Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche aegyptiaca Seed Germination
Abstract
:1. Introduction
2. Results
2.1. De Novo Assembly of P. aegyptiaca Transcriptome
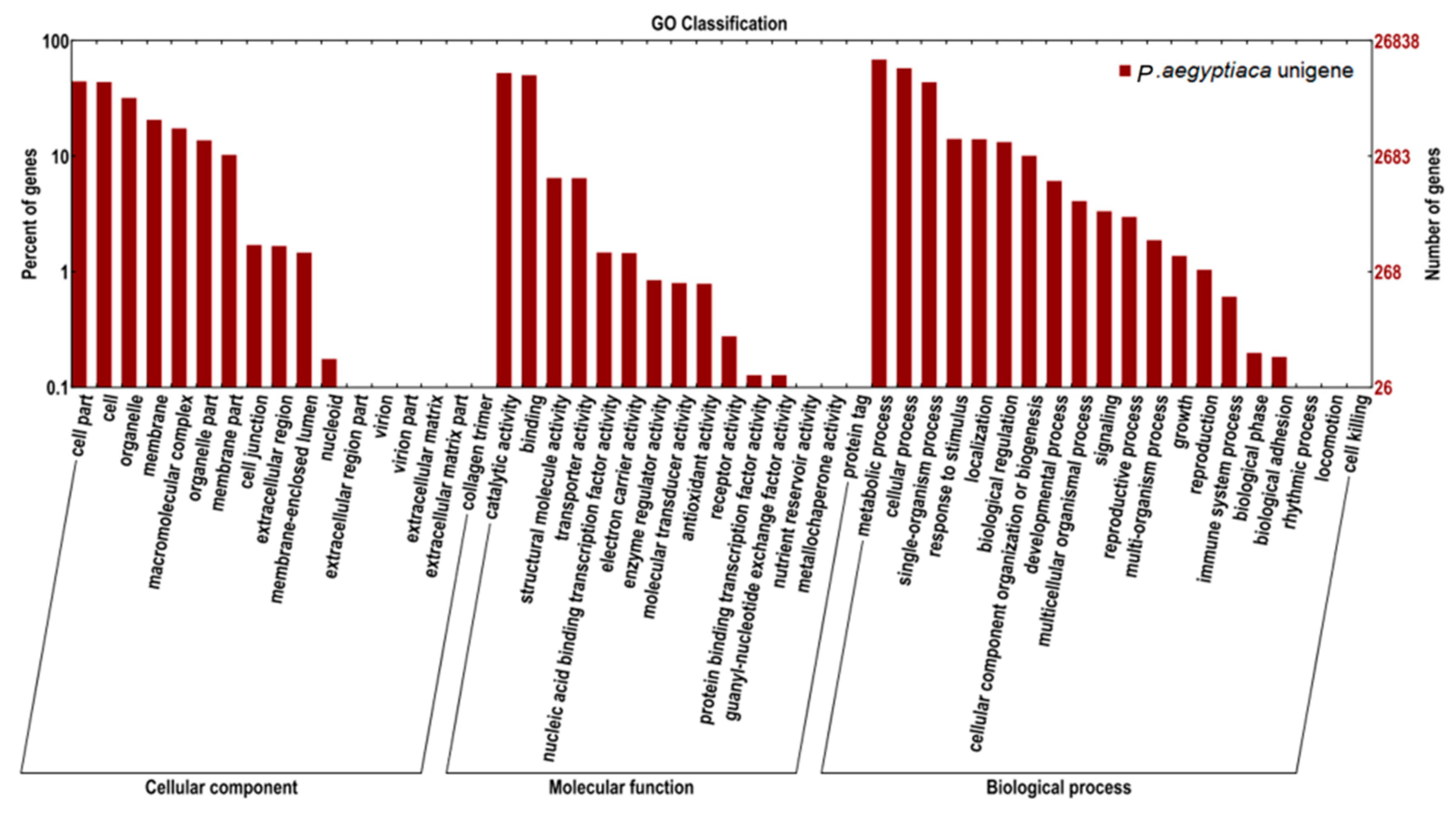
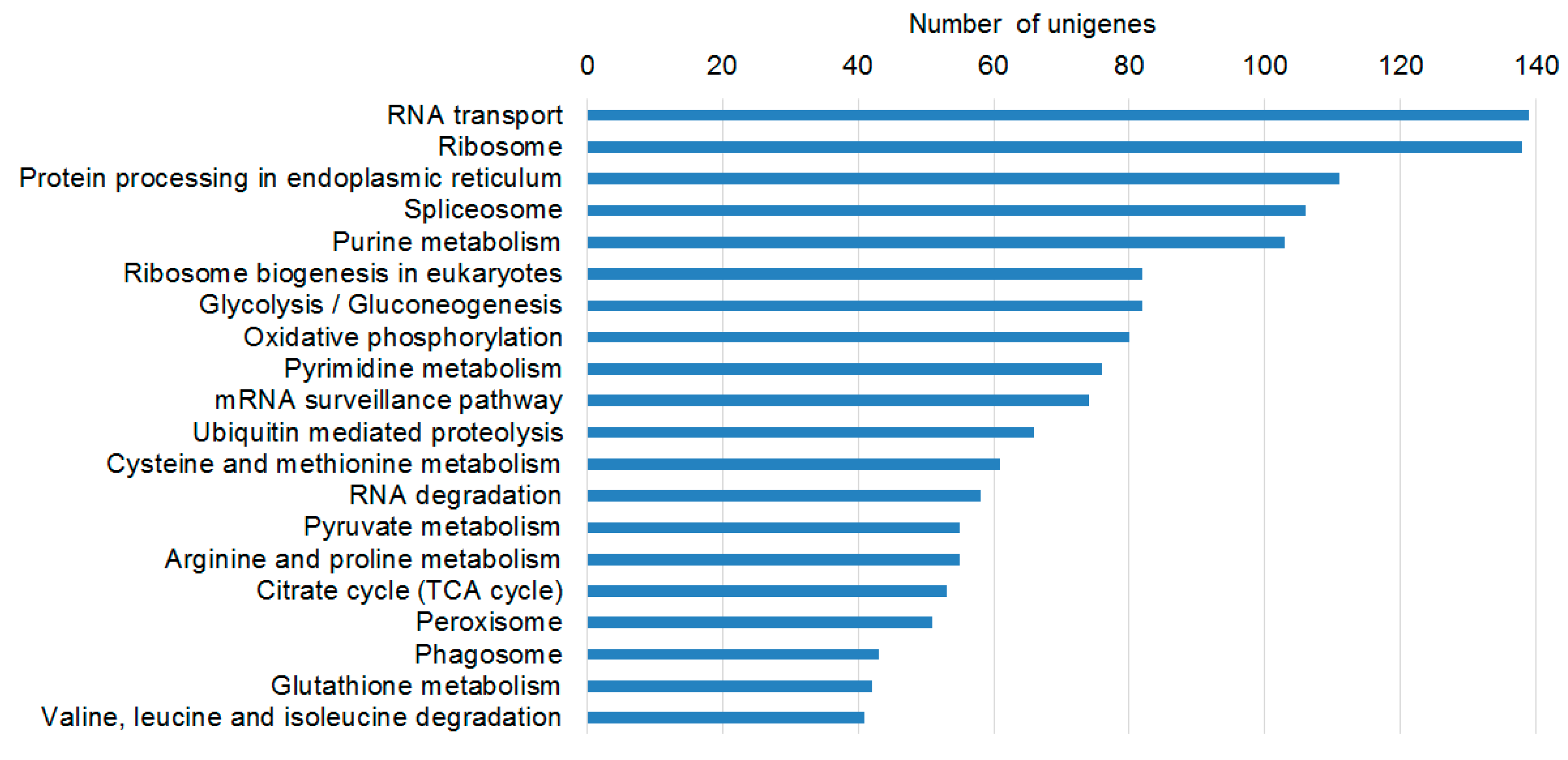
2.2. Transcriptome Functional Annotation
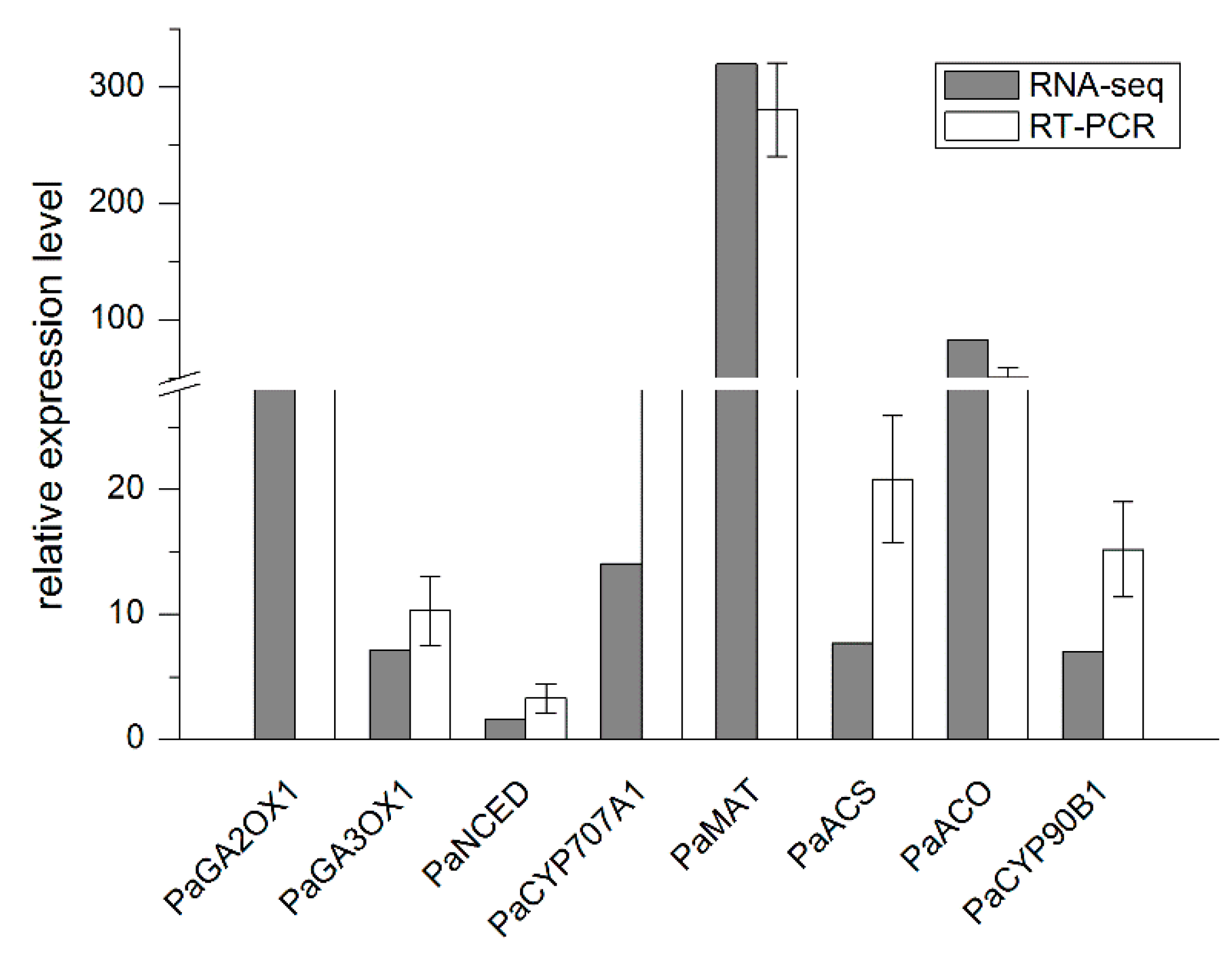
2.3. qRT-PCR Validation
2.4. Comparative Analysis of Differential Expression during Germination
2.5. Gene Change during P. aegyptiaca Seed Germination
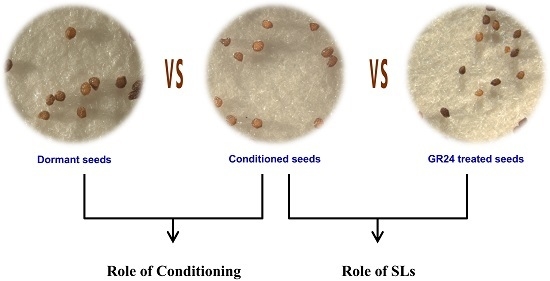
2.6. Endogenous Hormone Levels and Germination Test
3. Discussion
3.1. Transcriptome Assembly and Annotation
3.2. DEGs Associated with Broomrape Seed Germination
3.2.1. DNA, RNA, and Protein
3.2.2. Energy Metabolism
3.2.3. Phytohormone Activity
4. Materials and Methods
4.1. Sample Preparation
4.2. RNA-seq Library Preparation and Sequencing
4.3. Preprocessing of Illumina Reads
4.4. De Novo Transcriptome Assembly
4.5. Functional Annotation and Analysis
4.6. Differential Expression Analysis
4.7. qRT-PCR Validation
4.8. Endogenous Hormone Level Analysis
4.9. Seed Hormone Treatments
4.10. Accession Numbers
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Parker, C. Parasitic weeds: A world challenge. Weed Sci. 2012, 60, 269–276. [Google Scholar] [CrossRef]
- Yoder, J.I.; Scholes, J.D. Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr. Opin. Plant Biol. 2010, 13, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Ghannam, I.; Al-Masri, M.; Barakat, R. The effect of herbicides on the egyptian broomrape (Orobanche aegyptiaca) in tomato fields. Am. J. Plant Sci. 2012, 3, 346. [Google Scholar] [CrossRef]
- Westwood, J.H.; Yoder, J.I.; Timko, M.P. The evolution of parasitism in plants. Trends Plant Sci. 2010, 15, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Parker, C. Observations on the current status of Orobanche and Striga problems worldwide. Pest. Manag. Sci. 2009, 65, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Joel, D.; Hershenhorn, J.; Eizenberg, H.; Aly, R.; Ejeta, G.; Rich, P.; Ransom, J.; Sauerborn, J.; Rubiales, D. Biology and management of weedy root parasites. Horticult. Rev. Westport N. Y. 2007, 33, 267. [Google Scholar]
- Parker, C.; Riches, C.R. Parasitic Weeds of the World: Biology and Control; CAB International: Wallingford, UK, 1993. [Google Scholar]
- Press, M.C.; Graves, J.D. Parasitic Plants; Chapman & Hall Ltd: London, UK, 1995. [Google Scholar]
- Joel, D.M. The long-term approach to parasitic weeds control: Manipulation of specific developmental mechanisms of the parasite. Crop Prot. 2000, 19, 753–758. [Google Scholar] [CrossRef]
- Xie, X.; Yoneyama, K.; Yoneyama, K. The strigolactone story. Annu. Rev. Phytopathol. 2010, 48, 93–117. [Google Scholar] [CrossRef] [PubMed]
- Koltai, H. Receptors, repressors, pins: A playground for strigolactone signaling. Trends Plant Sci. 2014, 19, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Waldie, T.; Hayward, A.; Beveridge, C.A. Axillary bud outgrowth in herbaceous shoots: How do strigolactones fit into the picture? Plant Mol. Biol. 2010, 73, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Ruyter-Spira, C.; Al-Babili, S.; van der Krol, S.; Bouwmeester, H. The biology of strigolactones. Trends Plant Sci. 2013, 18, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Babaei, S.; Alizadeh, H.; Jahansouz, M.R.; Mashhadi, H.R.; Moeini, M.M. Management of Phelipanche aegyptiaca pomel: Using trap crops in rotation with tomato (Solanum lycopersicom L.). Aust. J. Crop Sci. 2010, 4, 437–442. [Google Scholar]
- Grenz, J.; Iştoc, V.; Manschadi, A.; Sauerborn, J. Interactions of sunflower (Helianthus annuus) and sunflower broomrape (Orobanche cumana) as affected by sowing date, resource supply and infestation level. Field Crop. Res. 2008, 107, 170–179. [Google Scholar] [CrossRef]
- Kannan, C.; Aditi, P.; Zwanenburg, B. Quenching the action of germination stimulants using borax and thiourea, a new method for controlling parasitic weeds: A proof of concept. Crop Prot. 2015, 70, 92–98. [Google Scholar] [CrossRef]
- MÜLLER-STÖVER, D.; Kohlschmid, E.; Sauerborn, J. A novel strain of Fusarium oxysporum from germany and its potential for biocontrol of Orobanche ramosa. Weed Res. 2009, 49, 175–182. [Google Scholar] [CrossRef]
- Sillero, J.C.; Rojas-Molina, M.M.; Ávila, C.M.; Rubiales, D. Induction of systemic acquired resistance against rust, ascochyta blight and broomrape in faba bean by exogenous application of salicylic acid and benzothiadiazole. Crop Prot. 2012, 34, 65–69. [Google Scholar] [CrossRef]
- López-Ráez, J.A.; Matusova, R.; Cardoso, C.; Jamil, M.; Charnikhova, T.; Kohlen, W.; Ruyter-Spira, C.; Verstappen, F.; Bouwmeester, H. Strigolactones: Ecological significance and use as a target for parasitic plant control. Pest Manag. Sci. 2009, 65, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Dor, E.; Yoneyama, K.; Wininger, S.; Kapulnik, Y.; Yoneyama, K.; Koltai, H.; Xie, X.; Hershenhorn, J. Strigolactone deficiency confers resistance in tomato line SL-ORT1 to the parasitic weeds Phelipanche and Orobanche spp. Phytopathology 2011, 101, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Walter, M.H.; Giavalisco, P.; Lytovchenko, A.; Kohlen, W.; Charnikhova, T.; Simkin, A.J.; Goulet, C.; Strack, D.; Bouwmeester, H.J. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 2010, 61, 300–311. [Google Scholar] [CrossRef] [PubMed]
- López-Ráez, J.A.; Charnikhova, T.; Fernández, I.; Bouwmeester, H.; Pozo, M.J. Arbuscular mycorrhizal symbiosis decreases strigolactone production in tomato. J. Plant Physiol. 2011, 168, 294–297. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, B.; Mwakaboko, A.S.; Kannan, C. Perspective of suicidal germination for parasitic weeds control. Pest Manag. Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Westwood, J.H.; Depamphilis, C.W.; Das, M.; Fernándezaparicio, M.; Honaas, L.A.; Timko, M.P.; Wafula, E.K.; Wickett, N.J.; Yoder, J.I. The parasitic plant genome project: New tools for understanding the biology of Orobanche and Striga. Weed Sci. 2012, 60, 295–306. [Google Scholar] [CrossRef]
- Ruiu, F.; Picarella, M.; Imanishi, S.; Mazzucato, A. A transcriptomic approach to identify regulatory genes involved in fruit set of wild-type and parthenocarpic tomato genotypes. Plant Mol. Biol. 2015, 89, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Honaas, L.A.; Wafula, E.K.; Yang, Z.; Der, J.P.; Wickett, N.J.; Altman, N.S.; Taylor, C.G.; Yoder, J.I.; Timko, M.P.; Westwood, J.H. Functional genomics of a generalist parasitic plant: Laser microdissection of host-parasite interface reveals host-specific patterns of parasite gene expression. BMC Plant Biol. 2013, 13, 9. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Ichihashi, Y.; Farhi, M.; Zumstein, K.; Townsley, B.; David-Schwartz, R.; Sinha, N.R. De novo assembly and characterization of the transcriptome of the parasitic weed dodder identifies genes associated with plant parasitism. Plant Physiol. 2014, 166, 1186–1199. [Google Scholar] [CrossRef] [PubMed]
- Wickett, N.J.; Honaas, L.A.; Wafula, E.K.; Das, M.; Huang, K.; Wu, B.; Landherr, L.; Timko, M.P.; Yoder, J.; Westwood, J.H. Transcriptomes of the parasitic plant family orobanchaceae reveal surprising conservation of chlorophyll synthesis. Curr. Biol. 2011, 21, 2098–2104. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E. A comparative transcriptome analysis on growth regulation between seeds and buds of arabidopsis. Available online: https://pag.confex.com/pag/xxiii/webprogram/Paper15834.html (accessed on 13 July 2016).
- Tao, T.; Zhao, L.; Lv, Y.; Chen, J.; Hu, Y.; Zhang, T.; Zhou, B. Transcriptome sequencing and differential gene expression analysis of delayed gland morphogenesis in Gossypium australe during seed germination. PLoS ONE 2013, 8, e75323. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Guo, G.; Lv, D.; Hu, Y.; Li, J.; Li, X.; Yan, Y. Transcriptome analysis during seed germination of elite chinese bread wheat cultivar Jimai 20. BMC Plant Biol. 2014, 14, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-Length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Verdejo, C.I.; Barandiaran, X.; Moreno, M.T.; Cubero, J.I.; di Pietro, A. An improved axenic system for studying pre-infection development of the parasitic plant Orobanche ramosa. Ann. Bot.-Lond. 2005, 96, 1121–1127. [Google Scholar] [CrossRef] [PubMed]
- Kusumoto, D.; Chae, S.H.; Mukaida, K.; Yoneyama, K.; Yoneyama, K.; Joel, D.M.; Takeuchi, Y. Effects of fluridone and norflurazon on conditioning and germination of Striga asiatica seeds. Plant Growth Regul. 2006, 48, 73–78. [Google Scholar] [CrossRef]
- Joel, D.; Steffens, J.; Matthews, D. Germination of weedy root parasites. Seed Dev. Germination 1995, 1, 567–597. [Google Scholar]
- Tsuchiya, Y.; Yoshimura, M.; Sato, Y.; Kuwata, K.; Toh, S.; Holbrook-Smith, D.; Zhang, H.; McCourt, P.; Itami, K.; Kinoshita, T. Probing strigolactone receptors in Striga hermonthica with fluorescence. Science 2015, 349, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, W.; Wu, Y.; Chen, C.; Lei, J. De novo transcriptome assembly in chili pepper (Capsicum frutescens) to identify genes involved in the biosynthesis of capsaicinoids. PLoS ONE 2013, 8, e48156. [Google Scholar] [CrossRef] [PubMed]
- Nakasugi, K.; Crowhurst, R.N.; Bally, J.; Wood, C.C.; Hellens, R.P.; Waterhouse, P.M. De novo transcriptome sequence assembly and analysis of rna silencing genes of Nicotiana benthamiana. PLoS ONE 2013, 8, e59534. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C.; Zhang, Y.; Zhang, X.; Wang, Y.; Hua, W. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dal Degan, F.; Rocher, A.; Cameron-Mills, V.; Von Wettstein, D. The expression of serine carboxypeptidases during maturation and germination of the barley grain. Proc. Natl. Acad. Sci. USA 1994, 91, 8209–8213. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, R.A.; Maslyar, D.J.; Heupel, R.C.; Harada, J.J. Spatial patterns of gene expression in Brassica napus seedlings: Identification of a cortex-specific gene and localization of mrnas encoding isocitrate lyase and a polypeptide homologous to proteinases. Plant Cell 1989, 1, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.A.; Narsai, R.; Carroll, A.; Ivanova, A.; Lohse, M.; Usadel, B.; Millar, A.H.; Whelan, J. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of rna instability in the germination process. Plant Physiol. 2009, 149, 961–980. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chua, N.-H. Arabidopsis decapping 5 is required for mrna decapping, p-body formation, and translational repression during postembryonic development. Plant Cell 2009, 21, 3270–3279. [Google Scholar] [CrossRef] [PubMed]
- Hunt, L.; Holdsworth, M.J.; Gray, J.E. Nicotinamidase activity is important for germination. Plant J. 2007, 51, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Macovei, A.; Balestrazzi, A.; Confalonieri, M.; Faé, M.; Carbonera, D. New insights on the barrel medic MtOOG1 and MtFPQ functions in relation to oxidative stress response in planta and during seed imbibition. Plant Physiol. Biochem. 2011, 49, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Rajjou, L.; Gallardo, K.; Debeaujon, I.; Vandekerckhove, J.; Job, C.; Job, D. The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mrnas during germination. Plant Physiol. 2004, 134, 1598–1613. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Linneweber, C.; Small, I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008, 13, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D.; Bradford, K.J.; Hilhorst, H.W.; Nonogaki, H. Development and maturation. In Seeds; Springer: Berlin, Germany, 2013; pp. 27–83. [Google Scholar]
- Ogé, L.; Bourdais, G.; Bove, J.; Collet, B.; Godin, B.; Granier, F.; Boutin, J.-P.; Job, D.; Jullien, M.; Grappin, P. Protein repair l-isoaspartyl methyltransferase1 is involved in both seed longevity and germination vigor in Arabidopsis. Plant Cell 2008, 20, 3022–3037. [Google Scholar] [CrossRef] [PubMed]
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [PubMed]
- Wind, J.J.; Peviani, A.; Snel, B.; Hanson, J.; Smeekens, S.C. Abi4: Versatile activator and repressor. Trends Plant Sci. 2013, 18, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Guo, Q.; Gu, Z. Gaba shunt and polyamine degradation pathway on γ-aminobutyric acid accumulation in germinating fava bean (Vicia faba L.) under hypoxia. Food Chem. 2013, 136, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Linkies, A.; Leubner-Metzger, G. Beyond gibberellins and abscisic acid: How ethylene and jasmonates control seed germination. Plant Cell Rep. 2012, 31, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Chae, S.H.; Yoneyama, K.; Takeuchi, Y.; Joel, D.M. Fluridone and norflurazon, carotenoid-biosynthesis inhibitors, promote seed conditioning and germination of the holoparasite Orobanche minor. Physiol. Plant. 2004, 120, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Omigawa, Y.; Ogasawara, M.; Yoneyama, K.; Konnai, M.; Worsham, A.D. Effects of brassinosteroids on conditioning and germination of clover broomrape (Orobanche minor) seeds. Plant Growth Regul. 1995, 16, 153–160. [Google Scholar] [CrossRef]
- Zehhar, N.; Ingouff, M.; Bouya, D.; Fer, A. Possible involvement of gibberellins and ethylene in Orobanche ramosa germination. Weed Res. 2002, 42, 464–469. [Google Scholar] [CrossRef]
- Lechat, M.-M.; Pouvreau, J.-B.; Péron, T.; Gauthier, M.; Montiel, G.; Véronési, C.; Todoroki, Y.; Le Bizec, B.; Monteau, F.; Macherel, D. PrCYP707A1, an aba catabolic gene, is a key component of Phelipanche ramosa seed germination in response to the strigolactone analogue GR24. J. Exp. Bot. 2012, 63, 5311–5322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Lee, K.; Seo, P. The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 2015, 87, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Lechat, M.-M.; Brun, G.; Montiel, G.; Véronési, C.; Simier, P.; Thoiron, S.; Pouvreau, J.-B.; Delavault, P. Seed response to strigolactone is controlled by abscisic acid-independent DNA methylation in the obligate root parasitic plant, Phelipanche ramosa L. Pomel. J. Exp. Bot. 2015, 66, 1329–1340. [Google Scholar] [CrossRef] [PubMed]
- Matusova, R.; Rani, K.; Verstappen, F.W.; Franssen, M.C.; Beale, M.H.; Bouwmeester, H.J. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. Are derived from the carotenoid pathway. Plant Physiol. 2005, 139, 920–934. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Zhou, W.; Jin, Z.; Cao, D.; Joel, D.; Takeuchi, Y.; Yoneyama, K. Germination response of Orobanche seeds subjected to conditioning temperature, water potential and growth regulator treatments. Weed Res. 2005, 45, 467–476. [Google Scholar] [CrossRef]
- Song, W.; Zhou, W.; Jin, Z.; Zhang, D.; Yoneyama, K.; Takeuchi, Y.; Joel, D. Growth regulators restore germination of Orobanche seeds that are conditioned under water stress and suboptimal temperature. Crop Pasture Sci. 2006, 57, 1195–1201. [Google Scholar] [CrossRef]
- Joel, D.; Back, A.; Kleifeld, Y.; Gepstein, S. Seed conditioning and its role in orobanche seed germination: Inhibition by paclobutrazol. In Progress in Orobanche Research, Proceedings of the International Workshop on Orobanche Research, Obermarchtal, Germany, 19–22 August 1989.
- Uematsu, K.; Nakajima, M.; Yamaguchi, I.; Yoneyama, K.; Fukui, Y. Role of camp in gibberellin promotion of seed germination in Orobanche minor Smith. J. Plant Growth Regul. 2007, 26, 245–254. [Google Scholar] [CrossRef]
- Krizek, D.T.; Mandava, N.B. Influence of spectral quality on the growth response of intact bean plants to brassinosteroid, a growth-promoting steroidal lactone. I. Stem elongation and morphogenesis. Physiol. Plant. 1983, 57, 317–323. [Google Scholar] [CrossRef]
- Graeber, K.; Linkies, A.; Müller, K.; Wunchova, A.; Rott, A.; Leubner-Metzger, G. Cross-Species approaches to seed dormancy and germination: Conservation and biodiversity of aba-regulated mechanisms and the brassicaceae DOG1 genes. Plant Mol. Biol. 2010, 73, 67–87. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.; Linkies, A.; Müller, K.; Oracz, K.; Wang, X.; Lynn, J.R.; Leubner-Metzger, G.; Finch-Savage, W.E. Regulation of seed germination in the close arabidopsis relative Lepidium sativum: A global tissue-specific transcript analysis. Plant Physiol. 2011, 155, 1851–1870. [Google Scholar] [CrossRef] [PubMed]
- Kucera, B.; Cohn, M.A.; Leubner-Metzger, G. Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 2005, 15, 281–307. [Google Scholar] [CrossRef]
- Linkies, A.; Müller, K.; Morris, K.; Turečková, V.; Wenk, M.; Cadman, C.S.; Corbineau, F.; Strnad, M.; Lynn, J.R.; Finch-Savage, W.E. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: A comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 2009, 21, 3803–3822. [Google Scholar] [CrossRef] [PubMed]
- Matilla, A.; Matilla-Vazquez, M. Involvement of ethylene in seed physiology. Plant Sci. 2008, 175, 87–97. [Google Scholar] [CrossRef]
- Chun, D.; Wilhelm, S.; Sagen, J.; Musselman, L.; Worsham, A.; Eplee, R. Components of record germination in vitro of branched broomrape, orobanche ramosa L. In Supplement to the Proceedings of the Second International Symposium on Parasitic Weeds, Raleigh, NC, USA, 16–19 July 1979.
- Bebawi, F.F.; Eplee, R.E. Efficacy of ethylene as a germination stimulant of Striga hermonthica seed. Weed Sci. 1986, 34, 694–698. [Google Scholar]
- Egley, G.; Dale, J. Ethylene, 2-chloroethylphosphonic acid, and witchweed germination. Weed Sci. 1970, 18, 586–589. [Google Scholar]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.; Holbrook-Smith, D.; Stogios, P.J.; Onopriyenko, O.; Lumba, S.; Tsuchiya, Y.; Savchenko, A.; McCourt, P. Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 2015, 350, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Westwood, J.H. Characterization of the Orobanche–Arabidopsis system for studying parasite–host interactions. Weed Sci. 2009, 48, 742–748. [Google Scholar]
- Fernández-Aparicio, M.; Rubiales, D.; Bandaranayake, P.C.; Yoder, J.I.; Westwood, J.H. Transformation and regeneration of the holoparasitic plant Phelipanche aegyptiaca. Plant Methods 2011, 7, 683–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.-J.; Chen, Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2013, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J. topGo: Enrichment analysis for gene ontology. Available online: http://bioconductor.uib.no/2.7/bioc/html/topGO.html (accessed on 13 July 2016).
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. Kaas: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed]
- Leng, N.; Dawson, J.; Kendziorski, C. Ebseq: An R package for gene and isoform differential expression analysis of rna-seq data, R package version 1.5.3. Available online: https://www.biostat.wisc.edu/~kendzior/EBSEQ/ (accessed on 13 July 2016).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.A.; Sinha, R.; Schlauch, D.; Quackenbush, J. Rna-seq analysis in mev. Bioinformatics 2011, 27, 3209–3210. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, A.J. Java treeview—extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Verdejo, C.; Die, J.; Nadal, S.; Jimenez-Marin, A.; Moreno, M.; Roman, B. Selection of housekeeping genes for normalization by real-time RT-PCR: Analysis of Or-MYB1 gene expression in Orobanche ramosa development. Anal. Biochem. 2008, 379, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Durgbanshi, A.; Arbona, V.; Pozo, O.; Miersch, O.; Sancho, J.V.; Gómez-Cadenas, A. Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography-electrospray tandem mass spectrometry. J. Agric. Food Chem. 2005, 53, 8437–8442. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-W.; Hwang, J.-Y.; Kim, Y.-S.; Joo, S.-H.; Chang, S.C.; Lee, J.S.; Takatsuto, S.; Kim, S.-K. Arabidopsis CYP85A2, a cytochrome P450, mediates the baeyer-villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 2005, 17, 2397–2412. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Xu, M.; Wu, L.; Shen, C.; Ma, H.; Lin, J. CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacum by antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol. Biol. 2014, 85, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, S.M.; Woltering, E.; Hermans, C.; Harren, F.J.; te Lintel Hekkert, S. Research tools: Ethylene detection. In Ethylene in Plants; Springer: Berlin, Germany, 2015; pp. 263–286. [Google Scholar]
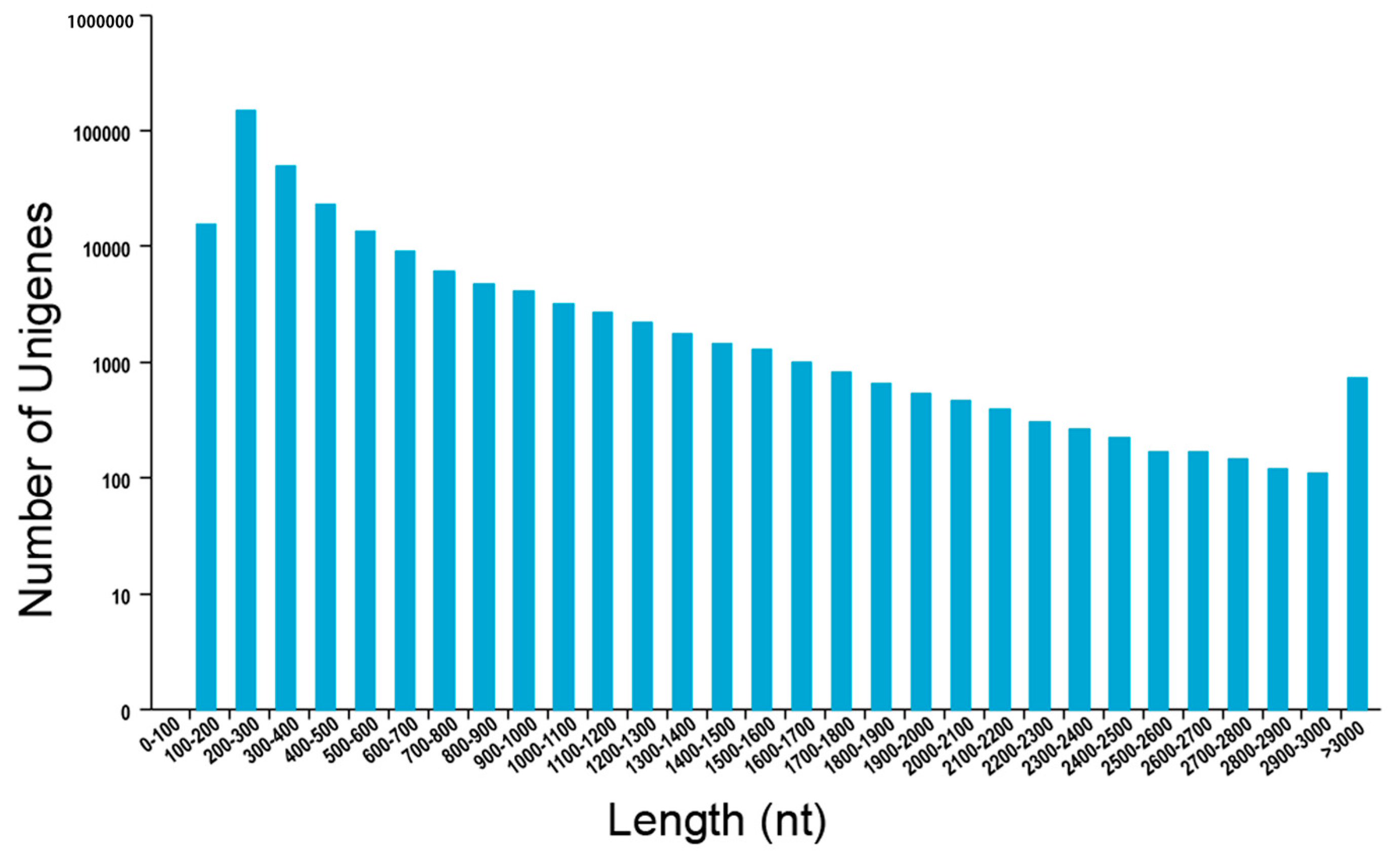



| Length Range | Contig | Transcript | Unigene |
|---|---|---|---|
| Total number | 11,469,131 | 274,964 | 94,419 |
| Total length (bp) | 670,369,345 | 293,096,156 | 66,808,988 |
| N50 (bp) | 53 | 1558 | 836 |
| Average (bp) | 58.45 | 1065.94 | 707.58 |
| Treatment | Germination (%) | ||
|---|---|---|---|
| No Conditioned | Conditioned | Conditioned and GR24 Treatment | |
| BR | 0 a | 0 a | 83.3 ± 3.8 fg |
| GA3 | 0 a | 2.2 ± 2.2 a | 94.4 ± 1.1 h |
| ABA | 0 a | 0 a | 0 a |
| Ethephon | 0 a | 0 a | 90.0 ± 1.9 gh |
| Fluridone | 26.5 ± 1.8 b | 7.8 ± 2.9 a | 86.7 ± 5.7 gh |
| GA3 + Fluridone | 35.6 ± 1.1 c | 10.0 ± 3.3 a | 91.1 ± 2.2 gh |
| Ethephon + Fluridone | 24.4 ± 2.2 b | 7.8 ± 1.1 a | 80.0 ± 5.0 f |
| GA3 + Ethephon + Fluridone | 47.8 ± 2.9 e | 20.0 ± 1.9 b | 91.1 ± 4.0 gh |
| Water | 0 a | 0 a | 75.6 ± 9.1 f |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Z.; Tian, F.; Cao, X.; Xu, Y.; Chen, M.; Xiang, B.; Zhao, S. Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche aegyptiaca Seed Germination. Int. J. Mol. Sci. 2016, 17, 1139. https://doi.org/10.3390/ijms17071139
Yao Z, Tian F, Cao X, Xu Y, Chen M, Xiang B, Zhao S. Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche aegyptiaca Seed Germination. International Journal of Molecular Sciences. 2016; 17(7):1139. https://doi.org/10.3390/ijms17071139
Chicago/Turabian StyleYao, Zhaoqun, Fang Tian, Xiaolei Cao, Ying Xu, Meixiu Chen, Benchun Xiang, and Sifeng Zhao. 2016. "Global Transcriptomic Analysis Reveals the Mechanism of Phelipanche aegyptiaca Seed Germination" International Journal of Molecular Sciences 17, no. 7: 1139. https://doi.org/10.3390/ijms17071139