Potential Metabolic Biomarkers to Identify Interstitial Lung Abnormalities
Abstract
:1. Introduction
2. Results
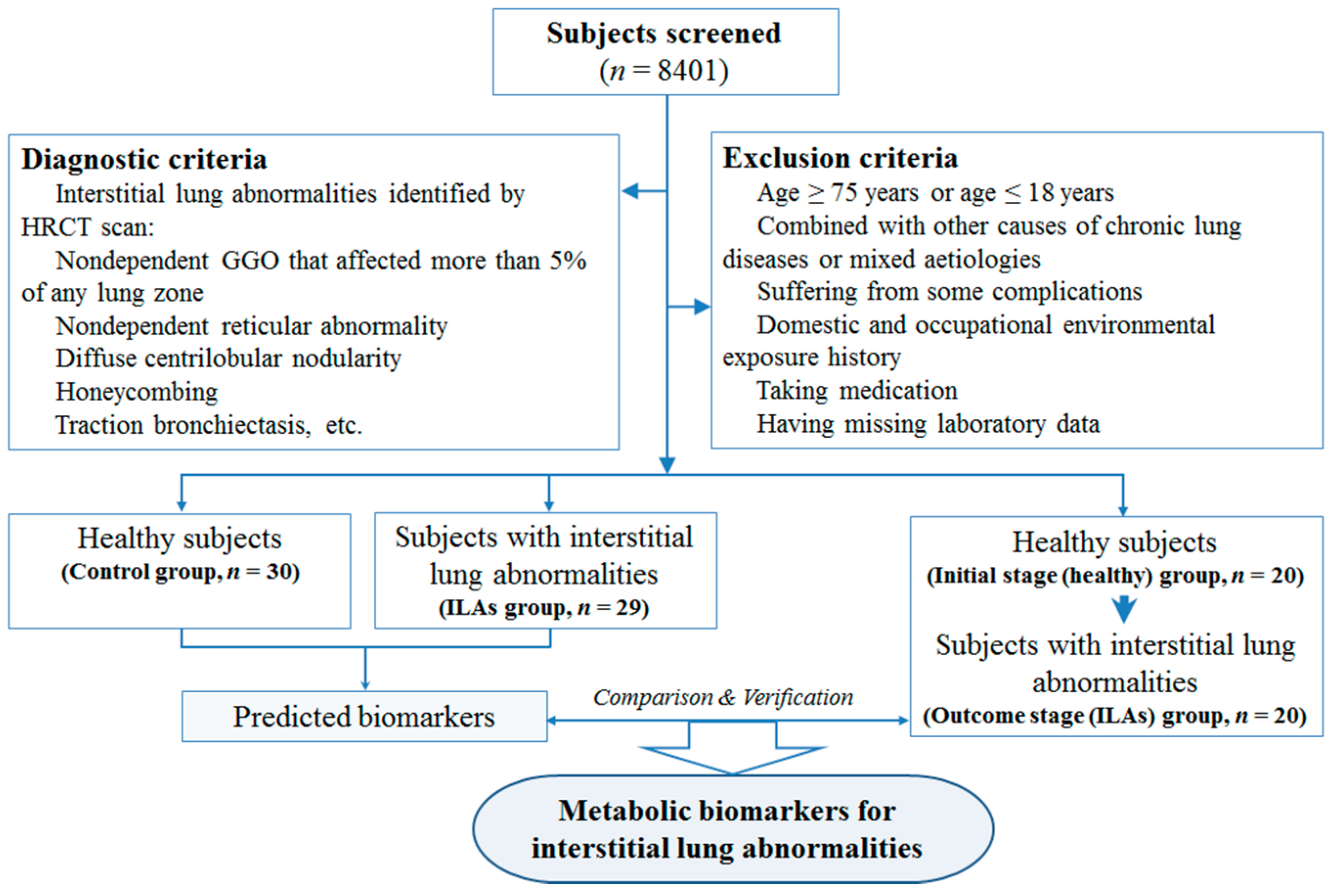
2.1. Baseline Characteristics of Study Subjects
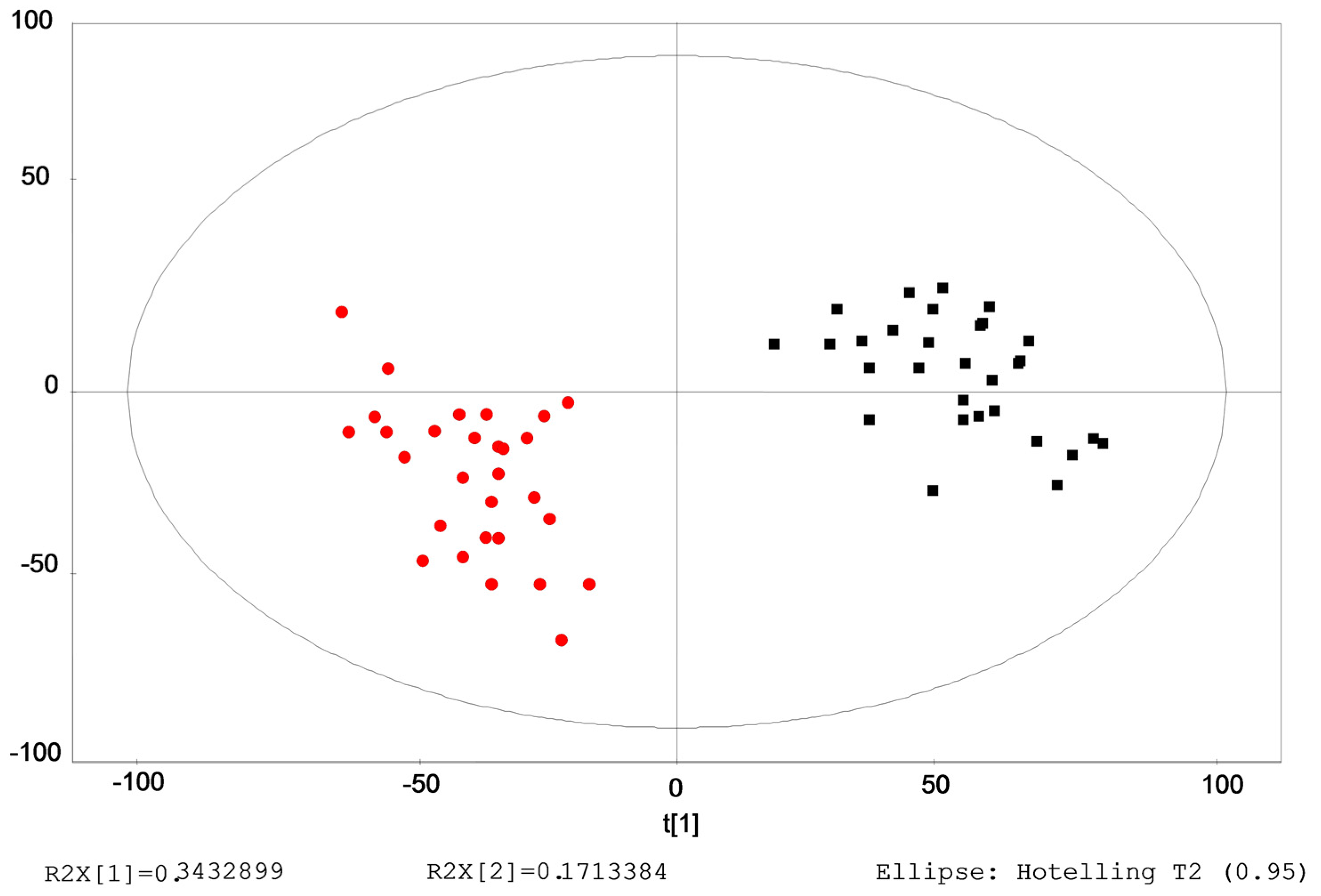
2.2. Evaluation of the Repeatability and Stability of the LC–Q–TOF–MS Method
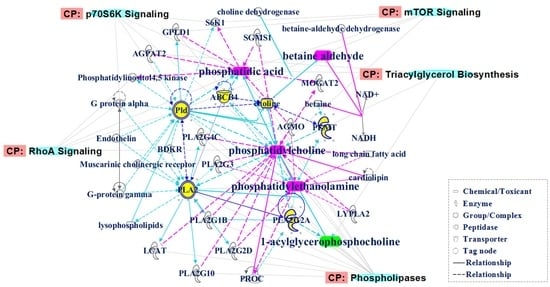
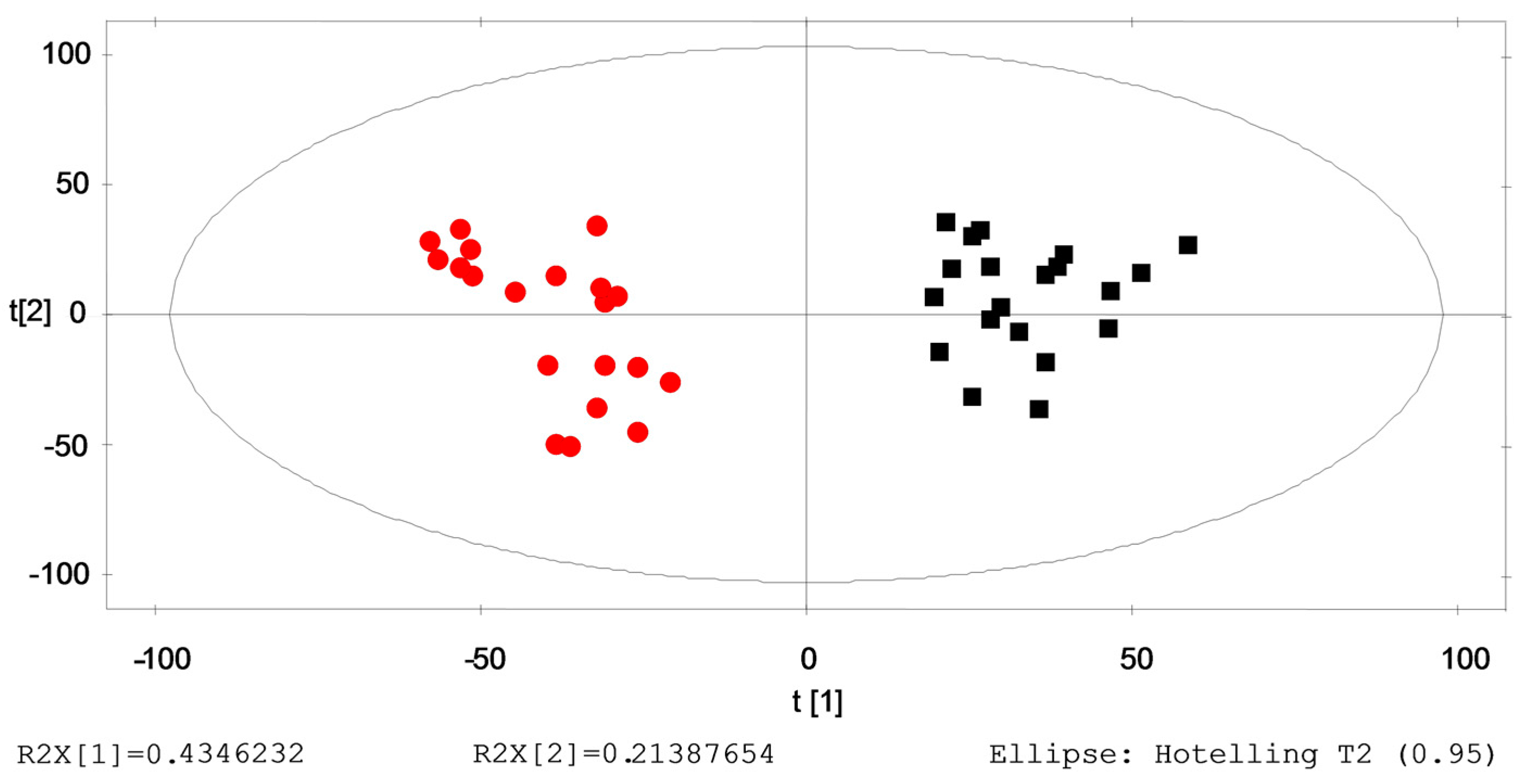
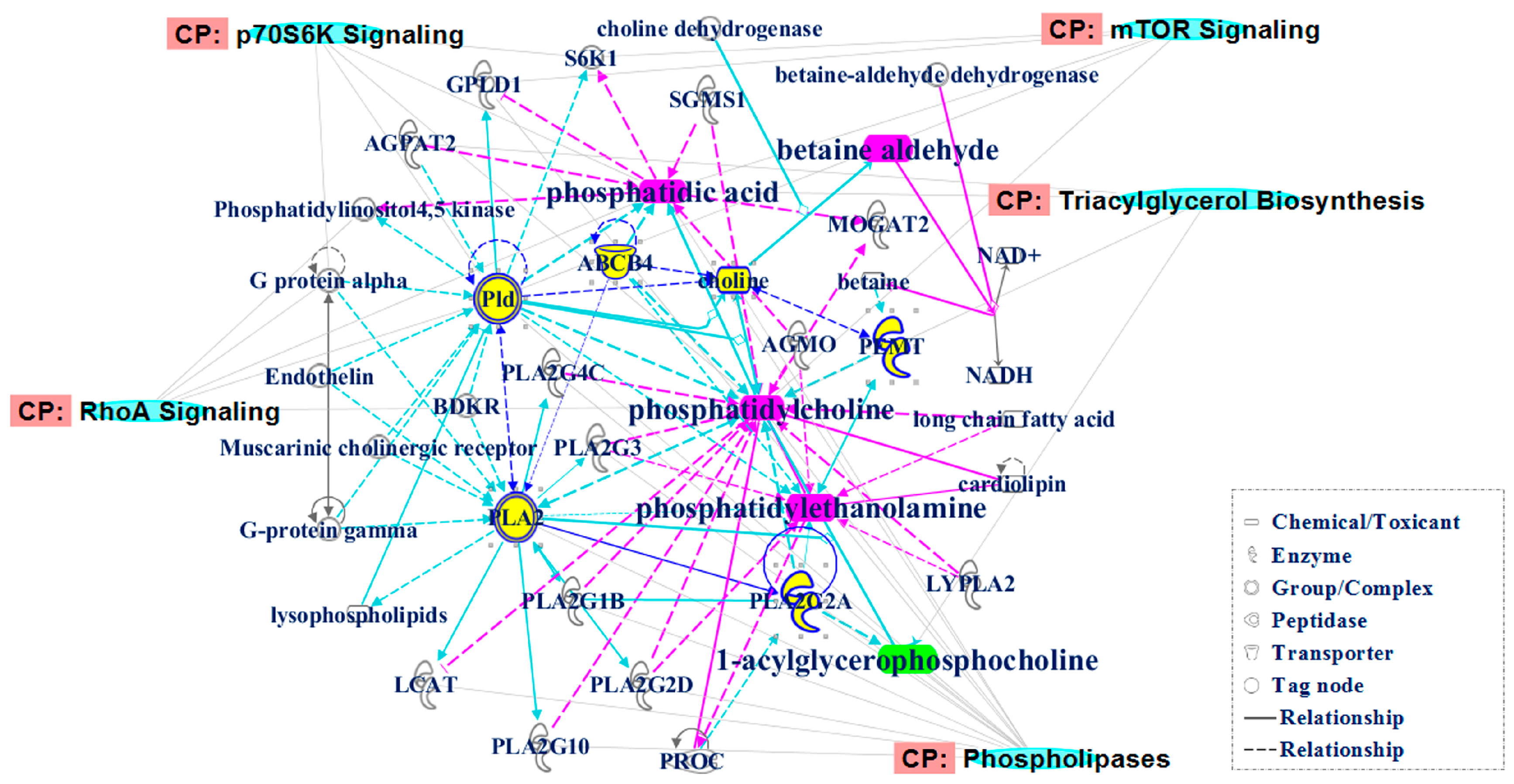
2.3. Identification of the Differential Metabolites in Interstitial Lung Abnormalities (ILAs)
2.4. Verification of the Identified Metabolites by Healthy→ILAs Subjects
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Questionnaire
4.3. Peripheral Blood Sampling and Biochemical Testing
4.4. High-Resolution Computed Tomography (HRCT) Examinations
4.5. LC–Q–TOF–MS Analysis
4.6. Sequence Analysis
4.7. Data Processing and Statistical Analysis
4.8. Molecular Network Analysis
4.9. Prediction of Metabolites Indication Ability
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 1-Acyl-GPC | 1-Acylglycerophosphocholine |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| AUC | Area under the curve |
| BA | Betaine aldehyde |
| BPCs | Base peak chromatograms |
| COPD | Chronic obstructive pulmonary disease |
| CRE | Creatinine |
| EICs | Extracted ion chromatograms |
| EMI | Enzyme-metabolite interaction |
| ESI | Electrospray ionization |
| HGB | Hemoglobin count |
| GGO | Ground-glass opacity |
| Healthy→ILAs subjects | The subjects who were disease-free initially and then one year later suffered from ILAs |
| HRCT | High-resolution computed tomography |
| ILAs | Interstitial lung abnormalities |
| ILD | Interstitial lung disease |
| IPA | Ingenuity pathway analysis |
| IPF | Idiopathic pulmonary fibrosis |
| LC-Q-TOF-MS | Liquid chromatography quadruple time-of-flight mass spectrometry |
| LY | Lymphocyte count |
| mTOR | Mammalian target of rapamycin |
| NE | Neutrophil count |
| NSCLC | Non-small-cell lung cancer |
| PA | Phosphatidic acid |
| PC | Phosphatidylcholine |
| PCA | Principal component analysis |
| PE | Phosphatidylethanolamine |
| PI3K | Phosphatidyl inositol 3-kinase |
| PLA | Phospholipase A |
| PLD | Phospholipase D |
| PLS-DA | Partial least squares discriminant analysis |
| PLT | Blood platelet count |
| PPI | Protein-protein interaction |
| QC | Quality control |
| RBC | Red blood cell count |
| RhoA | Ras homolog family member A |
| ROC | Receiver operating characteristic |
| RSDs | Relative standard derivations |
| RV | Representative value |
| sPLA | Secretory phospholipase A |
| SUA | Serum uric acid |
| WBC | White blood cell count |
References
- Lederer, D.J.; Enright, P.L.; Kawut, S.M.; Hoffman, E.A.; Hunninghake, G.; van Beek, E.J.; Austin, J.H.; Jiang, R.; Lovasi, G.S.; Barr, R.G. Cigarette smoking is associated with subclinical parenchymal lung disease: The Multi-Ethnic Study of Atherosclerosis (MESA)-lung study. Am. J. Respir. Crit. Care Med. 2009, 180, 407–414. [Google Scholar] [CrossRef] [PubMed]
- El-Chemaly, S.; Ziegler, S.G.; Calado, R.T.; Wilson, K.A.; Wu, H.P.; Haughey, M.; Peterson, N.R.; Young, N.S.; Gahl, W.A.; Moss, J.; et al. Natural history of pulmonary fibrosis in two subjects with the same telomerase mutation. Chest 2011, 139, 1203–1209. [Google Scholar] [CrossRef] [PubMed]
- Tsushima, K.; Sone, S.; Yoshikawa, S.; Yokoyama, T.; Suzuki, T.; Kubo, K. The radiological patterns of interstitial change at an early phase: Over a 4-year follow-up. Respir. Med. 2010, 104, 1712–1221. [Google Scholar] [CrossRef] [PubMed]
- Rosas, I.O.; Ren, P.; Avila, N.A.; Chow, C.K.; Franks, T.J.; Travis, W.D.; McCoy, J.P., Jr.; May, R.M.; Wu, H.P.; Nguyen, D.M.; et al. Early interstitial lung disease in familial pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2007, 176, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Araki, T.; Nishino, M.; Zazueta, O.E.; Gao, W.; Dupuis, J.; Okajima, Y.; Latourelle, J.C.; Rosas, I.O.; Murakami, T.; O’Connor, G.T.; et al. Paraseptal emphysema: Prevalence and distribution on CT and association with interstitial lung abnormalities. Eur. J. Radiol. 2015, 84, 1413–1418. [Google Scholar] [CrossRef] [PubMed]
- Washko, G.R.; Hunninghake, G.M.; Fernandez, I.E.; Nishino, M.; Okajima, Y.; Yamashiro, T.; Ross, J.C.; Estepar, R.S.; Lynch, D.A.; Brehm, J.M.; et al. Lung volumes and emphysema in smokers with interstitial lung abnormalities. N. Engl. J. Med. 2011, 364, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Paulin, F.; Doyle, T.J.; Fletcher, E.A.; Ascherman, D.P.; Rosas, I.O. Rheumatoid Arthritis-Associated Interstitial Lung Disease and Idiopathic Pulmonary Fibrosis: Shared Mechanistic and Phenotypic Traits Suggest Overlapping Disease Mechanisms. Rev. Investig. Clin. 2015, 67, 280–286. [Google Scholar]
- Hunninghake, G.M.; Hatabu, H.; Okajima, Y.; Gao, W.; Dupuis, J.; Latourelle, J.C.; Nishino, M.; Araki, T.; Zazueta, O.E.; Kurugol, S.; et al. MUC5B promoter polymorphism and interstitial lung abnormalities. N. Engl. J. Med. 2013, 368, 2192–2200. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.J.; Washko, G.R.; Fernandez, I.E.; Nishino, M.; Okajima, Y.; Yamashiro, T.; Divo, M.J.; Celli, B.R.; Sciurba, F.C.; Silverman, E.K.; et al. Interstitial lung abnormalities and reduced exercise capacity. Am. J. Respir. Crit. Care Med. 2012, 185, 756–762. [Google Scholar] [CrossRef] [PubMed]
- Sverzellati, N.; Guerci, L.; Randi, G.; Calabro, E.; La Vecchia, C.; Marchiano, A.; Pesci, A.; Zompatori, M.; Pastorino, U. Interstitial lung diseases in a lung cancer screening trial. N. Engl. J. Med. 2011, 38, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.E.; Gao, W.; Levy, D.; Santhanakrishnan, R.; Araki, T.; Rosas, I.O.; Hatabu, H.; Latourelle, J.C.; Nishino, M.; Dupuis, J.; et al. Galectin-3 is Associated with Restrictive Lung Disease and Interstitial Lung Abnormalities. Am. J. Respir. Crit. Care Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.J.; Hunninghake, G.M.; Rosas, I.O. Subclinical interstitial lung disease: Why you should care. Am. J. Respir. Crit. Care Med. 2012, 185, 1147–1153. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.Y.; Lynch, D.; Chawla, A.; Garg, K.; Tammemagi, M.C.; Sahin, H.; Misumi, S.; Kwon, K.S. Interstitial lung abnormalities in a CT lung cancer screening population: Prevalence and progression rate. Radiology 2013, 268, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.D.; Hutchison, P.J.; Nicholson, T.T. Risk factors for development of acute lung injury, physicians’ decision-making roles in critical illness, and prevalence of interstitial lung abnormalities in lung cancer screening populations. Am. J. Respir. Crit. Care Med. 2014, 189, 1273–1274. [Google Scholar] [CrossRef] [PubMed]
- Nishino, M.; Cardarella, S.; Dahlberg, S.E.; Araki, T.; Lydon, C.; Jackman, D.M.; Rabin, M.S.; Hatabu, H.; Johnson, B.E. Interstitial lung abnormalities in treatment-naive advanced non-small-cell lung cancer patients are associated with shorter survival. Eur. J. Radiol. 2015, 84, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, D.A.; Ebrahim, S.; Smith, G.D. Associations of measures of lung function with insulin resistance and Type 2 diabetes: Findings from the British Women’s Heart and Health Study. Diabetologia 2004, 47, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.Y.; Yao, C.A.; Wang, H.C.; Huang, K.C. Impaired lung function is associated with obesity and metabolic syndrome in adults. Obesity 2006, 14, 1654–1661. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, R.; Sparrow, D.; Weiss, S.T. Baseline ventilatory function predicts the development of higher levels of fasting insulin and fasting insulin resistance index: The Normative Aging Study. Eur. Respir. J. 1998, 12, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Obata, F.; Kuranaga, E.; Tomioka, K.; Ming, M.; Takeishi, A.; Chen, C.H.; Soga, T.; Miura, M. Necrosis-driven systemic immune response alters SAM metabolism through the FOXO-GNMT axis. Cell Rep. 2014, 7, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Im, S.S.; Yousef, L.; Blaschitz, C.; Liu, J.Z.; Edwards, R.A.; Young, S.G.; Raffatellu, M.; Osborne, T.F. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 2011, 13, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C. Systems biology: Metabonomics. Nature 2008, 455, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- German, J.B.; Hammock, B.D.; Watkins, S.M. Metabolomics: Building on a century of biochemistry to guide human health. Metabolomics 2005, 1, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Dutta, M.; Joshi, M.; Srivastava, S.; Lodh, I.; Chakravarty, B.; Chaudhury, K. A metabonomics approach as a means for identification of potential biomarkers for early diagnosis of endometriosis. Mol. Biosyst. 2012, 8, 3281–3287. [Google Scholar] [CrossRef] [PubMed]
- Coen, M.; O’Sullivan, M.; Bubb, W.A.; Kuchel, P.W.; Sorrell, T. Proton nuclear magnetic resonance-based metabonomics for rapid diagnosis of meningitis and ventriculitis. Clin. Infect. Dis. 2005, 41, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, C.; Liu, F.; Lu, Y.; Cheng, J. Metabonomics revealed xanthine oxidase-induced oxidative stress and inflammation in the pathogenesis of diabetic nephropathy. Anal. Bioanal. Chem. 2015, 407, 2569–2579. [Google Scholar] [CrossRef] [PubMed]
- Amaral, J.J.; Antunes, L.C.; de Macedo, C.S.; Mattos, K.A.; Han, J.; Pan, J.; Candea, A.L.; Henriques, M.; Ribeiro-Alves, M.; Borchers, C.H.; et al. Metabonomics reveals drastic changes in anti-inflammatory/pro-resolving polyunsaturated fatty acids-derived lipid mediators in leprosy disease. PLoS Negl. Trop. Dis. 2013, 7, e2381. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tang, Y.; Liu, S.; Mao, S.; Ling, Y.; Liu, D.; He, X.; Wang, X. Metabonomic profiling of serum and urine by 1H-NMR-based spectroscopy discriminates patients with chronic obstructive pulmonary disease and healthy individuals. PLoS ONE 2013, 8, e65675. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Chen, Y.; Zhang, R.; Song, Y.; Sun, J.; He, J.; Bai, J.; Dong, L.; Zhan, Q.; Abliz, Z. Integrated ionization approach for RRLC–MS/MS-based metabonomics: Finding potential biomarkers for lung cancer. J. Proteom. Res. 2010, 9, 4071–4081. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lu, C.; Liu, X.; Su, J.; Dai, W.; Yan, S.; Lu, A.; Zhang, W. Global and targeted metabolomics reveal that Bupleurotoxin, a toxic type of polyacetylene, induces cerebral lesion by inhibiting GABA receptor in mice. J. Proteom. Res. 2014, 13, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Li, J.; Liu, X.; Ko, J.; He, X.; Lu, C.; Liu, Z.; Zhao, H.; Xiao, C.; Niu, X.; et al. Deciphering the differential toxic responses of Radix aconiti lateralis praeparata in healthy and hydrocortisone-pretreated rats based on serum metabolic profiles. J. Proteom. Res. 2013, 12, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Liscovitch, M.; Czarny, M.; Fiucci, G.; Tang, X. Phospholipase D: Molecular and cell biology of a novel gene family. Biochem. J. 2000, 345, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; You, Y.; Ryan, A.J.; Mallampalli, R.K. Upregulation of surfactant synthesis triggers ABCA1-mediated basolateral phospholipid efflux. J. Lipid Res. 2004, 45, 1758–1767. [Google Scholar] [CrossRef] [PubMed]
- Kurabe, N.; Hayasaka, T.; Igarashi, H.; Mori, H.; Sekihara, K.; Tao, H.; Yamada, H.; Kahyo, T.; Onishi, I.; Tsukui, H.; et al. Visualization of phosphatidylcholine (16:0/16:0) in type II alveolar epithelial cells in the human lung using imaging mass spectrometry. Pathol. Int. 2013, 63, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, W.; Hoffmann, S.; Dombrowsky, H.; Rau, G.A.; Kamlage, A.; Kappler, M.; Haitsma, J.J.; Freihorst, J.; von der Hardt, H.; Poets, C.F. Phosphatidylcholine molecular species in lung surfactant: Composition in relation to respiratory rate and lung development. Am. J. Respir. Cell Mol. Biol. 2001, 25, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Grothe, J.; Riethmuller, J.; Tschurtz, S.M.; Raith, M.; Pynn, C.J.; Stoll, D.; Bernhard, W. Plasma phosphatidylcholine alterations in cystic fibrosis patients: Impaired metabolism and correlation with lung function and inflammation. Cell. Physiol. Biochem. 2015, 35, 1437–1453. [Google Scholar] [CrossRef] [PubMed]
- Machado-Aranda, D.; Wang, Z.; Yu, B.; Suresh, M.V.; Notter, R.H.; Raghavendran, K. Increased phospholipase A2 and lyso-phosphatidylcholine levels are associated with surfactant dysfunction in lung contusion injury in mice. Surgery 2013, 153, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Foster, D.A.; Xu, L. Phospholipase D in cell proliferation and cancer. Mol. Cancer Res. 2003, 1, 789–800. [Google Scholar] [PubMed]
- Ha, K.S.; Exton, J.H. Activation of actin polymerization by phosphatidic acid derived from phosphatidylcholine in IIC9 fibroblasts. J. Cell Biol. 1993, 123, 1789–1796. [Google Scholar] [CrossRef] [PubMed]
- Oude Weernink, P.A.; Lopez de Jesus, M.; Schmidt, M. Phospholipase D signaling: Orchestration by PIP2 and small GTPases. Naunyn Schmiedeb. Arch. Pharmacol. 2007, 374, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.; Parinandi, N.; Wang, L.; Usatyuk, P.; Natarajan, V. Phospholipase D/phosphatidic acid signal transduction: Role and physiological significance in lung. Mol. Cell. Biochem. 2002, 234–235, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.B.; Kotha, S.R.; Sherwani, S.I.; Sliman, S.M.; Gurney, T.O.; Loar, B.; Butler, S.O.; Morris, A.J.; Marsh, C.B.; Parinandi, N.L. Pulmonary fibrosis inducer, bleomycin, causes redox-sensitive activation of phospholipase D and cytotoxicity through formation of bioactive lipid signal mediator, phosphatidic acid, in lung microvascular endothelial cells. Int. J. Toxicol. 2011, 30, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Abraham, E.; Bursten, S.; Shenkar, R.; Allbee, J.; Tuder, R.; Woodson, P.; Guidot, D.M.; Rice, G.; Singer, J.W.; Repine, J.E. Phosphatidic acid signaling mediates lung cytokine expression and lung inflammatory injury after hemorrhage in mice. J. Exp. Med. 1995, 181, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Mazie, A.R.; Spix, J.K.; Block, E.R.; Achebe, H.B.; Klarlund, J.K. Epithelial cell motility is triggered by activation of the EGF receptor through phosphatidic acid signaling. J. Cell Sci. 2006, 119, 1645–1654. [Google Scholar] [CrossRef] [PubMed]
- Watts, K.L.; Cottrell, E.; Hoban, P.R.; Spiteri, M.A. RhoA signaling modulates cyclin D1 expression in human lung fibroblasts; implications for idiopathic pulmonary fibrosis. Respir. Res. 2006, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Park, H.J.; Park, Y.S.; Lee, S.M.; Yim, J.J.; Yoo, C.G.; Han, S.K.; Kim, Y.W. Clinical significance of mTOR, ZEB1, ROCK1 expression in lung tissues of pulmonary fibrosis patients. BMC Pulm. Med. 2014, 14, 168. [Google Scholar] [CrossRef] [PubMed]
- Chitra, P.; Saiprasad, G.; Manikandan, R.; Sudhandiran, G. Berberine inhibits Smad and non-Smad signaling cascades and enhances autophagy against pulmonary fibrosis. J. Mol. Med. 2015, 93, 1015–1031. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Sabatini, D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005, 17, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Vilella-Bach, M.; Bachmann, R.; Flanigan, A.; Chen, J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 2001, 294, 1942–1945. [Google Scholar] [CrossRef] [PubMed]
- Isotani, S.; Hara, K.; Tokunaga, C.; Inoue, H.; Avruch, J.; Yonezawa, K. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase α in vitro. J. Biol. Chem. 1999, 274, 34493–34498. [Google Scholar] [CrossRef] [PubMed]
- Dennis, P.B.; Jaeschke, A.; Saitoh, M.; Fowler, B.; Kozma, S.C.; Thomas, G. Mammalian TOR: A homeostatic ATP sensor. Science 2001, 294, 1102–1105. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Lehman, N.; Ledford, B.; di Fulvio, M.; Frondorf, K.; McPhail, L.C.; Gomez-Cambronero, J. Phospholipase D2-derived phosphatidic acid binds to and activates ribosomal p70 S6 kinase independently of mTOR. FASEB J. 2007, 21, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Rustow, B.; Kunze, D. Synthesis of phosphatidylcholine and phosphatidylethanolamine in relation to the concentration of membrane-bound diacylglycerols of rat lung microsomes. Biochim. Biophys. Acta 1984, 793, 372–378. [Google Scholar] [CrossRef]
- Yu, G.; Huang, B.; Chen, G.; Mi, Y. Phosphatidylethanolamine-binding protein 4 promotes lung cancer cells proliferation and invasion via PI3K/Akt/mTOR axis. J. Thorac. Dis. 2015, 7, 1806–1816. [Google Scholar] [PubMed]
- Triggiani, M.; Giannattasio, G.; Calabrese, C.; Loffredo, S.; Granata, F.; Fiorello, A.; Santini, M.; Gelb, M.H.; Marone, G. Lung mast cells are a source of secreted phospholipases A2. J. Allergy Clin. Immunol. 2009, 124, 558–565.e3. [Google Scholar] [CrossRef] [PubMed]
- Granata, F.; Nardicchi, V.; Loffredo, S.; Frattini, A.; Ilaria Staiano, R.; Agostini, C.; Triggiani, M. Secreted phospholipases A2: A proinflammatory connection between macrophages and mast cells in the human lung. Immunobiology 2009, 214, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.J. Perceptual decision-making. Watching neurons discriminate. Nature 1989, 341, 20–21. [Google Scholar] [CrossRef] [PubMed]
| Indicators/Groups | ILAs | Initial Stage (Healthy) | Outcome Stage (ILAs) | Control |
|---|---|---|---|---|
| Sex (M/F) | 22/7 | 16/4 | 16/4 | 22/8 |
| Age (years) | 63.90 ± 6.04 | 60.70 ± 10.00 | 61.20 ± 9.47 | 61.11 ± 7.53 |
| Smoking rate (%) | 48.28 * | 50.00 * | 50.00 * | 18.95 |
| WBC (109/L) | 6.31 ± 1.25 | 6.51 ± 1.21 | 6.45 ± 1.38 | 5.92 ± 1.21 |
| LY (109/L) | 1.93 ± 0.54 | 1.91 ± 0.50 | 1.93 ± 0.62 | 1.82 ± 0.46 |
| NE (109/L) | 4.29 ± 0.97 | 4.29 ± 0.96 | 4.41 ± 1.19 | 3.79 ± 1.07 |
| RBC (1012/L) | 4.72 ± 0.33 | 4.79 ± 0.47 | 4.80 ± 0.39 | 4.68 ± 0.45 |
| HGB (g/L) | 141.97 ± 11.61 | 145.05 ± 12.69 | 141.95 ± 13.29 | 141.38 ± 10.96 |
| PLT (109/L) | 190.14 ± 41.78 | 200.50 ± 52.00 | 203.35 ± 51.09 | 205.82 ± 46.46 |
| ALT (mmol/L) | 18.79 ± 7.61 | 21.85 ± 6.68 | 16.90 ± 5.15 | 18.15 ± 8.48 |
| AST (mmol/L) | 23.31 ± 6.48 | 22.15 ± 3.56 | 22.05 ± 3.30 | 25.00 ± 14.16 |
| CRE (mmol/L) | 76.52 ± 19.56 | 79.2 ± 11.22 | 68.45 ± 11.39 | 70.38 ± 13.02 |
| SUA (μmol/L) | 329.79 ± 70.61 | 341.15 ± 85.76 | 318.30 ± 85.42 | 304.54 ± 57.58 |
| n | Rt (min) | Exact Mass | Formula | Compound | Fold Changes | RV |
|---|---|---|---|---|---|---|
| 1 | 7.3660 | 675.4839 | C36H70NO8P | Phosphatidylcholine (PC)(28:1) | 7.4118 | 0.0511 |
| 2 | 11.6928 | 674.4886 | C37H71O8P | Phosphatidic acid (PA)(34:1) | 12.0640 | 0.0437 |
| 3 | 11.798 | 467.3012 | C22H46NO7P | 1-Acylglycerophosphocholine (1-acyl-GPC) | −17.2178 | 0.0283 |
| 4 | 8.8195 | 776.7257 | C50H96O5 | Triacylglycerol | 8.9617 | 0.0187 |
| 5 | 11.8360 | 715.5152 | C39H74NO8P | Phosphatidylethanolamine (PE)(34:2) | 3.5377 | 0.0166 |
| 6 | 12.3754 | 148.0194 | C5H8O3S | 2-Keto-4-methylthiobutyric acid | −19.2816 | 0.0033 |
| 7 | 9.3124 | 116.0837 | C6H12O2 | Caproic acid | 2.83241 | 0.0021 |
| 8 | 8.5394 | 100.0888 | C6H12O | Caproaldehyde | 3.6839 | 0.0013 |
| 9 | 8.7894 | 128.1201 | C8H16O | Octanal | 4.2036 | 0.0012 |
| 10 | 7.5562 | 102.0919 | C5H12NO | Betaine aldehyde (BA) | 2.9631 | 0.0005 |
| 11 | 7.6113 | 72.0575 | C4H8O | 2-Butanone | 8.4656 | 0 |
| n | Rt (min) | Exact Mass | Formula | Compound | Fold Changes |
|---|---|---|---|---|---|
| 1 | 14.4294 | 690.5223 | C45H70O5 | Diacylglycerol | −9.9068 |
| 2 | 11.6928 | 674.4886 | C37H71O8P | PA(34:1) | 1.4748 |
| 3 | 11.8360 | 715.5152 | C39H74NO8P | PE(34:2) | 7.3538 |
| 4 | 7.3660 | 675.4839 | C36H70NO8P | PC(28:1) | 1.6725 |
| 5 | 11.798 | 467.3012 | C22H46NO7P | 1-Acylglycerophosphocholine (1-acyl-GPC) | −12.9007 |
| 6 | 1.9809 | 274.2297 | C19H30O | 3-Oxosteroid | 11.3056 |
| 7 | 6.7841 | 121.0891 | C8H11N | Phenylethylamine | −3.3337 |
| 8 | 7.5562 | 102.0919 | C5H12NO | Betaine aldehyde | 1.1937 |
| 9 | 8.7372 | 202.1205 | C10H18O4 | Sebacic acid | 10.5327 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Y.; Jia, D.; Lin, Z.; Guo, B.; He, B.; Lu, C.; Xiao, C.; Liu, Z.; Zhao, N.; Bian, Z.; et al. Potential Metabolic Biomarkers to Identify Interstitial Lung Abnormalities. Int. J. Mol. Sci. 2016, 17, 1148. https://doi.org/10.3390/ijms17071148
Tan Y, Jia D, Lin Z, Guo B, He B, Lu C, Xiao C, Liu Z, Zhao N, Bian Z, et al. Potential Metabolic Biomarkers to Identify Interstitial Lung Abnormalities. International Journal of Molecular Sciences. 2016; 17(7):1148. https://doi.org/10.3390/ijms17071148
Chicago/Turabian StyleTan, Yong, Dongmei Jia, Zhang Lin, Baosheng Guo, Bing He, Cheng Lu, Cheng Xiao, Zhongdi Liu, Ning Zhao, Zhaoxiang Bian, and et al. 2016. "Potential Metabolic Biomarkers to Identify Interstitial Lung Abnormalities" International Journal of Molecular Sciences 17, no. 7: 1148. https://doi.org/10.3390/ijms17071148