Mitochondrial Protein Profile in Mice with Low or Excessive Selenium Diets
Abstract
:1. Introduction
2. Results
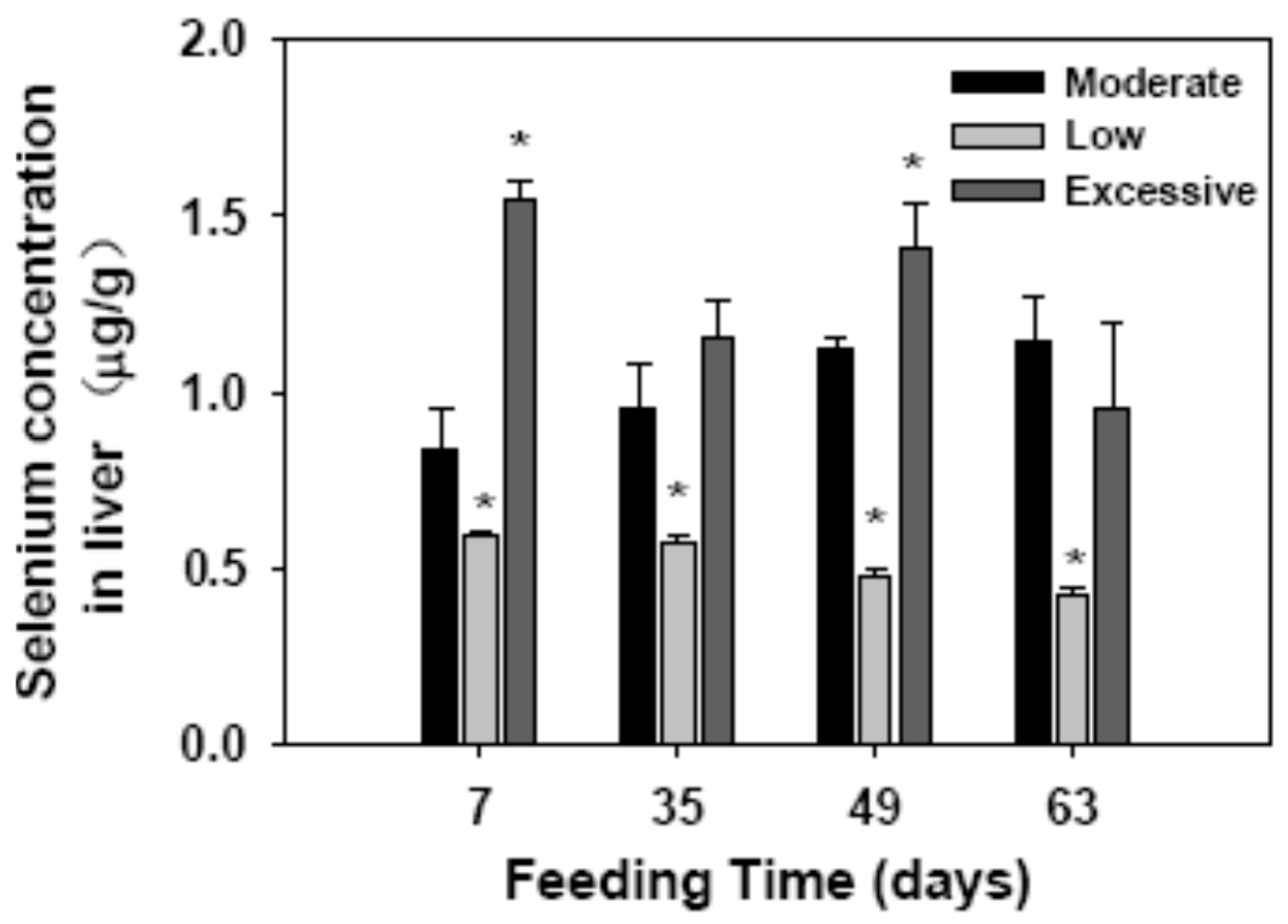
2.1. Selenium Concentration in Liver
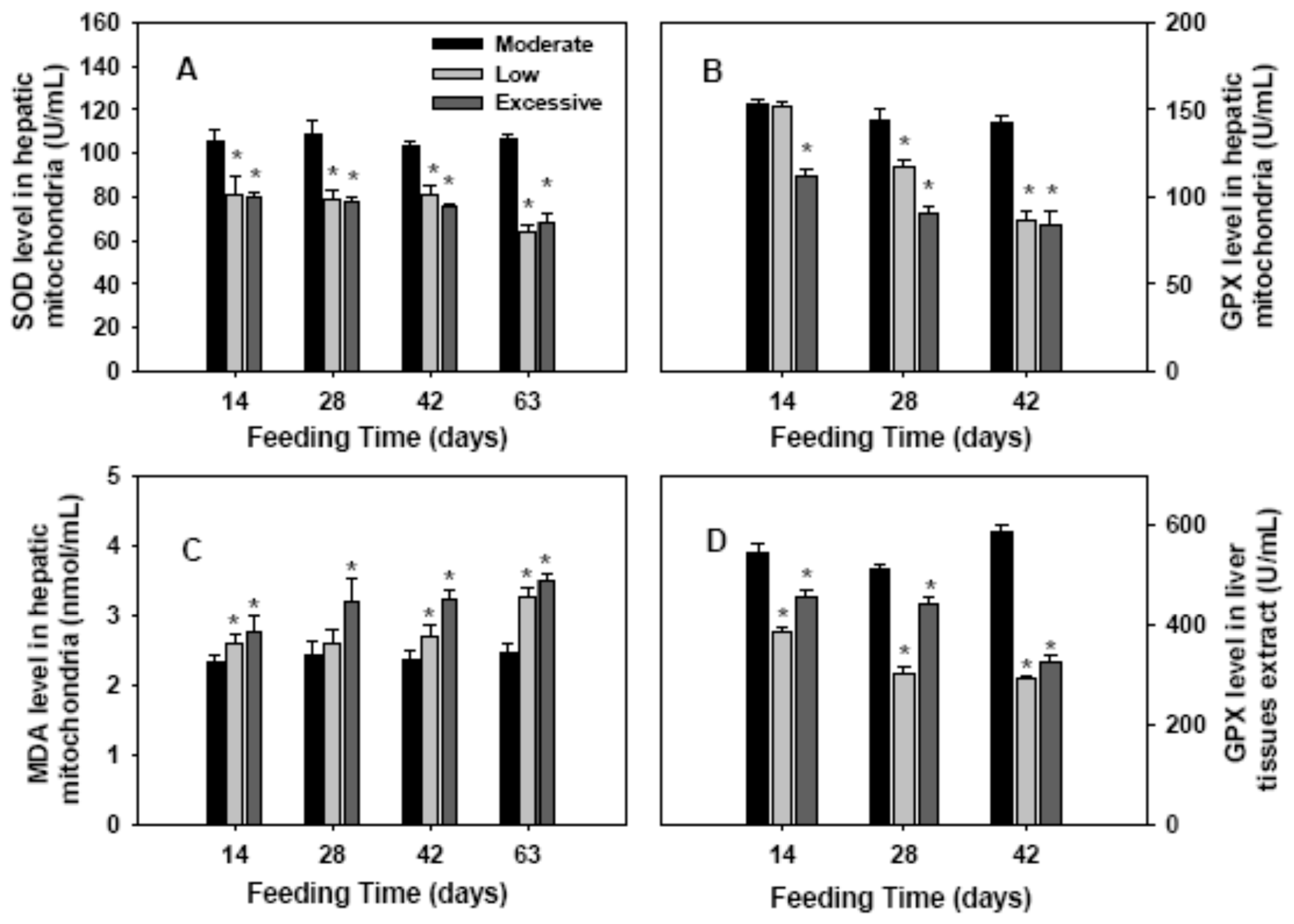
2.2. Antioxidant Enzymes Activities and Lipid Perioxidation in Hepatic Mitochondria
2.3. Differentially-Expressed Hepatic Mitochondria Proteins in Different Groups of Mice
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Establishing a Mouse Model
4.3. Determination of the Selenium Concentration in Liver
4.4. Sample Preparation and Biochemical Analyses
4.5. Isolation of Hepatic Mitochondria
4.6. Sample Preparation for Two-Dimensional Gel Electrophoresis
4.7. Electrophoresis and Spot Detection
4.8. Mass Spectrum Analyses
4.9. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 17β-HSD | 17β hydroxysteroid dehydrogenase |
| ALT | alanine aminotransferase |
| AST | aspartate aminotransferase |
| ATPsyn-d | ATP synthase subunit d |
| ATPsyn-α | ATP synthase subunit α |
| CPS1 | carbamoyl phosphate synthetase 1 |
| EF2 | elongation factor 2 |
| GPX | glutathione peroxidase |
| HSP90 | heat shock protein 90 kDa |
| LC-MS/MS | liquid chromatography mass spectrometry |
| MALDI-TOF | matrix-assisted laser desorption/ionization tandem time-of-flight |
| MDA | malondialdehyde |
| PMSF | phenylmethanesulfonyl fluoride |
| Se | Selenium |
| SOD | superoxide dismutase |
| SUCLA2 | subunit β of mitochondrial succinyl-CoA ligase |
References
- Kieliszek, M.; Blazejak, S. Selenium: Significance, and outlook for supplementation. Nutrition 2013, 29, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohe, R.; Maiorino, M. Glutathione peroxidase. Biochim. Biophys. Acta. 2013, 30, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Weeks, B.S.; Hanna, M.S.; Cooperstein, D. Dietary selenium and selenoprotein function. Med. Sci. Monit. 2012, 18, RA127–RA132. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, V.; Tomblin, J.; Yeager Armistead, M.; Murray, E. Selenium (sodium selenite) causes cytotoxicity and apoptotic mediated cell death in PLHC-1 fish cell line through DNA and mitochondrial membrane potential damage. Ecotoxicol. Environ. Saf. 2013, 87, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.J.; Rathinasabapathi, B.; Wu, B.; Luo, J.; Pu, L.P.; Ma, L.Q. Arsenic and selenium toxicity and their interactive effects in humans. Environ. Int. 2014, 69, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Yang, C.; Liu, J.; Ong, C. Dual role of glutathione in selenite-induced oxidative stress and apoptosis in human hepatoma cells. Free Radic. Biol. Med. 2000, 28, 1115–1124. [Google Scholar] [CrossRef]
- Kim, T.S.; Yun, B.Y.; Kim, I.Y. Induction of the mitochondrial permeability transition by selenium compounds mediated by oxidation of the protein thiol groups and generation of the superoxide. Biochem. Pharmacol. 2003, 66, 2301–2311. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.F.; Hill, K.E. Regulation of selenium metabolism and transport. Annu. Rev. Nutr. 2015, 35, 109–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, W.; Chen, H.; Liao, N.; Wang, Z.; Zhang, X.; Hai, C. High selenium impairs hepatic insulin sensitivity through opposite regulation of ROS. Toxicol. Lett. 2014, 224, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Guo, S.; Liao, S.; Su, R.; Pan, J.; Lin, Y.; Tang, Z. Influence of selenium on hepatic mitochondrial antioxidant capacity in ducklings intoxicated with aflatoxin B1. Biol. Trace Element Res. 2012, 145, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Saied, N.M.; Hamza, A.A. Selenium ameliorates isotretinoin-induced liver injury and dyslipidemia via antioxidant effect in rats. Toxicol. Mech. Methods 2014, 24, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Liao, S.; Guo, S.; Li, H.; Yang, M.; Tang, Z. Protective effects of selenium on aflatoxin B1-induced mitochondrial permeability transition, DNA damage, and histological alterations in duckling liver. Biol. Trace Element Res. 2015, 163, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Taskin, E.; Dursun, N. Recovery of adriamycin induced mitochondrial dysfunction in liver by selenium. Cytotechnology 2015, 67, 977–986. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Cao, H.; Pan, J.; Li, C.; Chen, Y.; Tang, Z. The Protective Roles of Selenium on Hepatic Tissue Ultrastructure and Mitochondrial Antioxidant Capacity in Copper-Overloaded Rats. Biol. Trace Element Res. 2015, 167, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, J.; Guo, Y.; Ye, H.; Yu, C.; Xu, C.; Xu, L.; Wu, S.; Sun, W.; Wei, H.; et al. Functional proteomic analysis of nonalcoholic fatty liver disease in rat models: Enoyl-coenzyme a hydratase down-regulation exacerbates hepatic steatosis. Hepatology 2010, 51, 1190–1199. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lu, D.Z.; Li, Y.M.; Zhang, X.Q.; Zhou, X.X.; Jin, X. Proteomic analysis of liver mitochondria from rats with nonalcoholic steatohepatitis. World J. Gastroenterol. 2014, 20, 4778–4786. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, A.; Alvarez-Vijande, R.; Capdevila, S.; Alcoberro, J.; Alcaraz, A. Antioxidant patterns (superoxide dismutase, glutathione reductase, and glutathione peroxidase) in kidneys from non-heart-beating-donors: Experimental study. Transplant. Proc. 2007, 39, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Liu, Q.; Xu, H.B.; Zhu, Y.S.; Yang, X.L. Effects of Selenium overexposure on glutathione peroxidase and thioredoxin reductase gene expressions and activity. Biol. Trace Element Res. 2002, 89, 165–175. [Google Scholar] [CrossRef]
- Krohn, R.M.; Lemaire, M.; Silva, L.F.N.; Lemarie, C.; Bolt, A.; Mann, K.K.; Smits, J.E. High-selenium lentil diet protects against arsenic-induced atherosclerosis in a mouse model. J. Nutr. Biochem. 2016, 27, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.R.; Huang, Y.H.; Wang, G.X.; Wu, Y.X.; Xian, J.A.; Wang, A.L.; Cao, J.M. Deficient and excess dietary selenium levels affect growth performance, blood cells apoptosis and liver HSP70 expression in juvenile yellow catfish Pelteobagrus fulvidraco. Fish Physiol. Biochem. 2016, 42, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Touat-Hamici, Z.; Legrain, Y.; Bulteau, A.L.; Chavatte, L. Selective up-regulation of human selenoproteins in response to oxidative stress. J. Biol. Chem. 2014, 289, 14750–14761. [Google Scholar] [CrossRef] [PubMed]
- Kaul, G.; Pattan, G.; Rafeequi, T. Eukaryotic elongation factor-2 (eEF2): Its regulation and peptide chain elongation. Cell Biochem. Funct. 2011, 29, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Arguelles, S.; Camandola, S.; Hutchison, E.R.; Cutler, R.G.; Ayala, A.; Mattson, M.P. Molecular control of the amount, subcellular location, and activity state of translation elongation factor 2 in neurons experiencing stress. Free Radic. Biol. Med. 2013, 61, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Prehn, C.; Moller, G.; Adamski, J. Recent advances in 17β-hydroxysteroid dehydrogenases. J. Steroid Biochem. Mol. Biol. 2009, 114, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Wang, Y.; Jia, X.; Wu, W.; Li, L.; Tian, X.; Li, S.; Wang, C.; Xu, H.; Cao, J.; et al. Comparative proteomic study reveals 17β-HSD13 as a pathogenic protein in nonalcoholic fatty liver disease. Proc. Natl. Acad. Sci. USA 2014, 111, 11437–11442. [Google Scholar] [CrossRef] [PubMed]
- Hekman, C.; Tomich, J.M.; Hatefi, Y. Mitochondrial ATP synthase complex. Membrane topography and stoichiometry of the F0 subunits. J. Biol. Chem. 1991, 266, 13564–13571. [Google Scholar] [PubMed]
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione peroxidase-1 in health and disease: From molecular mechanisms to herapeutic opportunities. Antioxid. Redox Signal. 2011, 15, 1957–1997. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Li, D.; Alesi, G.N.; Fan, J.; Kang, H.B.; Lu, Z.; Boggon, T.J.; Jin, P.; Yi, H.; Wright, E.R.; et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell 2015, 27, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Valencia, A.P.; Schappal, A.E.; Matthew Morris, E.; Thyfault, J.P.; Lowe, D.A.; Spangenburg, E.E. The presence of the ovary prevents hepatic mitochondrial oxidative stress in young and aged female mice through glutathione peroxidase 1. Exp. Gerontol. 2016, 73, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.N.; Jeong, J.H.; Kim, D.J.; Chung, Y.H.; Shin, E.J.; Nguyen, L.T.; Nam, Y.; Lee, Y.J.; Cho, E.H.; Nah, S.Y.; et al. Genetic overexpressing of GPx-1 attenuates cocaine-induced renal toxicity via induction of anti-apoptotic factors. Clin. Exp. Pharmacol. Physiol. 2016, 43, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Huang, X.; Wang, L.; Li, Q.; Xu, J.; Jia, G.; Liu, G.; Chen, X.; Shang, H.; Zhao, H. Supranutritional dietary selenium depressed expression of selenoprotein genes in three immune organs of broilers. Anim. Sci. J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Alam, Q.; Alam, M.Z.; Azhar, E.I.; Sait, K.H.; Anfina, N.; Mushtaq, G.; Kamal, M.A.; Rasool, M. Current understanding of HSP90 as a novel therapeutic target: An emerging approach for the treatment of cancer. Curr. Pharm. Des. 2016, in press. [Google Scholar] [CrossRef]
- Castro, J.P.; Reeg, S.; Botelho, V.; Almeida, H.; Grune, T. HSP90 cleavage associates with oxidized proteins accumulation after oxidative stress. Free Radic. Biol. Med. 2014, 75 (Suppl. 1), S24–S25. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.; Aponte, A.M.; French, S.A.; Chess, D.J.; Balaban, R.S. Succinyl-CoA synthetase is a phosphate target for the activation of mitochondrial metabolism. Biochemistry 2009, 48, 7140–7149. [Google Scholar] [CrossRef] [PubMed]
- Ostergaard, E. Disorders caused by deficiency of succinate-CoA ligase. J. Inherit. Metab. Dis. 2008, 31, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Preuss, B.; Berg, C.; Dengjel, J.; Stevanovic, S.; Klein, R. Relevance of the inner mitochondrial membrane enzyme F1F0-ATPase as an autoantigen in autoimmune liver disorders. Liver Int. 2012, 32, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Christoffels, V.M.; Habets, P.E.; Das, A.T.; Clout, D.E.; van Roon, M.A.; Moorman, A.F.; Lamers, W.H. A single regulatory module of the carbamoylphosphate synthetase I gene executes its hepatic program of expression. J. Biol. Chem. 2000, 275, 40020–40027. [Google Scholar] [CrossRef] [PubMed]
- Reeves, P.G.; Nielsen, F.H.; Fahey, G.C. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993, 123, 1939–1951. [Google Scholar] [PubMed]
- Tang, Z.; Iqbal, M.; Cawthon, D.; Bottje, W.G. Heart and breast muscle mitochondrial dysfunction in pulmonary hypertension syndrome in broilers (Gallus domesticus). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2002, 132, 527–540. [Google Scholar] [CrossRef]

| Spot | Description | Accession | Score | Mr | Sequence Coverage | pI | Expression * |
|---|---|---|---|---|---|---|---|
| A04 | mCG116926 | gi|148671376 | 80 | 35,753 | 24 | 7.72 | ↑ |
| A05 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit d | gi|159572410 | 76 | 15,929 | 37 | 5.16 | ↑ |
| A07 | Hydroxysteroid (17-β) dehydrogenase 4 | gi|148677986 | 107 | 32,826 | 17 | 8.76 | ↑ |
| B01 | Eukaryotic elongation factor 2 | gi|192989 | 137 | 30,212 | 30 | 6.2 | ↓ |
| Spot | Description | Accession | Score | Mr | Sequence Coverage | pI | Expression * |
|---|---|---|---|---|---|---|---|
| E03 | HSP90 | gi|6755863 | 65 | 92,703 | 12 | 4.74 | ↓ |
| E06 | SUCLA2 | gi|28175163 | 147 | 38,384 | 8 | 5.05 | ↓ |
| F10 | CPS1 | gi|187466221 | 101 | 128,299 | 5 | 6.22 | ↑ |
| F13 | mCG121563 | gi|148667830 | 208 | 158,788 | 5 | 6.59 | ↑ |
| F26 | GPX1 | gi|84871986 | 137 | 22,544 | 27 | 6.74 | ↓ |
| F27 | ATP synthase, H+ transporting, mitochondrial F1 complex, α subunit | gi|148677503 | 203 | 30,708 | 35 | 8.98 | ↑ |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Wang, C.; Zhang, Q.; Yan, H.; Li, Y.; Pan, J.; Tang, Z. Mitochondrial Protein Profile in Mice with Low or Excessive Selenium Diets. Int. J. Mol. Sci. 2016, 17, 1137. https://doi.org/10.3390/ijms17071137
Hu L, Wang C, Zhang Q, Yan H, Li Y, Pan J, Tang Z. Mitochondrial Protein Profile in Mice with Low or Excessive Selenium Diets. International Journal of Molecular Sciences. 2016; 17(7):1137. https://doi.org/10.3390/ijms17071137
Chicago/Turabian StyleHu, Lianmei, Congcong Wang, Qin Zhang, Hao Yan, Ying Li, Jiaqiang Pan, and Zhaoxin Tang. 2016. "Mitochondrial Protein Profile in Mice with Low or Excessive Selenium Diets" International Journal of Molecular Sciences 17, no. 7: 1137. https://doi.org/10.3390/ijms17071137
APA StyleHu, L., Wang, C., Zhang, Q., Yan, H., Li, Y., Pan, J., & Tang, Z. (2016). Mitochondrial Protein Profile in Mice with Low or Excessive Selenium Diets. International Journal of Molecular Sciences, 17(7), 1137. https://doi.org/10.3390/ijms17071137