Chronic Heat Stress Induces Immune Response, Oxidative Stress Response, and Apoptosis of Finishing Pig Liver: A Proteomic Approach
Abstract
:1. Introduction
2. Results
2.1. Liver Weight
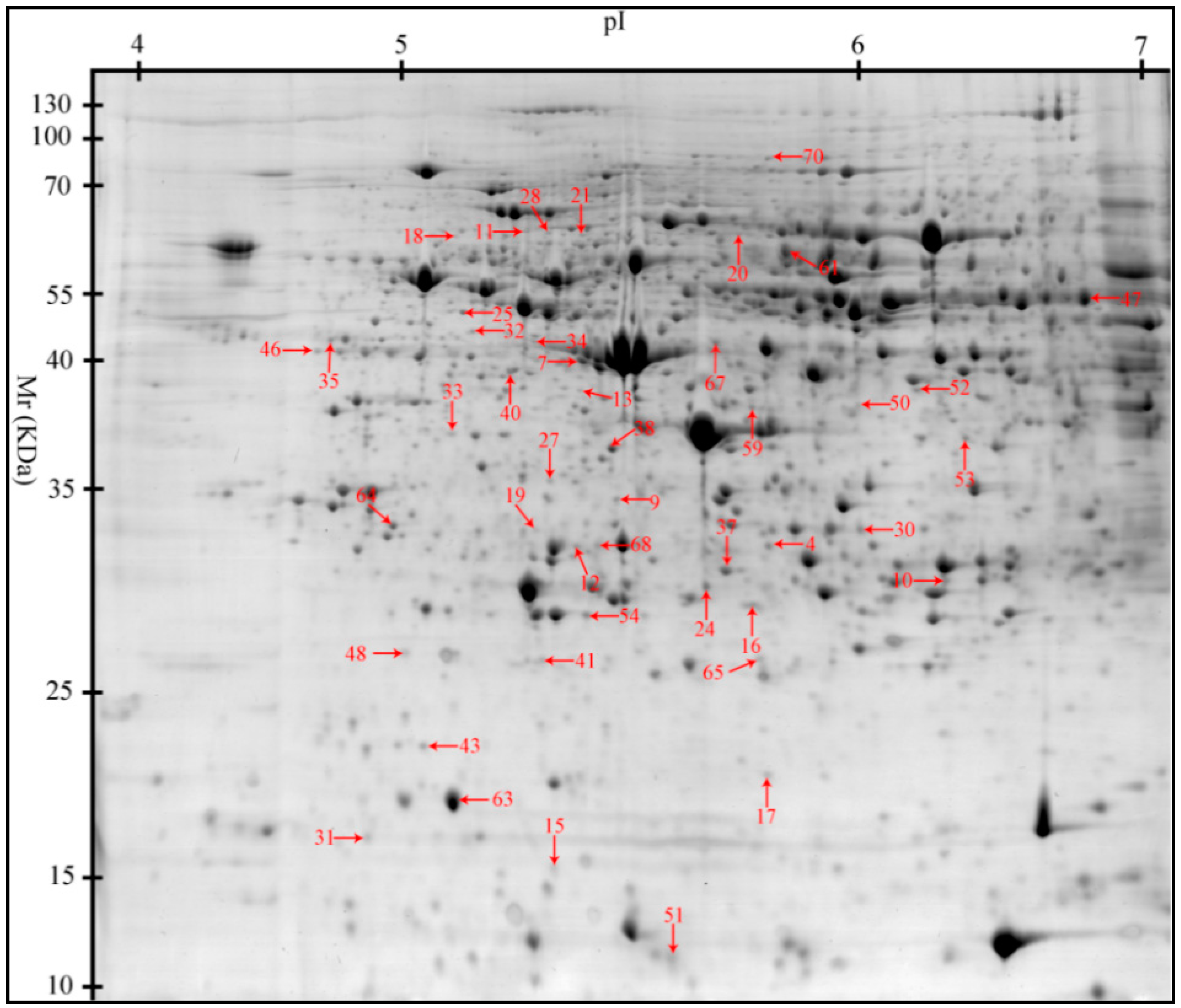
2.2. Proteomic Changes to the Liver in Response to Heat Stress (HS)
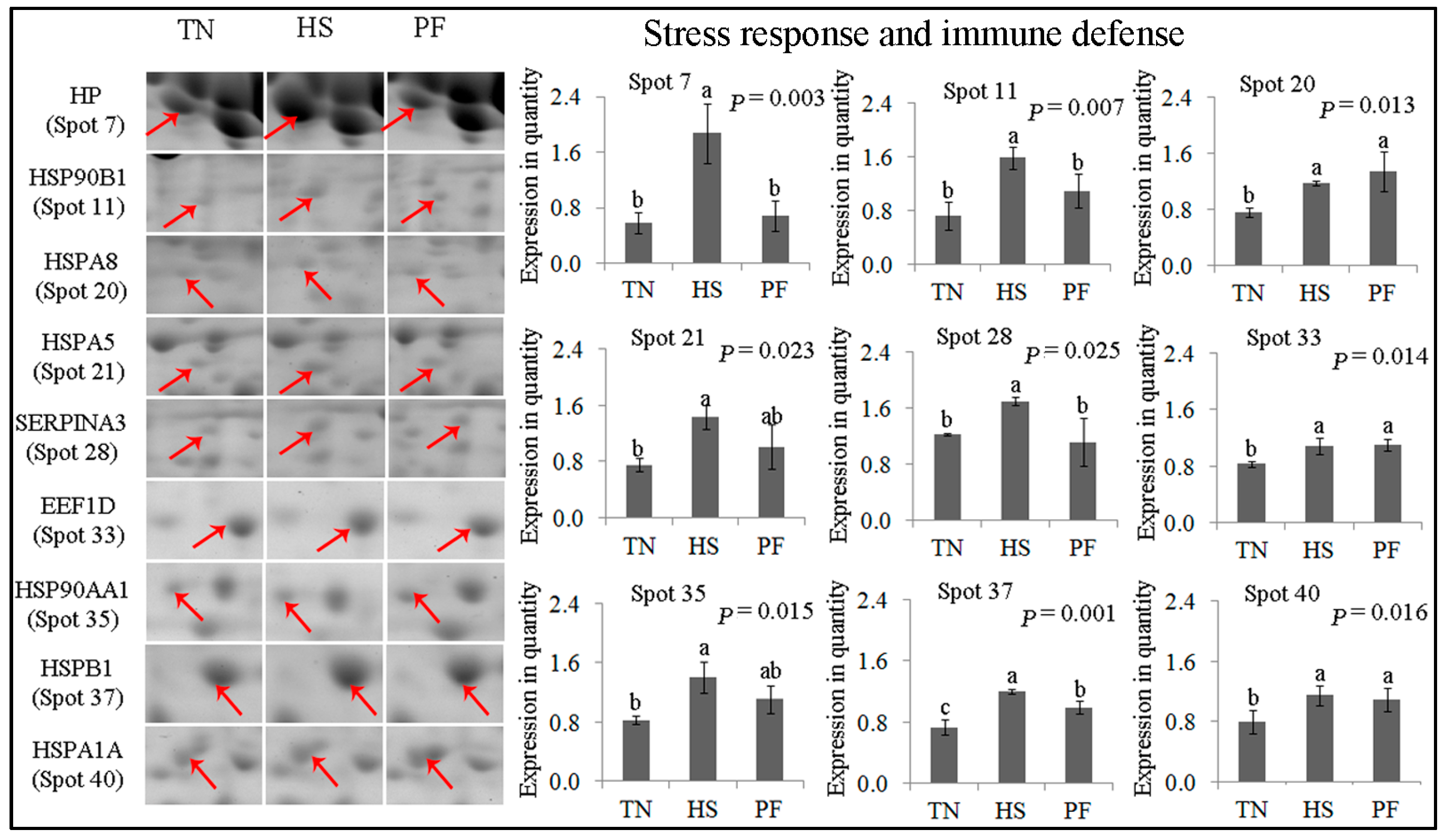
2.2.1. Stress Response and Immune Defense
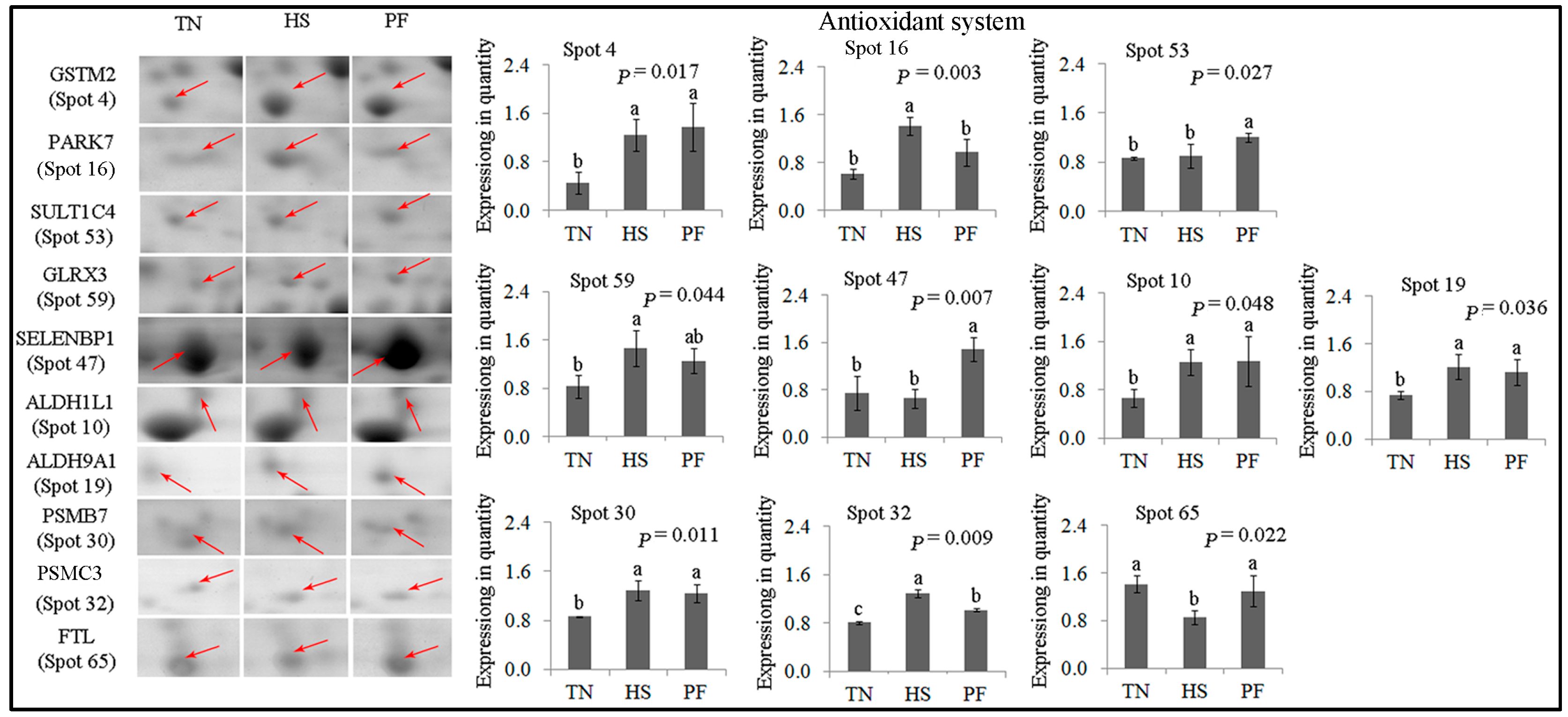
2.2.2. Antioxidant System
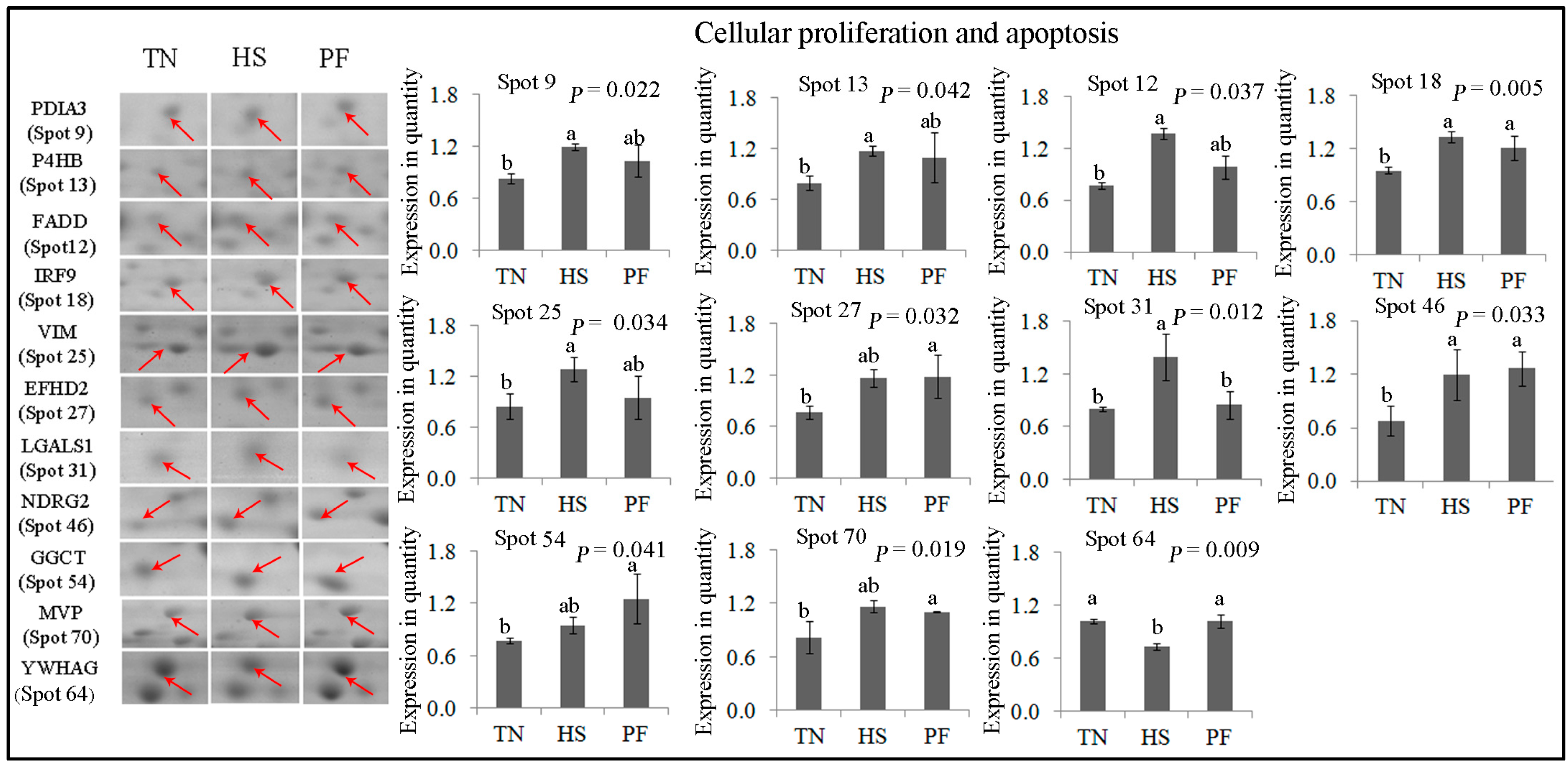
2.2.3. Cellular Proliferation and Apoptosis
2.2.4. Metabolism
2.2.5. Signal Transduction
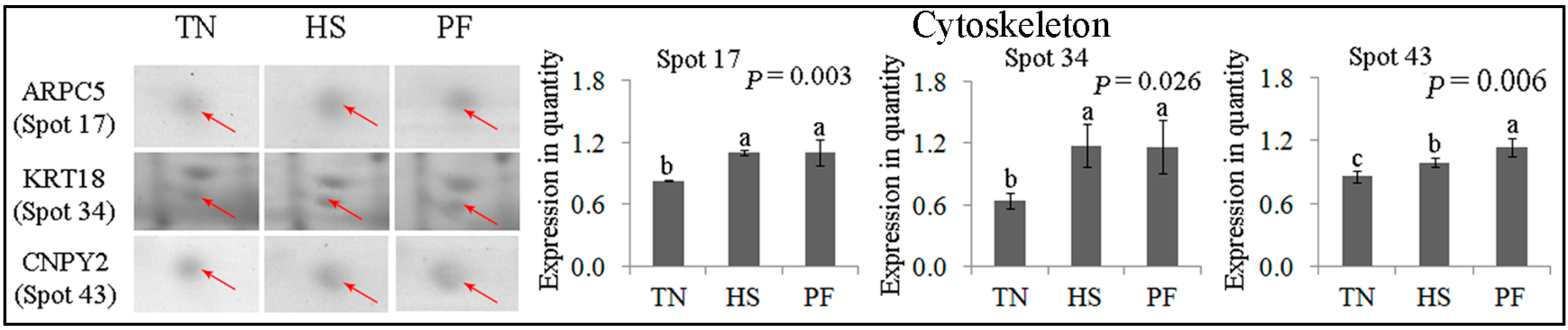
2.2.6. Cytoskeleton
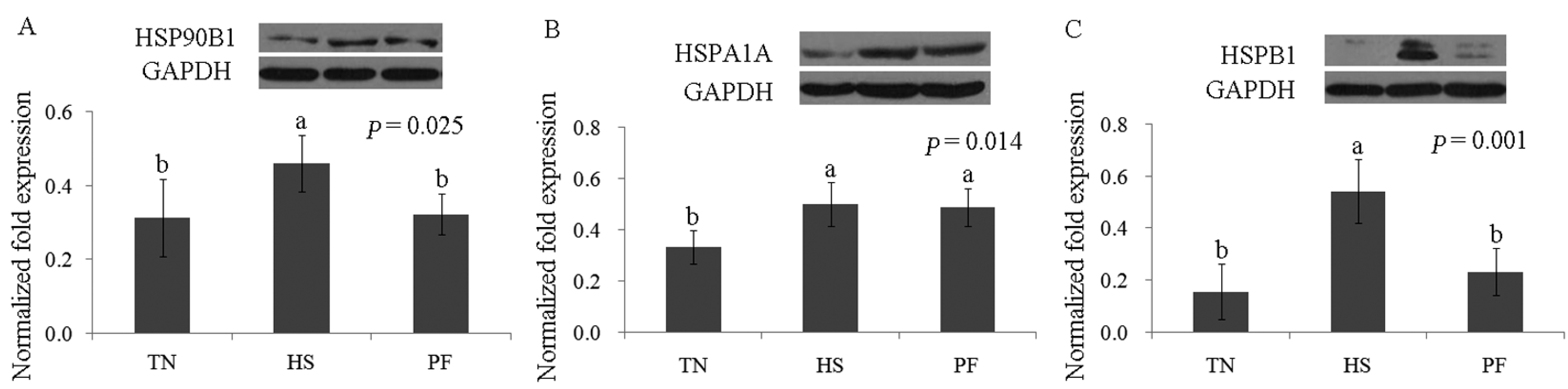
2.3. Confirmation of Differentially Abundant Proteins by Western Blot
2.4. Changes to the Redox State and Related Enzymes
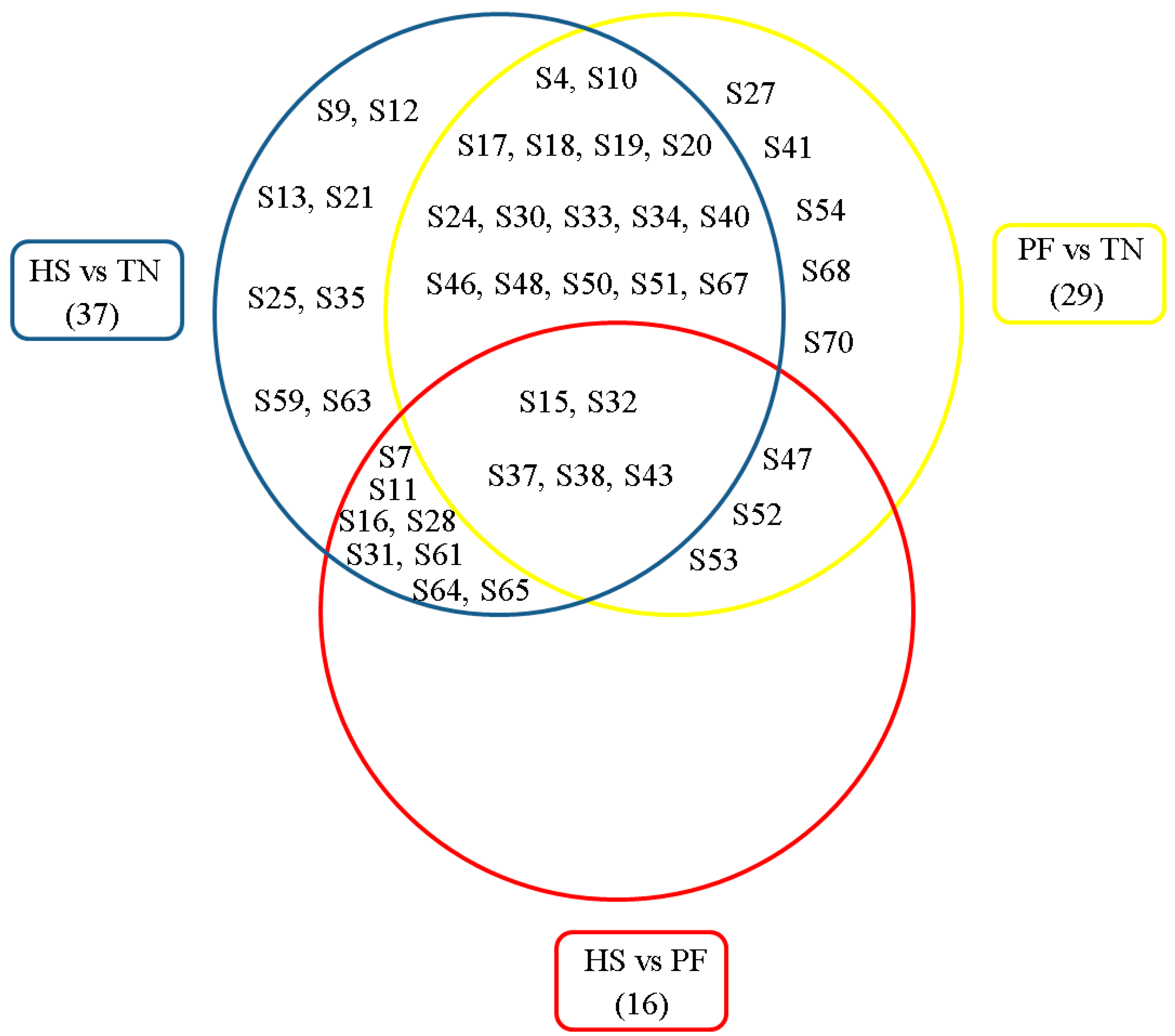
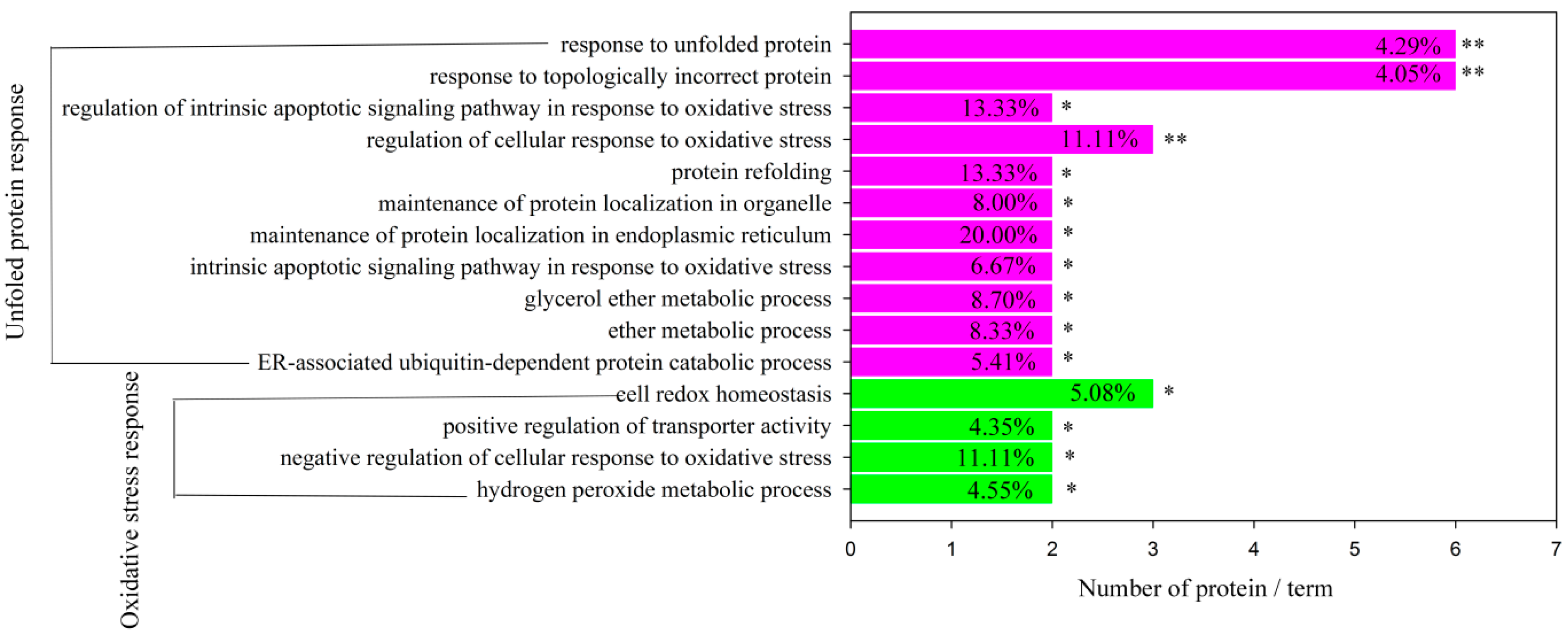
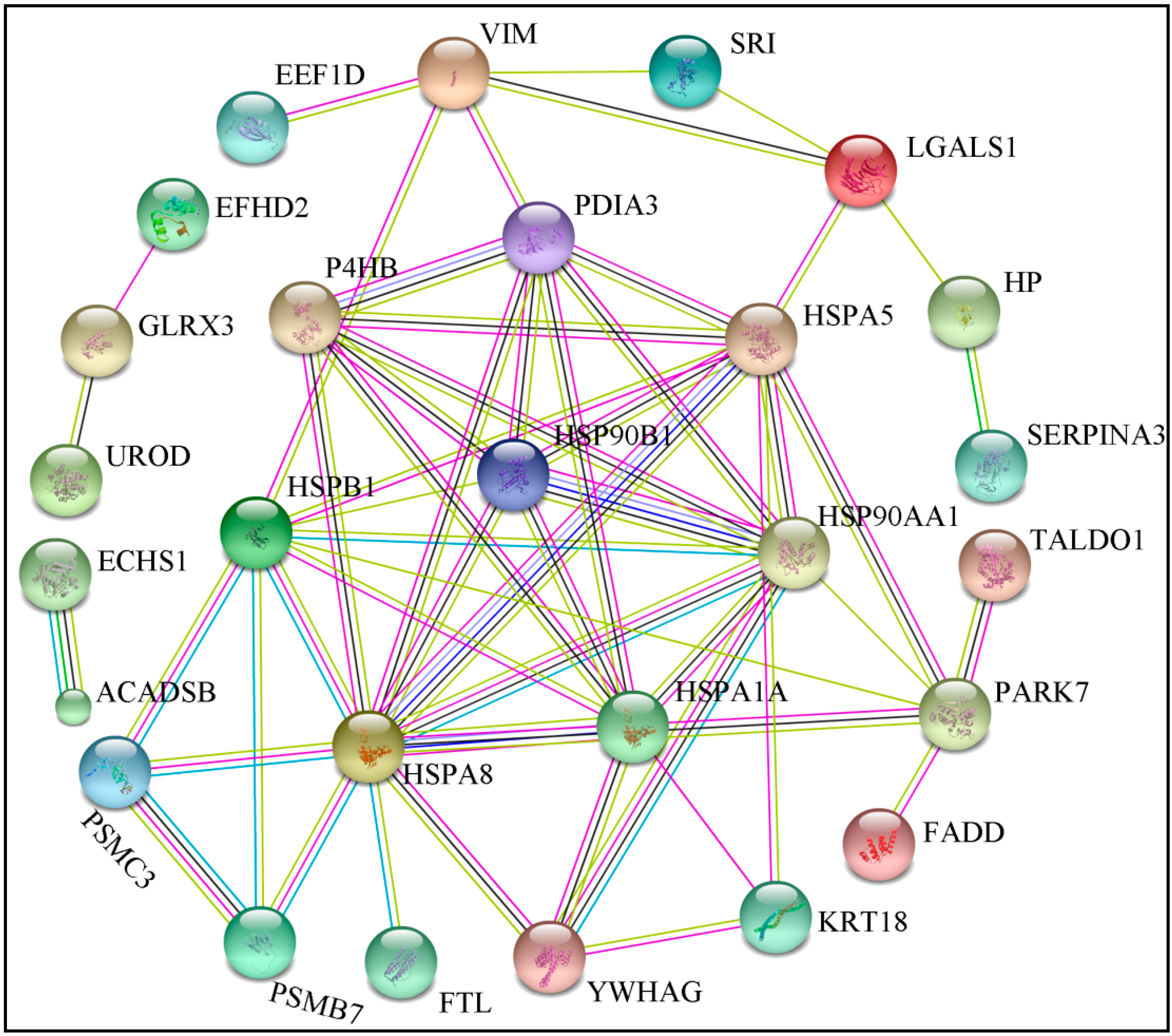
2.5. Bioinformatics Analysis of Differentially Expressed Proteins
3. Discussion
4. Materials and Methods
4.1. Animals and Experimental Design
4.2. Biological Samples
4.3. Proteomic Analysis
4.3.1. Liver Protein Extraction
4.3.2. Two-Dimensional Gel Electrophoresis
4.3.3. Image Acquisition and Analysis
4.3.4. Trypsin Digestion of Protein and Identification of Differentially Expressed Proteins by MS
4.4. Bioinformatic Approach
4.5. Validation of Differentially Expressed Proteins by Western Blot
4.6. Determination of Anti-oxidant Enzyme Activities in Finishing Pig Liver
4.7. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Coffey, R.D.; Parker, G.R.; Laurent, K.M. Feeding growing-finishing pigs to maximize lean growth rate. In ASC-147, Cooperative Extension Service; University of Kentucky College of Agriculture: Lexington, KY, USA, 1995. [Google Scholar]
- Dallaire, S.; Drolet, R.; Brodeur, D. Sow mortality associated with high ambient temperatures. Can. Vet. J. Rev. Vet. Can. 1996, 37, 237–239. [Google Scholar]
- Pearce, S.C.; Gabler, N.K.; Ross, J.W.; Escobar, J.; Patience, J.F.; Rhoads, R.P.; Baumgard, L.H. The effects of heat stress and plane of nutrition on metabolism in growing pigs. J. Anim. Sci. 2013, 91, 2108–2118. [Google Scholar] [CrossRef] [PubMed]
- Cruzen, S.; Boddicker, R.; Graves, K.; Johnson, T.; Arkfeld, E.; Baumgard, L.; Ross, J.; Safranski, T.; Lucy, M.; Lonergan, S. Effects of long term heat stress in utero or during finishing on pork carcass composition. Meat Sci. 2015, 101, 108. [Google Scholar] [CrossRef]
- St-Pierre, N.R.; Cobanov, B.; Schnitkey, G. Economic losses from heat stress by US livestock industries. J. Dairy Sci. 2003, 86, E52–E77. [Google Scholar] [CrossRef]
- Zhang, H.J.; Drake, V.J.; Morrison, J.P.; Oberley, L.W.; Kregel, K.C. Selected contribution: Differential expression of stress-related genes with aging and hyperthermia. J. Appl. Physiol. 2002, 92, 1762–1769. [Google Scholar] [CrossRef] [PubMed]
- Garcia, H.; Grageola, F.; Morales, A.; Araiza, A.; Arce, N.; Cervantes, M. Severe heat stress affects the expression of Hsp90, GLUT4 and b0,+, in liver and leptin in adipose tissue of pigs. FASEB J. 2012, 26, 651–653. [Google Scholar]
- Jin, Y.; Peng, Y.; Liu, F.; Cheng, G.; Guo, K.; An, L.; Zhu, X.; Luan, W.; Xu, J. Effect of heat stress on the porcine small intestine: A morphological and gene expression study. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2010, 156, 119–128. [Google Scholar]
- Pennarossa, G.; Maffei, S.; Rahman, M.M.; Berruti, G.; Brevini, T.A.L.; Gandolfi, F. Characterization of the constitutive pig ovary heat shock chaperone machinery and its response to acute thermal stress or to seasonal variations1. Biol. Reprod. 2012, 87, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Xiang-Hong, J.; Yan-Hong, Y.; Han-Jin, X.; Li-Long, A.; Ying-Mei, X.; Pei-Rong, J.; Ming, L. Selection of reference genes for gene expression studies in PBMC from Bama miniature pig under heat stress. Veterinary Immunol. Immunopathol. 2011, 144, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Speakman, J.R.; Krol, E. Maximal heat dissipation capacity and hyperthermia risk: Neglected key factors in the ecology of endotherms. J. Anim. Ecol. 2010, 79, 726–746. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.; Huff-Lonergan, E.; Lonergan, S.; Baumgard, L.; Gabler, N. Heat stress and reduced feed intake alter the intestinal proteomic profile. Faseb J. 2014, 28, 241–256. [Google Scholar]
- Yue, H.; Feng, Y.; Yang, P.; Feng, J.; Hai, L.; Gu, X. Nutritional and physiological responses of finishing pigs exposed to a permanent heat exposure during three weeks. Arch. Anim. Nutr. 2014, 68, 296–308. [Google Scholar]
- Cui, Y.; Gu, X. Proteomic changes of the porcine small intestine in response to chronic heat stress. J. Mol. Endocrinol. 2015, 55, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Morera, P.; Basiricò, L.; Hosoda, K.; Bernabucci, U. Chronic heat stress up-regulates leptin and adiponectin secretion and expression and improves leptin, adiponectin and insulin sensitivity in mice. J. Mol. Endocrinol. 2012, 48, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.; Xu, L.; Drake, V.; Oberley, L.; Oberley, T.; Moseley, P.; Kregel, K. Aging reduces adaptive capacity and stress protein expression in the liver after heat stress. J. Appl. Physiol. 2000, 89, 749–759. [Google Scholar] [PubMed]
- Horowitz, M.; Robinson, S.D.M. Heat shock proteins and the heat shock response during hyperthermia and its modulation by altered physiological conditions. Neurobiol. Hyperth. 2007, 162, 433–446. [Google Scholar]
- Horowitz, M. From molecular and cellular to integrative heat defense during exposure to chronic heat. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2000, 131, 475–483. [Google Scholar] [CrossRef]
- Aly, K.B.; Pipkin, J.L.; Hinson, W.G.; Feuers, R.J.; Duffy, P.H.; Lyn-Cook, L.; Hart, R.W. Chronic caloric restriction induces stress proteins in the hypothalamus of rats. Mech. Ageing Dev. 1994, 76, 11–23. [Google Scholar] [CrossRef]
- Sinclair, D.A. Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing Dev. 2005, 126, 987–1002. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y. Roles of heat shock protein gp96 in the ER quality control: Redundant or unique function? Mol. Cells 2005, 20, 173–182. [Google Scholar] [PubMed]
- Murata, H.; Shimada, N.; Yoshioka, M. Current research on acute phase proteins in veterinary diagnosis: An overview. Vet.J. 2004, 168, 28–40. [Google Scholar] [CrossRef]
- Ruslan, M. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar]
- Zhou, J.; An, H.; Xu, H.; Liu, S.; Cao, X. Heat shock up-regulates expression of Toll-like receptor-2 and Toll-like receptor-4 in human monocytes via p38 kinase signal pathway. Immunology 2005, 114, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.H.; Xu, H.J.; Yong, Y.H.; An, L.L.; Jiao, P.R.; Liao, M. Heat stress upregulation of Toll-like receptors 2/4 and acute inflammatory cytokines in peripheral blood mononuclear cell (PBMC) of Bama miniature pigs: An in vivo and in vitro study. Animal 2014, 8, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.J.; Xu, L.; Drake, V.J.; Xie, L.; Oberley, L.W.; Kregel, K.C. Heat-induced liver injury in old rats is associated with exaggerated oxidative stress and altered transcription factor activation. Faseb J. 2003, 17, 2293–2308. [Google Scholar] [CrossRef] [PubMed]
- Bhusari, S.; Hearne, L.B.; Spiers, D.E.; Lamberson, W.R.; Antoniou, E. Transcriptional profiling of mouse liver in response to chronic heat stress. J. Therm. Biol. 2008, 33, 157–167. [Google Scholar] [CrossRef]
- Mannick, J.B. Regulation of apoptosis by protein S-nitrosylation. Amino Acids 2007, 32, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, E.L.; Iwasaki, K.; Tsuji, Y. Intracellular iron transport and storage: From molecular mechanisms to health implications. Antioxid. Redox Signal. 2008, 10, 997–1030. [Google Scholar] [CrossRef] [PubMed]
- Bacic, G.; Spasojevic, I.; Secerov, B.; Mojovic, M. Spin-trapping of oxygen free radicals in chemical and biological systems: New traps, radicals and possibilities. Spectrochim. Acta AMol. Biomol. Spectrosc. 2008, 69, 1354–1366. [Google Scholar] [CrossRef] [PubMed]
- Aw, T.Y. Intestinal glutathione: Determinant of mucosal peroxide transport, metabolism, and oxidative susceptibility. Toxicol. Appl. Pharmacol. 2005, 204, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Galli, F. Amino acid and protein modification by oxygen and nitrogen species. Amino Acids 2007, 32, 497–499. [Google Scholar] [CrossRef]
- Zhu, Y.; Carvey, P.M.; Ling, Z. Age-related changes in glutathione and glutathione-related enzymes in rat brain. Brain Res. 2006, 1090, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Haunhorst, P.; Berndt, C.; Eitner, S.; Godoy, J.R.; Lillig, C.H. Characterization of the human monothiol glutaredoxin 3 (PICOT) as iron-sulfur protein. Biochem. Biophys. Res. Commun. 2010, 394, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Shendelman, S.; Jonason, A.; Martinat, C.; Leete, T.; Abeliovich, A. DJ-1 is a redox-dependent molecular chaperone that inhibits α-synuclein aggregate formation. PLoS Biol. 2004, 2, 1764–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mceleny, K.; Coffey, R.; Morrissey, C.; Williamson, K.; Zangemeister, W.U.; Fitzpatrick, J.M.; Watson, R.W. An antisense oligonucleotide to cIAP-1 sensitizes prostate cancer cells to fas and TNFα mediated apoptosis. Prostate 2004, 59, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.J. Degradation of oxidized proteins by the 20S proteasome. Biochimie 2001, 83, 301–310. [Google Scholar] [CrossRef]
- Mi-Kyoung, K.; Nobunao, W.; Jennifer, L.G.; Masayuki, Y.; Thomas W, K. Antioxidants Enhance Mammalian Proteasome Expression through the Keap1-Nrf2 Signaling Pathway. Mol. Cell. Biol. 2003, 23, 8786–8794. [Google Scholar]
- Vasiliou, V. Role of human aldehyde dehydrogenases in Endobiotic and Xenobiotic Metabolism. Drug Metab. Rev. 2004, 36, 279–299. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Lee, A.S. Role of the unfolded protein response, GRP78 and GRP94 in Organ Homeostasis. J. Cell. Physiol. 2015, 230, 1413–1420. [Google Scholar] [CrossRef] [PubMed]
- Turano, C.; Coppari, S.; Altieri, F.; Ferraro, A. Proteins of the PDI family: Unpredicted non-ER locations and functions. J. Cell. Physiol. 2002, 193, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Hoffstrom, B.G.; Kaplan, A.; Letso, R.; Schmid, R.S.; Turmel, G.J.; Lo, D.C.; Stockwell, B.R. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nat. Chem. Biol. 2010, 6, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Yeh, W.C.; de la Pompa, J.L.; McCurrach, M.E.; Shu, H.B.; Elia, A.J.; Shahinian, A.; Ng, M.; Wakeham, A.; Khoo, W.; Mitchell, K.; et al. FADD: Essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 1998, 279, 1954–1958. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.; Shu, H.B.; Pan, M.G.; Goeddel, D.V. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 1996, 84, 299–308. [Google Scholar] [CrossRef]
- Byun, Y.; Chen, F.; Chang, R.; Trivedi, M.; Green, K.J.; Cryns, V.L. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 2001, 8, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Jungyeon, W.; Doo Yeon, K.; Muhnho, L.; Doyeun, K.; Meadows, G.G.; Joe, C.O. Cleavage of 14-3-3 protein by caspase-3 facilitates bad interaction with Bcl-x(L) during apoptosis. J. Biol. Chem. 2003, 278, 19347–19351. [Google Scholar]
- Tsuno, T.; Mejido, J.; Zhao, T.; Schmeisser, H.; Morrow, A.; Zoon, K.C. IRF9 is a key factor for eliciting the antiproliferative activity of IFN-α. J. Immunother. 2009, 32, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Yun, Z.; Wei, H.; Zhang, C.; Xing, G.; Yu, Y.; He, F. Characterization and expression of three novel differentiation-related genes belong to the human NDRG gene family. Mol. Cell. Biochem. 2002, 229, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Zhang, J.; Liu, X. NDRG2: A Myc-repressed gene involved in cancer and cell stress. Acta Biochim. Et Biophys. Sin. 2008, 40, 625–635. [Google Scholar] [CrossRef]
- Wang, L.; Liu, N.; Yao, L.; Li, F.; Zhang, J.Y.; Liu, J.; Ji, S.; Yang, A.; Han, H. NDRG2 is a new HIF-1 target gene necessary for hypoxia-induced apoptosis in A549 cells. Cell. Physiol. Biochem. 2008, 21, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Na, L.; Lifeng, W.; Xia, L.; Qi, Y.; Xinping, L.; Jing, Z.; Jian, Z.; Yousheng, W.; Shaoping, J.; Yingqi, Z. N-Myc downstream-regulated gene 2 is involved in p53-mediated apoptosis. Nucleic Acids Res. 2008, 36, 5335–5349. [Google Scholar]
- Zhou, Q.Q.; Yang, D.Z.; Luo, Y.J.; Li, S.Z.; Liu, F.Y.; Wang, G.S. Over-starvation aggravates intestinal injury and promotes bacterial and endotoxin translocation under high-altitude hypoxic environment. World J. Gastroenterol. 2011, 17, 1584–1593. [Google Scholar] [CrossRef] [PubMed]
- Grasl-Kraupp, B.; Bursch, W.; Ruttkay-Nedecky, B.; Wagner, A.; Lauer, B.; Schulte-Hermann, R. Food restriction eliminates preneoplastic cells through apoptosis and antagonizes carcinogenesis in rat liver. Proc. Natl. Acad. Sci. USA 1994, 91, 9995–9999. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gulbahce, N.; Yu, H. Network-based methods for human disease gene prediction. Brief. Funct. Genom. 2011, 10, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, W.; Gang, L.; Li, D.; Wu, G.; Wang, J. Temporal proteomic analysis reveals continuous impairment of intestinal development in neonatal piglets with intrauterine growth restriction. J. Proteome Res. 2010, 9, 924–935. [Google Scholar] [CrossRef] [PubMed]
- Ramagli, L.S.; Rodriguez, L.V. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis 1985, 6, 559–563. [Google Scholar] [CrossRef]
- Saito, R.; Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Lotia, S.; Pico, A.R.; Bader, G.D.; Ideker, T. A travel guide to Cytoscape plugins. Nat. Methods 2012, 9, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucl. Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]






| Parameter | TN | HS | PF | p Value |
|---|---|---|---|---|
| ALW (kg) | 1.65 ± 0.10 a | 1.48 ± 0.11 b | 1.44 ± 0.05 b | 0.009 |
| RLW (%) | 2.01 ± 0.13 a | 1.75 ± 0.23 b | 1.84 ± 0.22 b | 0.04 |
| Spot No. a | Accession No. b | Protein Name | Short Name | Score c | PM d | Sequence Coverage (%) | Theoretical Mr(kDa)/pI e | Functions |
|---|---|---|---|---|---|---|---|---|
| Stress Response and Immune Defense (9) | ||||||||
| 7 | gi|545829329 | PREDICTED: haptoglobin isoform X1 | HP | 176.71 | 15 | 41.53 | 45.21/6.13 | Acute-phase response; immune defense |
| 11 | gi|47523016 | Endoplasmin precursor | HSP90B1 | 119.66 | 6 | 7.59 | 92.47/4.76 | Innate immune response; response to ER stress |
| 20 | gi|345441750 | Heat shock 70 kDa protein 8 | HSPA8 | 166.46 | 12 | 21.67 | 70.90/5.37 | Chaperone; stress response |
| 21 | gi|350579657 | PREDICTED: 78 kDa glucose-regulated protein | HSPA5 | 141.01 | 13 | 19.27 | 72.33/5.07 | Chaperone; stress response |
| 28 | gi|47523270 | α-1-Antichymotrypsin 2 precursor | SERPINA3 | 101.78 | 17 | 57.32 | 47.65/5.33 | Inflammatory response; acute-phase response |
| 33 | gi|350582856 | PREDICTED: elongation factor 1-delta isoformX2 | EEF1D | 102.57 | 8 | 29.69 | 31.12/4.90 | Transcription regulation; heat shock protein binding |
| 35 | gi|47522774 | Heat shock protein HSP 90-α | HSP90AA1 | 170.82 | 10 | 13.11 | 84.66/4.94 | Innate immune response; stress response |
| 37 | gi|55668280 | Hsp27 | HSPB1 | 204.53 | 14 | 54.11 | 22.78/5.98 | Chaperone; stress response |
| 40 | gi|39777368 | Heat shock protein 70.2 | HSPA1A | 160.30 | 17 | 20.75 | 70.05/5.47 | Chaperone; stress response |
| Antioxidant System (10) | ||||||||
| 4 | gi|118403788 | Glutathione S-transferase mu 2 | GSTM2 | 198.75 | 14 | 42.66 | 25.74/5.99 | Glutathione metabolic; cellular detoxification |
| 16 | gi|67038668 | DJ-1 protein | PARK7 | 228.29 | 16 | 57.68 | 19.89/6.32 | Inflammatory response; oxidative stress response |
| 53 | gi|545815980 | PREDICTED: sulfotransferase 1C4-like (EC 2.8.2) | SULT1C4 | 154.30 | 10 | 30.65 | 35.52/8.22 | Transferase |
| 59 | gi|345199274 | Glutaredoxin 3 | GLRX3 | 79.56 | 7 | 20.36 | 37.43/5.31 | Cell redox homeostasis |
| 47 | gi|194036227 | PREDICTED: selenium-binding protein 1 | SELENBP1 | 339.53 | 34 | 65.68 | 52.39/5.93 | Selenium binding |
| 10 | gi|347582636 | Aldehyde dehydrogenase 1 family, member L1 | ALDH1L1 | 154.87 | 13 | 16.31 | 98.83/5.63 | Cellular aldehyde metabolic process |
| 19 | gi|545822151 | 4-trimethylaminobutyraldehyde dehydrogenase | ALDH9A1 | 132.85 | 5 | 9.15 | 53.80/5.69 | Cellular aldehyde metabolic process |
| 30 | gi|160419232 | Proteasome subunit β type-7 | PSMB7 | 136.91 | 7 | 25.60 | 27.89/8.60 | Protease; hydrolase activity |
| 32 | gi|346421411 | Proteasome 26S subunit ATPase 3 | PSMC3 | 146.53 | 15 | 31.36 | 49.20/5.13 | Protease; hydrolase activity |
| 65 | gi|346421372 | Ferritin, light polypeptide | FTL | 107.45 | 6 | 41.08 | 21.30/5.70 | Cellular iron ion homeostasis |
| Cellular Proliferation and Apoptosis (11) | ||||||||
| 9 | gi|304365428 | Protein disulfide-isomerase A3 precursor | PDIA3 | 120.17 | 7 | 24.27 | 56.78/5.98 | Cell redox homeostasis; apoptosis |
| 13 | gi|358009193 | Prolyl 4-hydroxylase β polypeptide | P4HB | 235.16 | 6 | 14.76 | 57.12/4.76 | Cell redox homeostasis; apoptosis |
| 12 | gi|46397561 | Fas-associating death domain-containing protein | FADD | 174.58 | 12 | 38.39 | 23.28/5.48 | Innate immunity; apoptosis |
| 18 | gi|323145752 | Interferon-stimulated protein 60 | IRF9 | 78.77 | 4 | 9.43 | 43.70/5.58 | Signal transduction; apoptosis |
| 25 | gi|408360214 | RecName: Full = Vimentin | VIM | 202.05 | 29 | 50.64 | 53.65/5.05 | Programmed cell death |
| 27 | gi|311258550 | PREDICTED: EF-hand domain-containing protein D2 | EFHD2 | 184.90 | 17 | 54.59 | 26.70/5.15 | Apoptosis |
| 31 | gi|47716872 | Galectin-1, partial | LGALS1 | 100.62 | 5 | 41.04 | 14.72/5.30 | Regulation of apoptotic process |
| 46 | gi|115499496 | NDRG2 | NDRG2 | 110.55 | 3 | 8.69 | 40.80/5.08 | Cell differentiation |
| 54 | gi|335308924 | PREDICTED: γ-glutamylcyclotransferase-like isoformX1 (EC 2.3.2.4) | GGCT | 230.41 | 15 | 59.04 | 21.01/5.07 | Apoptosis; glutathione homeostasis |
| 70 | gi|335284397 | PREDICTED: major vault protein isoform 1 | MVP | 190.97 | 30 | 33.67 | 99.33/5.34 | Apoptosis |
| 64 | gi|350581487 | PREDICTED: 14-3-3 protein γ isoform X1 | YWHAG | 255.37 | 20 | 65.31 | 28.30/4.80 | Programmed cell death |
| Metabolism (10) | ||||||||
| 24 | gi|298104076 | Enoyl-CoA hydratase, mitochondrial | ECHS1 | 139.16 | 11 | 31.38 | 31.39/8.34 | Fatty acid metabolism |
| 52 | gi|311271975 | PREDICTED: short/branched chain specific acyl-CoA Dehydrogenase, mitochondrial | ACADSB | 187.41 | 14 | 32.71 | 47.49/6.53 | Fatty acid metabolism |
| 50 | gi|349732238 | Transaldolase | TALDO1 | 119.81 | 18 | 39.17 | 37.54/6.36 | Carbohydrate metabolism |
| 38 | gi|335298275 | Glyoxalase domain-containing protein 4 | GLOD4 | 236.08 | 12 | 45.45 | 34.79/5.40 | Carbohydrate metabolism |
| 63 | gi|2642486 | Cytochrome b5 | CYB5A | 252.85 | 17 | 69.15 | 15.33/4.86 | vitamin metabolic process |
| 61 | gi|1931 | Carboxylesterase precursor | CES1 | 298.47 | 36 | 48.27 | 62.52/6.15 | lipid catabolic process |
| 15 | gi|33323483 | Cellular retinol binding protein 1 | RBP1 | 152.93 | 7 | 38.52 | 15.85/4.99 | vitamin A metabolic process |
| 51 | gi|343780946 | D-dopachrome decarboxylase | DDT | 141.84 | 10 | 59.32 | 12.71/6.72 | melanin biosynthetic process |
| 67 | gi|349732258 | Uroporphyrinogen decarboxylase | UROD | 209.27 | 13 | 32.43 | 40.79/5.77 | Porphyrin biosynthesis |
| 48 | gi|335294970 | PREDICTED: tetratricopeptide repeat protein 36 | TTC36 | 86.78 | 5 | 12.84 | 20.90/5.02 | Amino acid metabllism |
| Signaltransduction (2) | ||||||||
| 41 | gi|346644814 | Sorcin isoform 2 | SRI | 111.12 | 7 | 11.65 | 21.68/5.32 | Calcium homeostasis |
| 68 | gi|116175265 | Regucalcin | RGN | 119.57 | 7 | 35.07 | 33.25/5.89 | Calcium homeostasis |
| Cytoskeleton (3) | ||||||||
| 17 | gi|195562237 | Actin related protein 2/3 complex subunit 5 | ARPC5 | 137.21 | 7 | 38.41 | 16.32/5.47 | Cell migration |
| 34 | gi|545825420 | PREDICTED: keratin, type I cytoskeletal 18, partial | KRT18 | 102.03 | 15 | 32.24 | 48.06/5.34 | Cell cycle |
| 43 | gi|297307133 | Protein canopy homolog 2 precursor | CNPY2 | 201.68 | 9 | 46.15 | 20.65/4.81 | Cytoskeletal structure |
| Pathway Name | Count | Protein | p Value | q Value |
|---|---|---|---|---|
| Ascorbate and aldarate metabolism | 2 | ALDH9A1, RGN | 8.45 × 10−3 | 4.94 × 10−2 |
| Fatty acid degradation | 3 | ACADSB, ALDH9A1, ECHS1 | 1.47 × 10−3 | 1.47 × 10−2 |
| Proteasome | 2 | PSMB7, PSMC3 | 7.17 × 10−3 | 7.89 × 10−2 |
| Unfolded protein response | 6 | HSPA5,HSPA8, P4HB, HSPA1A, PDIA3 HSP90B1, HSP90AA1 | 1.91 × 10−5 | 2.48 × 10−4 |
| Antigen processing and presentation | 5 | HSP90AA1, HSPA1A, HSPA5, HSPA8, PDIA3 | 5.78 × 10−5 | 6.93 × 10−4 |
| Item | Content |
|---|---|
| Ingredient | g/kg |
| Corn | 662.0 |
| Soybean meal, 42.8% CP | 200.0 |
| Wheat bran | 65.0 |
| Wheat middlings | 40.0 |
| Limestone | 10.0 |
| Dicalcium phosphate | 6.0 |
| Salt | 4.0 |
| Premix 1 | 10.0 |
| l-Lysine·HCl | 3.0 |
| Chemical composition 2 | g/kg |
| Digestive energy (MJ/kg) | 13.39 |
| Crude protein | 157.3 |
| Calcium | 6.5 |
| Total phosphorus | 4.1 |
| Available phosphorus | 1.7 |
| Lysine | 9.2 |
| Met + Cys | 5.4 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Hao, Y.; Li, J.; Bao, W.; Li, G.; Gao, Y.; Gu, X. Chronic Heat Stress Induces Immune Response, Oxidative Stress Response, and Apoptosis of Finishing Pig Liver: A Proteomic Approach. Int. J. Mol. Sci. 2016, 17, 393. https://doi.org/10.3390/ijms17050393
Cui Y, Hao Y, Li J, Bao W, Li G, Gao Y, Gu X. Chronic Heat Stress Induces Immune Response, Oxidative Stress Response, and Apoptosis of Finishing Pig Liver: A Proteomic Approach. International Journal of Molecular Sciences. 2016; 17(5):393. https://doi.org/10.3390/ijms17050393
Chicago/Turabian StyleCui, Yanjun, Yue Hao, Jielei Li, Weiguang Bao, Gan Li, Yanli Gao, and Xianhong Gu. 2016. "Chronic Heat Stress Induces Immune Response, Oxidative Stress Response, and Apoptosis of Finishing Pig Liver: A Proteomic Approach" International Journal of Molecular Sciences 17, no. 5: 393. https://doi.org/10.3390/ijms17050393