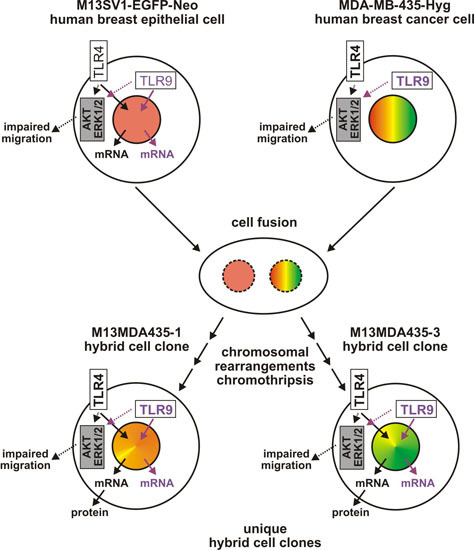
Hybrid Cells Derived from Human Breast Cancer Cells and Human Breast Epithelial Cells Exhibit Differential TLR4 and TLR9 Signaling
Abstract
:1. Introduction
2. Results
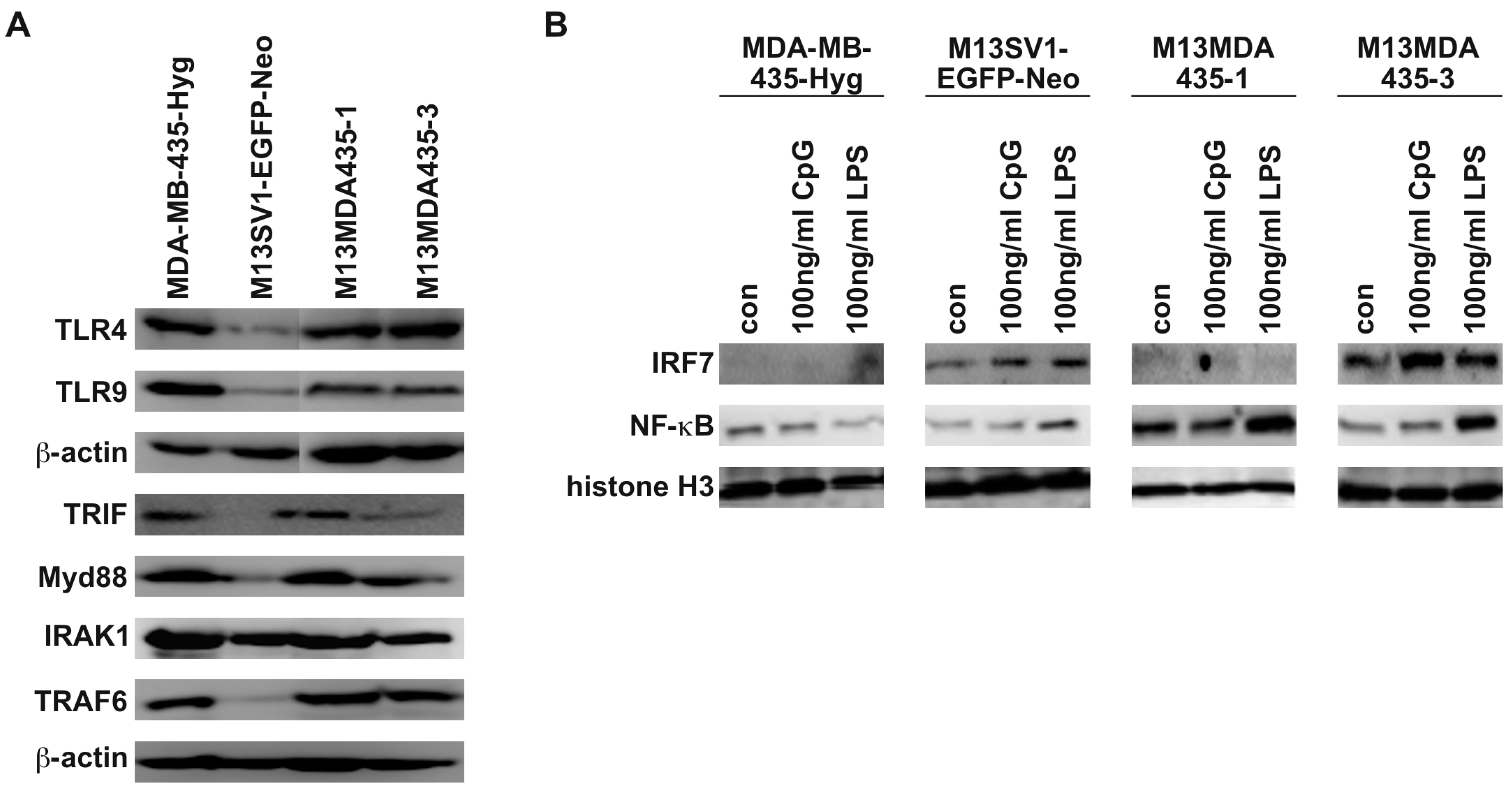
2.1. M13MDA435 Hybrid Cells Respond Differently to CpG-ODN and LPS Stimulation
2.2. M13MDA435 Hybrid Cells and Parental Cells Respond Differently to LPS and CpG-ODN Stimulation
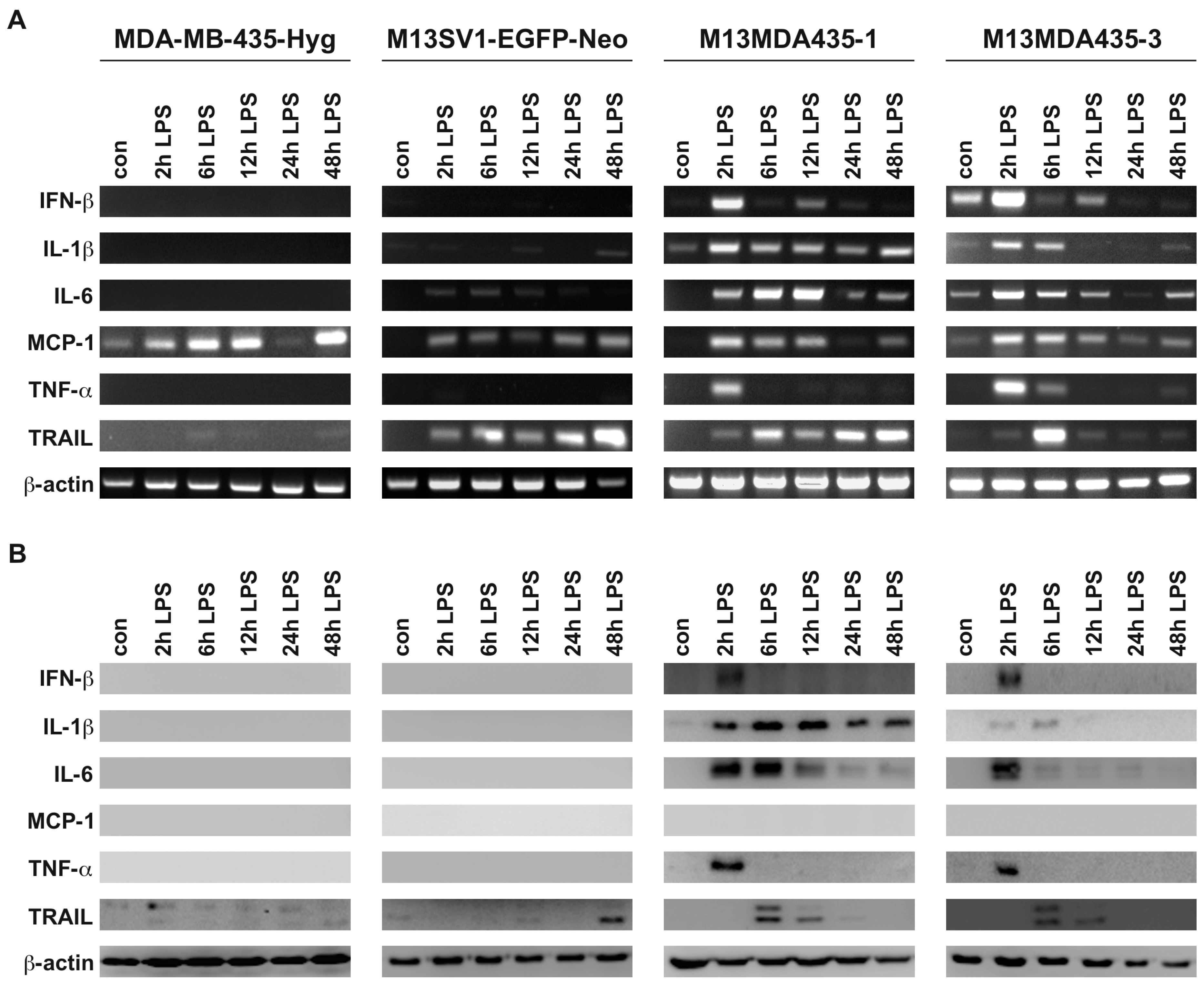
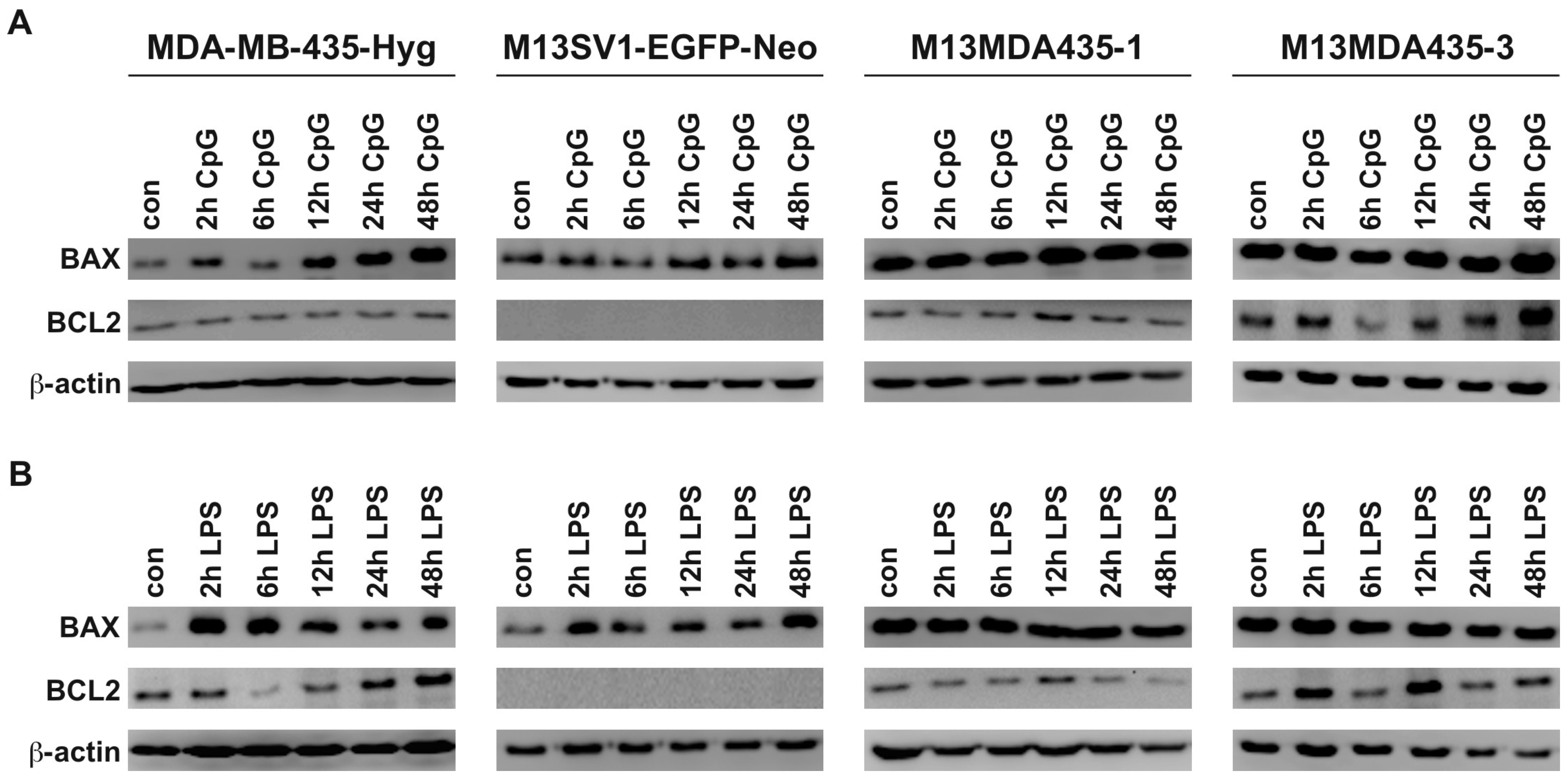
2.3. Expression of Pro-Inflammatory and Apoptosis-Inducing Cytokines in Hybrid Cells Does Not Correlate with BAX and BCL-2 Expression Levels
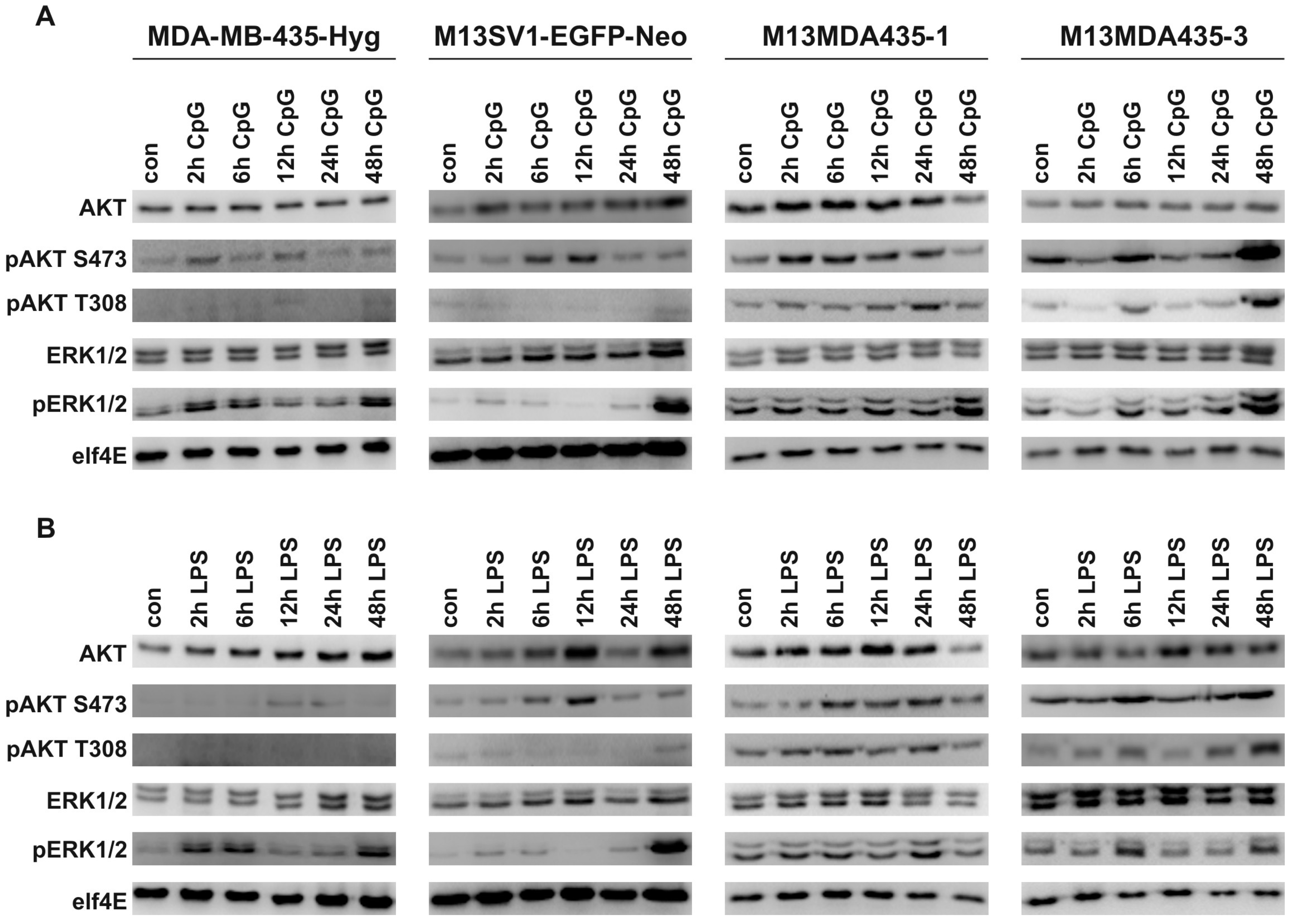
2.4. CpG-ODN and LPS Both Induce AKT and ERK1/2 Signaling in Parental Cells and Hybrid Cells
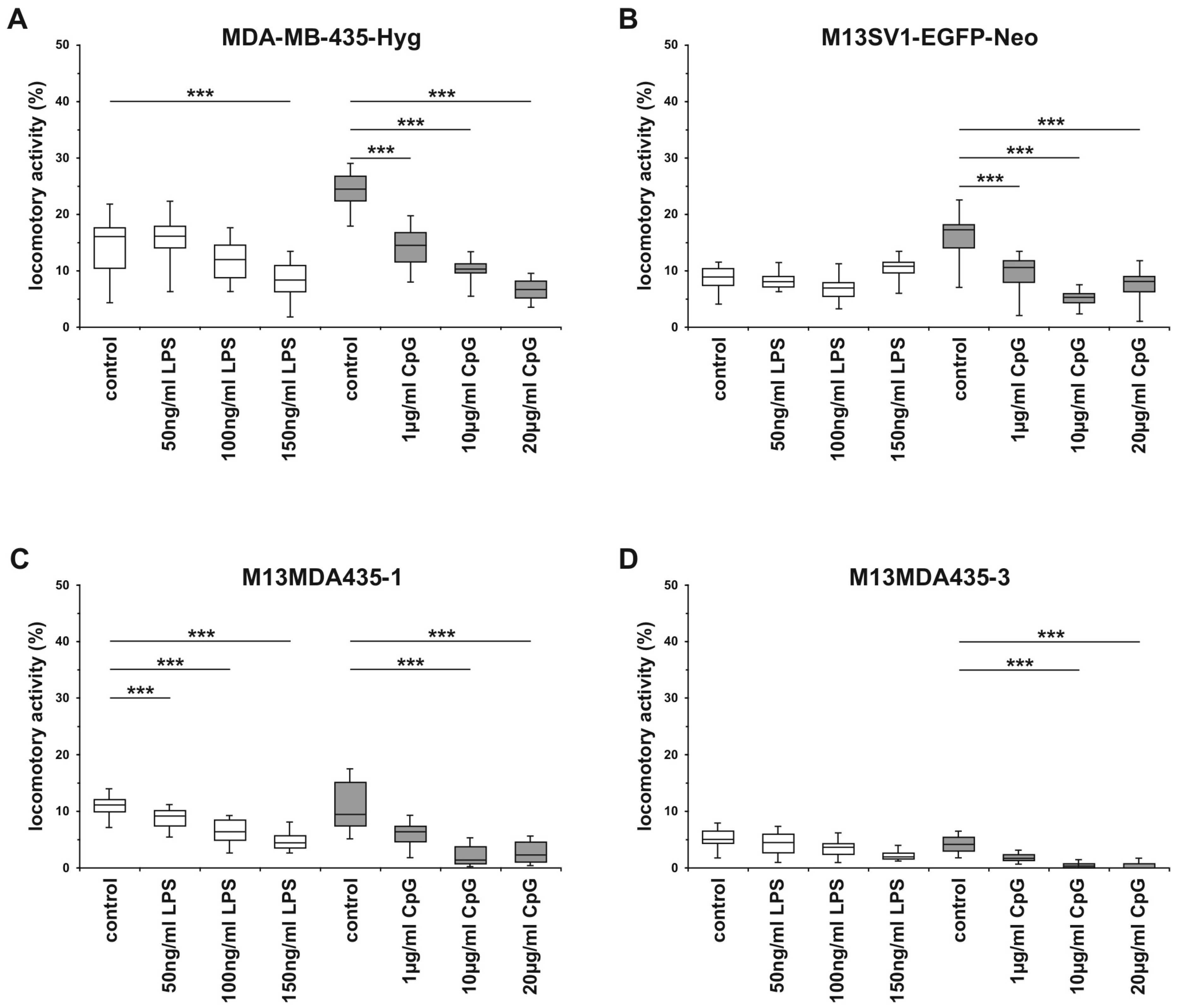
2.5. The Migratory Activity of Parental Cells and Hybrid Cells is Impaired by CpG-ODN in a Dose-Dependent Manner
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. RT-PCR
4.3. Extraction of Nuclear Proteins
4.4. Western Blot Analysis
4.5. 3D-Collagen Matrix Migration Assay
4.6. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CpG-ODN | CpG-oligodeoxynucleotide |
| DAMPs | danger-associated recognition patterns |
| HST | heterokaryon-to-synkaryon transition |
| lncRNA | long non-coding RNA |
| LPS | lipopolysaccharide |
| miRNA | microRNA |
| mTORC2 | mechanistic target of Rapamycin complex 2 |
| PAMPs | pathogen-associated recognition patterns |
| PDK1 | phosphoinositide-dependent kinase-1 |
| PI3K | phosphatidylinositol 3-kinase |
| PIP3 | phosphatidylinositol-3,4,5-triphosphate |
| TLRs | toll-like receptors |
References
- Akira, S.; Hemmi, H. Recognition of pathogen-associated molecular patterns by tlr family. Immunol. Lett. 2003, 85, 85–95. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- O'Neill, L.A.; Golenbock, D.; Bowie, A.G. The history of toll-like receptors—Redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, S.L.M.; Smits, A.I.P.M.; Driessen-Mol, A.; Baaijens, F.P.T.; Bouten, C.V.C. The immune response in in situ tissue engineering of aortic heart valves. In Calcific aortic Valve Disease; Aikawa, E., Ed.; InTech: Rijeka, Croatia, 2013. [Google Scholar]
- Goh, F.G.; Midwood, K.S. Intrinsic danger: Activation of toll-like receptors in rheumatoid arthritis. Rheumatology (Oxford) 2012, 51, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Singh, S.; Anang, V.; Bhatt, A.N.; Natarajan, K.; Dwarakanath, B.S. Pattern recognition receptors in cancer progression and metastasis. Cancer Growth Metastasis 2015, 8, 25–34. [Google Scholar] [PubMed]
- Dvorak, H.F. Tumors: Wounds that do not heal. Similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 1986, 315, 1650–1659. [Google Scholar] [PubMed]
- Carpentier, A.; Metellus, P.; Ursu, R.; Zohar, S.; Lafitte, F.; Barrie, M.; Meng, Y.; Richard, M.; Parizot, C.; Laigle-Donadey, F.; et al. Intracerebral administration of CpG oligonucleotide for patients with recurrent glioblastoma: A phase II study. Neuro Oncol. 2010, 12, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Chicoine, M.R.; Zahner, M.; Won, E.K.; Kalra, R.R.; Kitamura, T.; Perry, A.; Higashikubo, R. The in vivo antitumoral effects of lipopolysaccharide against glioblastoma multiforme are mediated in part by toll-like receptor 4. Neurosurgery 2007, 60, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Otto, F.; Schmid, P.; Mackensen, A.; Wehr, U.; Seiz, A.; Braun, M.; Galanos, C.; Mertelsmann, R.; Engelhardt, R. Phase ii trial of intravenous endotoxin in patients with colorectal and non-small cell lung cancer. Eur. J. Cancer 1996, 32A, 1712–1718. [Google Scholar] [CrossRef]
- Zhang, Y.; Lin, A.; Sui, Q.; Zhang, C.; Tian, Z.; Zhang, J. Phosphorothioate modification of the TLR9 ligand CpG odn inhibits poly(I:C)-induced apoptosis of hepatocellular carcinoma by entry blockade. Cancer Lett. 2014, 355, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Tian, Z.; Das, D.S.; Coffman, R.L.; Richardson, P.; Chauhan, D.; Anderson, K.C. A novel TLR-9 agonist C792 inhibits plasmacytoid dendritic cell-induced myeloma cell growth and enhance cytotoxicity of bortezomib. Leukemia 2014, 28, 1716–1724. [Google Scholar] [CrossRef] [PubMed]
- Harmey, J.H.; Bucana, C.D.; Lu, W.; Byrne, A.M.; McDonnell, S.; Lynch, C.; Bouchier-Hayes, D.; Dong, Z. Lipopolysaccharide-induced metastatic growth is associated with increased angiogenesis, vascular permeability and tumor cell invasion. Int. J. Cancer 2002, 101, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Pidgeon, G.P.; Harmey, J.H.; Kay, E.; Da Costa, M.; Redmond, H.P.; Bouchier-Hayes, D.J. The role of endotoxin/lipopolysaccharide in surgically induced tumour growth in a murine model of metastatic disease. Br. J. Cancer 1999, 81, 1311–1317. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, B.; Wang, T.; Xu, L.; He, C.; Wen, H.; Yan, J.; Su, H.; Zhu, X. Toll-like receptor 4 prompts human breast cancer cells invasiveness via lipopolysaccharide stimulation and is overexpressed in patients with lymph node metastasis. PLoS ONE 2014, 9, e109980. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.G.; Alvero, A.B.; Chen, R.; Silasi, D.A.; Abrahams, V.M.; Chan, S.; Visintin, I.; Rutherford, T.; Mor, G. TLR-4 signaling promotes tumor growth and paclitaxel chemoresistance in ovarian cancer. Cancer Res. 2006, 66, 3859–3868. [Google Scholar] [CrossRef] [PubMed]
- Volk-Draper, L.; Hall, K.; Griggs, C.; Rajput, S.; Kohio, P.; DeNardo, D.; Ran, S. Paclitaxel therapy promotes breast cancer metastasis in a TLR4-dependent manner. Cancer Res. 2014, 74, 5421–5434. [Google Scholar] [CrossRef] [PubMed]
- Ilvesaro, J.M.; Merrell, M.A.; Li, L.; Wakchoure, S.; Graves, D.; Brooks, S.; Rahko, E.; Jukkola-Vuorinen, A.; Vuopala, K.S.; Harris, K.W.; et al. Toll-like receptor 9 mediates CpG oligonucleotide-induced cellular invasion. Mol. Cancer Res. 2008, 6, 1534–1543. [Google Scholar] [CrossRef] [PubMed]
- Merrell, M.A.; Ilvesaro, J.M.; Lehtonen, N.; Sorsa, T.; Gehrs, B.; Rosenthal, E.; Chen, D.; Shackley, B.; Harris, K.W.; Selander, K.S. Toll-like receptor 9 agonists promote cellular invasion by increasing matrix metalloproteinase activity. Mol. Cancer Res. 2006, 4, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Tuomela, J.; Sandholm, J.; Kaakinen, M.; Patel, A.; Kauppila, J.H.; Ilvesaro, J.; Chen, D.; Harris, K.W.; Graves, D.; Selander, K.S. DNA from dead cancer cells induces TLR9-mediated invasion and inflammation in living cancer cells. Breast Cancer Res. Treat. 2013, 142, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Tuomela, J.; Sandholm, J.; Karihtala, P.; Ilvesaro, J.; Vuopala, K.S.; Kauppila, J.H.; Kauppila, S.; Chen, D.; Pressey, C.; Harkonen, P.; et al. Low TLR9 expression defines an aggressive subtype of triple-negative breast cancer. Breast Cancer Res. Treat. 2012, 135, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Pawelek, J.M.; Chakraborty, A.K. Fusion of tumour cells with bone marrow-derived cells: A unifying explanation for metastasis. Nat. Rev. Cancer 2008, 8, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Rachkovsky, M.; Sodi, S.; Chakraborty, A.; Avissar, Y.; Bolognia, J.; McNiff, J.M.; Platt, J.; Bermudes, D.; Pawelek, J. Melanoma X macrophage hybrids with enhanced metastatic potential. Clin. Exp. Metastasis 1998, 16, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Berndt, B.; Haverkampf, S.; Reith, G.; Keil, S.; Niggemann, B.; Zanker, K.S.; Dittmar, T. Fusion of CCL21 non-migratory active breast epithelial and breast cancer cells give rise to CCL21 migratory active tumor hybrid cell lines. PLoS ONE 2013, 8, e63711. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, T.; Schwitalla, S.; Seidel, J.; Haverkampf, S.; Reith, G.; Meyer-Staeckling, S.; Brandt, B.H.; Niggemann, B.; Zanker, K.S. Characterization of hybrid cells derived from spontaneous fusion events between breast epithelial cells exhibiting stem-like characteristics and breast cancer cells. Clin. Exp. Metastasis 2011, 28, 75–90. [Google Scholar] [CrossRef] [PubMed]
- Ozel, C.; Seidel, J.; Meyer-Staeckling, S.; Brandt, B.H.; Niggemann, B.; Zanker, K.S.; Dittmar, T. Hybrid cells derived from breast epithelial cell/breast cancer cell fusion events show a differential RAF-AKT crosstalk. Cell. Commun. Signal. 2012, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Berndt, B.; Zanker, K.S.; Dittmar, T. Cell fusion is a potent inducer of aneuploidy and drug resistance in tumor cell/ normal cell hybrids. Crit. Rev. Oncog. 2013, 18, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, T.; Nagler, C.; Niggemann, B.; Zänker, K.S. The dark side of stem cells: Triggering cancer progression by cell fusion. Curr. Mol. Med. 2013, 13, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, T.; Zanker, K.S. Tissue regeneration in the chronically inflamed tumor environment: Implications for cell fusion driven tumor progression and therapy resistant tumor hybrid cells. Int. J. Mol. Sci. 2015, 16, 30362–30381. [Google Scholar] [CrossRef] [PubMed]
- Dittmar, T.; Zänker, K.S. Cell Fusion in Health and Disease; Springer: Dordrecht, The Netherlands, 2011; Volume 2, pp. 1–202. [Google Scholar]
- Duelli, D.; Lazebnik, Y. Cell fusion: A hidden enemy? Cancer Cell 2003, 3, 445–448. [Google Scholar] [CrossRef]
- Dittmar, T.; Nagler, C.; Schwitalla, S.; Reith, G.; Niggemann, B.; Zanker, K.S. Recurrence cancer stem cells--made by cell fusion? Med. Hypotheses 2009, 73, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Fried, S.; Tosun, S.; Troost, G.; Keil, S.; Zaenker, K.S.; Dittmar, T. Lipopolysaccharide (LPS) promotes apoptosis in human breast epithelial x breast cancer hybrids, but not in parental cells. PLoS ONE 2016, 11, e0148438. [Google Scholar] [CrossRef] [PubMed]
- Rana, T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Sierra, A.; Castellsague, X.; Escobedo, A.; Lloveras, B.; Garcia-Ramirez, M.; Moreno, A.; Fabra, A. BCL-2 with loss of apoptosis allows accumulation of genetic alterations: A pathway to metastatic progression in human breast cancer. Int. J. Cancer 2000, 89, 142–147. [Google Scholar] [CrossRef]
- Bauerfeld, C.P.; Rastogi, R.; Pirockinaite, G.; Lee, I.; Huttemann, M.; Monks, B.; Birnbaum, M.J.; Franchi, L.; Nunez, G.; Samavati, L. TLR4-mediated AKT activation is MyD88/TRIF dependent and critical for induction of oxidative phosphorylation and mitochondrial transcription factor a in murine macrophages. J. Immunol. 2012, 188, 2847–2857. [Google Scholar] [CrossRef] [PubMed]
- Sester, D.P.; Brion, K.; Trieu, A.; Goodridge, H.S.; Roberts, T.L.; Dunn, J.; Hume, D.A.; Stacey, K.J.; Sweet, M.J. CPG DNA activates survival in murine macrophages through TLR9 and the phosphatidylinositol 3-kinase-AKT pathway. J. Immunol. 2006, 177, 4473–4480. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Coller, J. Rna in unexpected places: Long non-coding rna functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microrna biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.J.; Lin, Y.C.; Yao, P.L.; Yuan, A.; Chen, H.Y.; Shun, C.T.; Tsai, M.F.; Chen, C.H.; Yang, P.C. Tumor-associated macrophages: The double-edged sword in cancer progression. J. Clin. Oncol. 2005, 23, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Biswas, S.K.; Galdiero, M.R.; Sica, A.; Locati, M. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013, 229, 176–185. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Coussens, L.M. Inflammation and breast cancer. Balancing immune response: Crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Heo, T.H.; Wahler, J.; Suh, N. Potential therapeutic implications of IL-6/IL-6R/gp130-targeting agents in breast cancer. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Mohr, M.; Tosun, S.; Arnold, W.H.; Edenhofer, F.; Zanker, K.S.; Dittmar, T. Quantification of cell fusion events human breast cancer cells and breast epithelial cells using a cre-loxp-based double fluorescence reporter system. Cell. Mol. Life Sci. 2015, 72, 3769–3782. [Google Scholar] [CrossRef] [PubMed]
- Skokos, E.A.; Charokopos, A.; Khan, K.; Wanjala, J.; Kyriakides, T.R. Lack of tnf-alpha-induced MMP-9 production and abnormal E-cadherin redistribution associated with compromised fusion in MCP-1-null macrophages. Am. J. Pathol. 2011, 178, 2311–2321. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Zhu, F.; Zhang, H.Z.; Shang, Z.J. Tumor necrosis factor-alpha enhanced fusions between oral squamous cell carcinoma cells and endothelial cells via VCAM-1/VLA-4 pathway. Exp. Cell Res. 2012, 318, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.Y.; Lee, H.; Jung, B.Y.; Ock, J.; Lee, M.S.; Lee, W.H.; Suk, K. Tlr4, but not TLR2, signals autoregulatory apoptosis of cultured microglia: A critical role of IFN-β as a decision maker. J. Immunol. 2005, 174, 6467–6476. [Google Scholar] [CrossRef] [PubMed]
- Zen, K.; Karsan, A.; Stempien-Otero, A.; Yee, E.; Tupper, J.; Li, X.; Eunson, T.; Kay, M.A.; Wilson, C.B.; Winn, R.K.; et al. NF-κB activation is required for human endothelial survival during exposure to tumor necrosis factor-α but not to interleukin-1β or lipopolysaccharide. J. Biol. Chem. 1999, 274, 28808–28815. [Google Scholar] [CrossRef] [PubMed]
- Kirshenbaum, L.A. Bcl-2 intersects the nfkappab signalling pathway and suppresses apoptosis in ventricular myocytes. Clin. Investig. Med. 2000, 23, 322–330. [Google Scholar]
- Tuomela, J.M.; Sandholm, J.A.; Kaakinen, M.; Hayden, K.L.; Haapasaari, K.M.; Jukkola-Vuorinen, A.; Kauppila, J.H.; Lehenkari, P.P.; Harris, K.W.; Graves, D.E.; et al. Telomeric g-quadruplex-forming DNA fragments induce TLR9-mediated and LL-37-regulated invasion in breast cancer cells in vitro. Breast Cancer Res. Treat. 2016, 155, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Wang, J.H.; Redmond, H.P. Silencing of TLR4 increases tumor progression and lung metastasis in a murine model of breast cancer. Ann. Surg. Oncol. 2013, 20, S389–S396. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.F.; Zheng, L.; Kim, C.S.; Lee, K.J.; Kim, D.H.; Cai, D.Q.; Qin, J.W.; Yu, Y.H.; Wu, Z.; Kim, S.K. CPG oligodeoxynucleotide induces apoptosis and cell cycle arrest in a20 lymphoma cells via TLR9-mediated pathways. Mol. Immunol. 2013, 54, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Brignole, C.; Marimpietri, D.; Di Paolo, D.; Perri, P.; Morandi, F.; Pastorino, F.; Zorzoli, A.; Pagnan, G.; Loi, M.; Caffa, I.; et al. Therapeutic targeting of TLR9 inhibits cell growth and induces apoptosis in neuroblastoma. Cancer Res. 2010, 70, 9816–9826. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Saccani, A.; Borsatti, A.; Power, C.A.; Wells, T.N.; Luini, W.; Polentarutti, N.; Sozzani, S.; Mantovani, A. Bacterial lipopolysaccharide rapidly inhibits expression of C–C chemokine receptors in human monocytes. J. Exp. Med. 1997, 185, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Fox, J.M.; Letellier, E.; Oliphant, C.J.; Signoret, N. TLR2-dependent pathway of heterologous down-modulation for the CC chemokine receptors 1, 2, and 5 in human blood monocytes. Blood 2011, 117, 1851–1860. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Chandrasekaran, P.; Venkatesan, S. Tlr signaling paralyzes monocyte chemotaxis through synergized effects of p38 mapk and global RAP-1 activation. PLoS ONE 2012, 7, e30404. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Sun, W.; Cruz, A.; Saitoh, M.; Tai, M.H.; Trosko, J.E. A human breast epithelial cell type with stem cell characteristics as target cells for carcinogenesis. Radiat. Res. 2001, 155, 201–207. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Heyder, C.; Gloria-Maercker, E.; Hatzmann, W.; Niggemann, B.; Zanker, K.S.; Dittmar, T. Role of the beta1-integrin subunit in the adhesion, extravasation and migration of T24 human bladder carcinoma cells. Clin. Exp. Metastasis 2005, 22, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Balz, L.M.; Bartkowiak, K.; Andreas, A.; Pantel, K.; Niggemann, B.; Zanker, K.S.; Brandt, B.H.; Dittmar, T. The interplay of HER2/HER3/PI3K and EGFR/HER2/PLC-γ1 signalling in breast cancer cell migration and dissemination. J. Pathol. 2012, 227, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Rommerswinkel, N.; Niggemann, B.; Keil, S.; Zanker, K.S.; Dittmar, T. Analysis of cell migration within a three-dimensional collagen matrix. J. Vis. Exp. 2014, e51963. [Google Scholar] [CrossRef] [PubMed]
- Bjerkvig, R.; Tysnes, B.B.; Aboody, K.S.; Najbauer, J.; Terzis, A.J. Opinion: The origin of the cancer stem cell: Current controversies and new insights. Nat. Rev. Cancer 2005, 5, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Merchak, K.; Lee, W.; Grande, J.P.; Cascalho, M.; Platt, J.L. Cell fusion connects oncogenesis with tumor evolution. Am. J. Pathol. 2015, 185, 2049–2060. [Google Scholar] [CrossRef] [PubMed]
- Hatch, E.M.; Hetzer, M.W. Chromothripsis. Curr. Biol. 2015, 25, R397–R399. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, D.M.; Pavia, R.A.; Tsao, M.C. In vivo hybridisation of human tumour and normal hamster cells. Nature 1974, 250, 649–651. [Google Scholar] [CrossRef] [PubMed]

| Target | Primer Sequence 5′-3′ | Annealing Tempeature °C | Amplicon (bp) |
|---|---|---|---|
| IFN-β | F: AGTAGGCGACACTGTTCGTG | 60 | 174 |
| R: AGCCTCCCATTCAATTGCCA | |||
| IL-1β | F 1: AGCCATGGCAGAAGTACCTG | 54 | 219 |
| R: TCCATGGCCACAACAACTGA | |||
| IL-6 | F: ATGAACTCCTTCTCCACAAGCGC | 60 | 628 |
| R: GAAGAGCCCTCAGGCTGGACTG | |||
| MCP-1 | F: CCCCAGTCACCTGCTGTTAT | 60 | 135 |
| R: AGATCTCCTTGGCCACAATG | |||
| TNF-α | F: AACATCCAACCTTCCCAAACG | 54 | 109 |
| R: GACCCTAAGCCCCCAATTCTC | |||
| TRAIL | F: GAGCTGAAGCAGATGCAGGAC | 60 | 137 |
| R: TGAGGAGTTGCCACTTGACT | |||
| β-actin | F: GTGACGTTGACATCCGTAAAGACC | 55 | 290 |
| R: TCAGTAACAGTCCGCCTAGAAGCA |
| Antibody | Manufacturer |
|---|---|
| AKT, rabbit monoclonal | Cell Signaling 1 |
| pAKT S473, rabbit monoclonal | Cell Signaling 1 |
| pAKT T308, rabbit monoclonal | Cell Signaling 1 |
| BAX, rabbit monoclonal | Cell Signaling 1 |
| BCL-2, mouse monoclonal | Cell Signaling 1 |
| ERK1/2, rabbit polyclonal | Cell Signaling 1 |
| pERK1/2, rabbit polyclonal | Cell Signaling 1 |
| Histone H3, rabbit polyclonal | Abcam 2 |
| IFN-β, mouse monoclonal | Biozol 3 |
| IL-1β, rabbit monoclonal | Cell Signaling 1 |
| IL-6, rabbit monoclonal | Cell Signaling 1 |
| IRAK1, mouse monoclonal | Abgent Inc. 4 |
| IRF7, rabbit polyclonal | Cell Signaling 1 |
| MCP-1, rabbit polyclonal | Cell Signaling 1 |
| Myd88, rabbit monoclonal | Cell Signaling 1 |
| NF-κB p65, rabbit polyclonal | Santa Cruz Biotech 5 |
| TLR4, rabbit polyclonal | Santa Cruz Biotech 5 |
| TLR9, rabbit polyclonal | ProSci Inc. 6 |
| TNF-α, rabbit monoclonal | Cell Signaling 1 |
| TRAIL, rabbit monoclonal | Cell Signaling 1 |
| TRAF6, rabbit monoclonal | Cell Signaling 1 |
| TRIF, rabbit polyclonal | Cell Signaling 1 |
| elf4E, rabbit monoclonal | Cell Signaling 1 |
| β-actin, rabbit monoclonal | Cell Signaling 1 |
| anti-mouse-IgG-HRP-linked | Cell Signaling 1 |
| anti-rabbit-IgG-HRP-linked | Cell Signaling 1 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tosun, S.; Fried, S.; Niggemann, B.; Zänker, K.S.; Dittmar, T. Hybrid Cells Derived from Human Breast Cancer Cells and Human Breast Epithelial Cells Exhibit Differential TLR4 and TLR9 Signaling. Int. J. Mol. Sci. 2016, 17, 726. https://doi.org/10.3390/ijms17050726
Tosun S, Fried S, Niggemann B, Zänker KS, Dittmar T. Hybrid Cells Derived from Human Breast Cancer Cells and Human Breast Epithelial Cells Exhibit Differential TLR4 and TLR9 Signaling. International Journal of Molecular Sciences. 2016; 17(5):726. https://doi.org/10.3390/ijms17050726
Chicago/Turabian StyleTosun, Songül, Sabrina Fried, Bernd Niggemann, Kurt S. Zänker, and Thomas Dittmar. 2016. "Hybrid Cells Derived from Human Breast Cancer Cells and Human Breast Epithelial Cells Exhibit Differential TLR4 and TLR9 Signaling" International Journal of Molecular Sciences 17, no. 5: 726. https://doi.org/10.3390/ijms17050726