathways in modulating root architecture during beneficial plant–microorganism interactions. Plant Cell Environ. 2013, 36, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Felten, J.; Legue, V.; Ditengou, F.A. Lateral root stimulation in the early interaction between Arabidopsis thaliana and the ectomycorrhizal fungus Laccaria bicolor. Plant Signal. Behav. 2010, 5, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, L.; Zhang, N.; Li, Z.; Zhang, G.; Xu, Y.; Shen, Q.; Zhang, R. Plant-microbe communication enhances auxin biosynthesis by a root-associated bacterium, Bacillus amyloliquefaciens SQR9. Mol. Plant-Microbe Interact. 2016, 29, 324–330. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, P.J.; Schmelz, E.A.; Moussatche, P.; Lund, S.T.; Jones, J.B.; Klee, H.J. usceptible to intolerance—A range of hormonal actions in a susceptible Arabidopsis pathogen response. Plant J. 2003, 33, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Rampey, R.A.; LeClere, S.; Kowalczyk, M.; Ljung, K.; Sandberg, G.; Bartel, B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004, 135, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2010, 2, a001594. [Google Scholar] [CrossRef] [PubMed]
- LeClere, S.; Tellez, R.; Rampey, R.A.; Matsuda, S.P.; Bartel, B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002, 277, 20446–20452. [Google Scholar] [CrossRef] [PubMed]
- Bajguz, A.; Piotrowska, A. Conjugates of auxin and cytokinin. Phytochemistry 2009, 70, 957–969. [Google Scholar] [CrossRef] [PubMed]
- Riov, J.; Bangerth, F. Metabolism of auxin in tomato fruit tissue: Formation of high molecular weight conjugates of oxindole-3-acetic acid via the oxidation of indole-3-acetylaspartic acid. Plant Physiol. 1992, 100, 1396–1402. [Google Scholar] [CrossRef] [PubMed]
- Östin, A.; Kowalyczk, M.; Bhalerao, R.P.; Sandberg, G. Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 1998, 118, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, M.; Sandberg, G. Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol. 2001, 127, 1845–1853. [Google Scholar] [CrossRef] [PubMed]
- Eyer, L.; Vain, T.; Pařízková, B.; Oklestkova, J.; Barbez, E.; Kozubíková, H.; Pospíšil, T.; Wierzbicka, R.; Kleine-Vehn, J.; Fránek, M.; et al. 2,4-D and IAA amino acid conjugates show distinct metabolism in Arabidopsis. PLoS ONE 2016, 11, e0159269. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, C.; Dennis, E.G.; Booker, G.W.; Polyak, S.W.; Boss, P.K.; Davies, C. A novel tool for studying auxin-metabolism: The inhibition of grapevine indole-3-acetic acid-amido synthetases by a reaction intermediate analogue. PLoS ONE 2012, 7, e37632. [Google Scholar] [CrossRef] [PubMed]
- Bartel, B.; Fink, G. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 1995, 268, 1745–1748. [Google Scholar] [CrossRef] [PubMed]
- Davies, R.T.; Goetz, D.H.; Lasswell, J.; Anderson, M.N.; Bartel, B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 1999, 11, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Magidin, M.; Pittman, J.K.; Hirschi, K.D.; Bartel, B. ILR2, a novel gene regulating IAA conjugate sensitivity and metal transport in Arabidopsis thaliana. Plant J. 2003, 35, 523–534. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Gu, H.; Zhao, Y.; Ma, Z.; Shi, G.; Yang, Y.; Pichersky, E.; Chen, H.; Liu, M.; Chen, Z.; et al. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 2005, 17, 2693–2704. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, R.; Ma, C.-J.; Vlot, A.C.; Klessig, D.F.; Pichersky, E. Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the AtMES esterase family of Arabidopsis. Plant Physiol. 2008, 147, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
- Spiess, G.M.; Hausman, A.; Yu, P.; Cohen, J.D.; Rampey, R.A.; Zolman, B.K. Auxin input pathway disruptions are mitigated by changes in auxin biosynthetic gene expression in Arabidopsis. Plant Physiol. 2014, 165, 1092–1104. [Google Scholar] [CrossRef] [PubMed]
- Bertoni, G. Indolebutyric acid–derived auxin and plant development. Plant Cell 2011, 23, 845. [Google Scholar] [CrossRef]
- Frick, E.M.; Strader, L.C. Roles for IBA-derived auxin in plant development. J. Exp. Bot. 2017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Peer, W.A. Auxin homeostasis: The DAO of catabolism. J. Exp. Bot. 2017, 68, 3145–3154. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.M. Destruction of Auxin. Ann. Rev. Plant Physiol. 1958, 9, 81–118. [Google Scholar] [CrossRef]
- Pěnčík, A.; Simonovik, B.; Petersson, S.V.; Henyková, E.; Simon, S.; Greenham, K.; Zhang, Y.; Kowalczyk, M.; Estelle, M.; Zažímalová, E.; et al. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell 2013, 25, 3858–3870. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Alonso, J.M. Auxin catabolism unplugged: Role of IAA oxidation in auxin homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 10742–10744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lin, J.E.; Harris, C.; Campos Mastrotti Pereira, F.; Wu, F.; Blakeslee, J.J.; Peer, W.A. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, 11010–11015. [Google Scholar] [CrossRef] [PubMed]
- Porco, S.; Pěnčík, A.; Rashed, A.; Voß, U.; Casanova-Sáez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R.; Swarup, R.; Swarup, K.; et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M.; Ten Tusscher, K.H. Periodic lateral root priming: What makes it tick? Plant Cell 2017, 29, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, Y.; Liu, X.; Zhang, X.; Liu, S.; Yu, X.; Ren, Y.; Zheng, X.; Zhou, K.; Jiang, L.; et al. A Role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 2013, 27, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Butler, E.D.; Gallagher, T.F. Characterization of auxin-induced ARRO-1 expression in the primary root of Malus domestica. J. Exp. Bot. 2000, 51, 1765–1766. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Hayashi, K.-I.; Natsume, M.; Kamiya, Y.; Sakakibara, H.; Kawaide, H.; Kasahara, H. UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol. 2014, 55, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Saito, K. Functional genomics for plant natural product biosynthesis. Nat. Prod. Rep. 2009, 26, 1466–1487. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.H.; Ma, X.M.; Han, P.; Wang, B.; Sun, Y.G.; Zhang, G.Z.; Li, Y.J.; Hou, B.K. UGT74D1 Is a novel auxin glycosyltransferase from Arabidopsis thaliana. PLoS ONE 2013, 8, e61705. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Van Aken, O.; Morreel, K.; Vandenbroucke, K.; van de Cotte, B.; De Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W.; et al. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szerszen, J.; Szczyglowski, K.; Bandurski, R. iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 1994, 265, 1699–1701. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Baldauf, S.; Lim, E.K.; Bowles, D.J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Li, Y.; Lim, E.K.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, 1–6. [Google Scholar] [CrossRef]
- Jackson, R.G.; Lim, E.-K.; Li, Y.; Kowalczyk, M.; Sandberg, G.; Hoggett, J.; Ashford, D.A.; Bowles, D.J. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J. Biol. Chem. 2001, 276, 4350–4356. [Google Scholar] [CrossRef] [PubMed]
- Mravec, J.; Skupa, P.; Bailly, A.; Hoyerova, K.; Krecek, P.; Bielach, A.; Petrasek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.-D.; et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
| Biosynthetic Pathway | Biosynthetic Pathway | Biosynthetic Pathway | Biosynthetic Pathway |
|---|---|---|---|
| iaaM | Pseudomonas/Agrobacterium tryptophan-2-monooxygenase | Auxin biosynthesis in transgenic overexpression lines | Trp → IAM |
| iaaH | Pseudomonas/Agrobacterium IAM hydrolase | Auxin biosynthesis in transgenic overexpression lines | IAM → IAA |
| IPDC | IPA decarboxylase | Bacterial pathway | IPyA synthesis |
| ASA1/WEI2/JDL1/CKRC6/ASA2 ASB1/WEI7 | Subunits of anthranilate synthase | Root tip expression, inhibitor of sur1 Hormone interactions (cytokinin/ethylene/jasmonic acid) | Chorismate → anthranilate |
| IGS | Indole-3-glycerol phosphate synthase | Catalyzes formation of key intermediate of Trp-dependent and Trp-independent pathways | Anthranilate → indole-3-glycerol phosphate |
| TSA1/TSB1-2 | Trp synthase a and Trp synthase b | Trp synthesis | Indole-3-glycerol phosphate → indole → Trp |
| INS | TSA homolog | Early expression in embryo | |
| PDX1 | vitamin B6 biosynthesis mutant | Trp → IAA | |
| TAA1/TAR1-4 | Arabidopsis tryptophan aminotransferases | Stem cell niche specification De novo root organogenesis Control root architecture nitrogen dependently | Trp → IPyA |
| TAAI/WEI8/SAV3/TIR2 | Arabidopsis | Trp → IPyA | |
| TaTAR2 | wheat | Lateral root formation Nitrate and phosphate starvation | IPyA |
| FISHBONE | Rice Orthologue TAA1 | ||
| OsYUC1 OsCOW1 | Rice Flavin-containing monooxygenases | Post embryonic root development Crown and adventitious roots | IPyA |
| YUC1-11 | Arabidopsis Flavin-containing monooxygenases | Root embryogenesis RAM maintenance Drought resistance De novo root organogenesis Root growth in Al3+ stress | IPyA → IAA TAM → N-hydroxytryptamine? |
| YUC (ZM2G141383) | Maize Flavin-containing monooxygenases | Root cap-mediated gravitropic U-turn in response to light | IPyA → IAA |
| MYROSINASE AtTGG4/AtTGG5 | Arabidopsis | Local auxin distribution in the root | IG → IAN |
| Nitrilases NIT1-4 | Arabidopsis | Root development and growth | IAN → IAA |
| CYP79B2/CYP79B3 | Arabidopsis Cytochrome P450 | Auxin gradients in the root | Trp → IAOx |
| IBR1,3,10/ECH2 | INDOLE- 3-BUTYRIC ACID RESPONSE (IBR) ENOYL–COA HYDRATASE2 | IBA to IAA conversion Priming LR founder cells | IBA → IAA |
| AO | Aldehyde oxidase | IPyA to IAD pathway? | IDA → IAA |
| AMI1 | IAM HYDROLASE orthologue of iaaH | Required for BY2 growth on IAM medium | IAM → IAA PAM → PAA |
| SUR1/SUR2 | Glucosinolate biosynthesis genes SUPERROOT1/2 | Overproduction LR and AR | IAOx → IG |
| UGT74B1 | UDP-glycosyltransferase | High auxin levels in mutant Mutation in IG synthesis | |
| CYP71A13 | IAOx → IAN | ||
| ILR1/ILL2/IAR3 | IAA-leucine resistant1 IAA-leucine resistant-like 2 IAA-Ala Resistant3 hydrolase | Conjugated IAA → IAA | |
| GH3 | GRETCHEN HAGEN 3 (GH3) protein family | IAA-conjugation to amino acids | IAA → amide conjugated IAA |
| DAO | DIOXYGENASE FOR AUXIN OXIDATION 1 | IAA → oxIAA |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olatunji, D.; Geelen, D.; Verstraeten, I. Control of Endogenous Auxin Levels in Plant Root Development. Int. J. Mol. Sci. 2017, 18, 2587. https://doi.org/10.3390/ijms18122587
Olatunji D, Geelen D, Verstraeten I. Control of Endogenous Auxin Levels in Plant Root Development. International Journal of Molecular Sciences. 2017; 18(12):2587. https://doi.org/10.3390/ijms18122587
Chicago/Turabian StyleOlatunji, Damilola, Danny Geelen, and Inge Verstraeten. 2017. "Control of Endogenous Auxin Levels in Plant Root Development" International Journal of Molecular Sciences 18, no. 12: 2587. https://doi.org/10.3390/ijms18122587