Two ω-3 FADs Are Associated with Peach Fruit Volatile Formation
Abstract
:1. Introduction
2. Results and Discussion
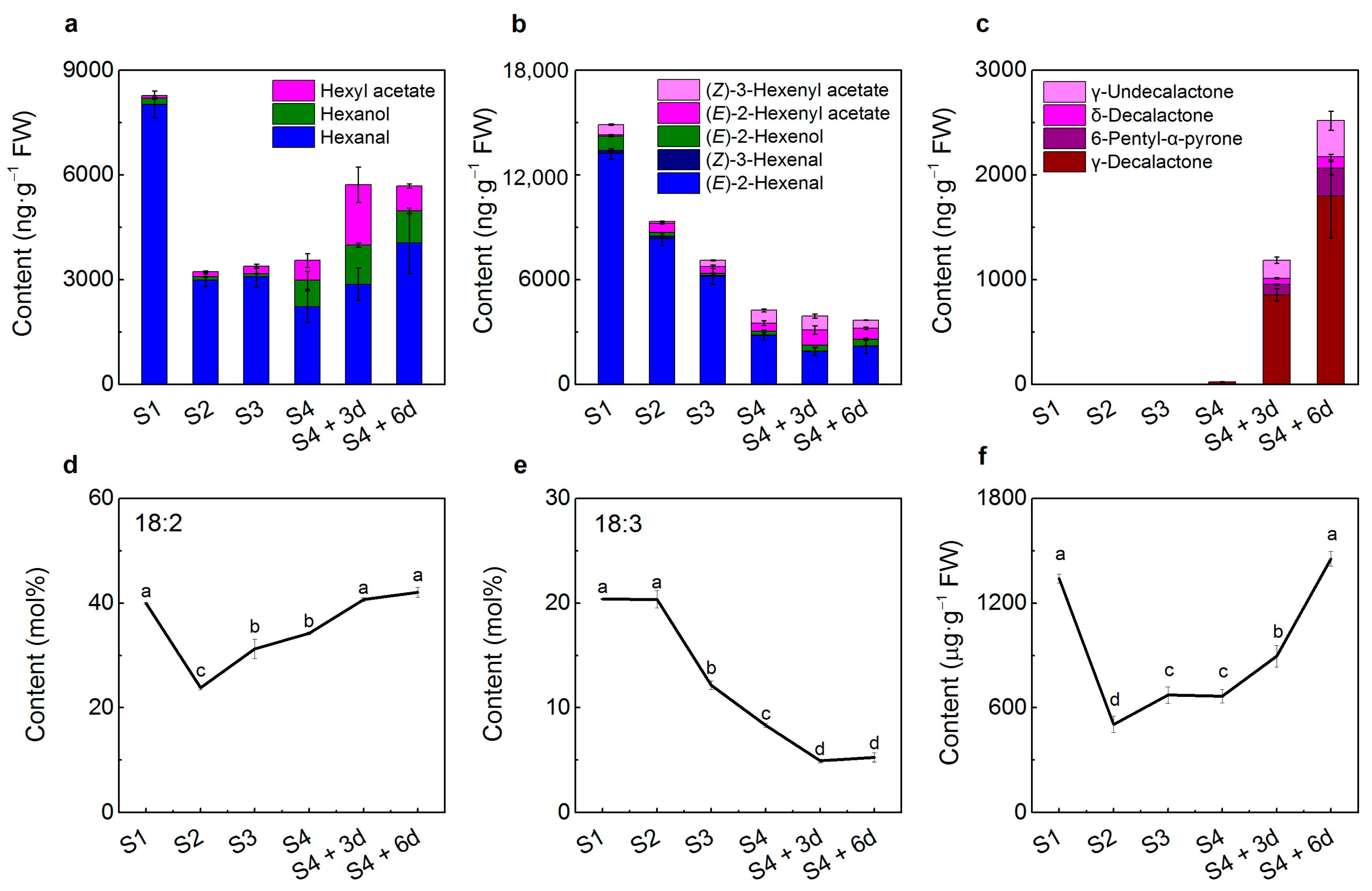
2.1. Volatile Production during Peach Fruit Development and Ripening
2.2. Peach Fruit Fatty Acid Compositions
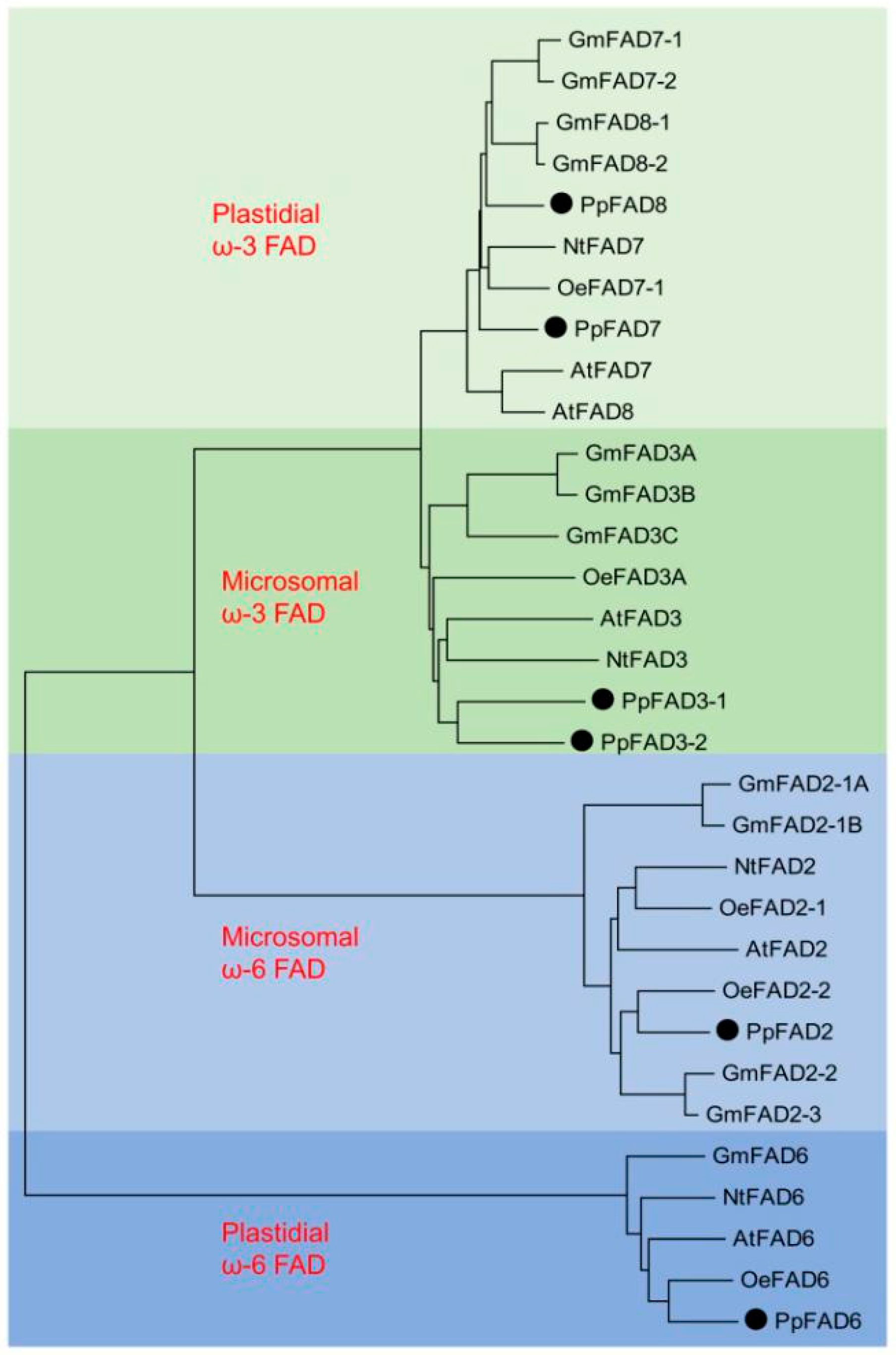
2.3. Cloning and Sequence Analysis of the Peach Fatty Acid Desaturases (FAD) Gene Family
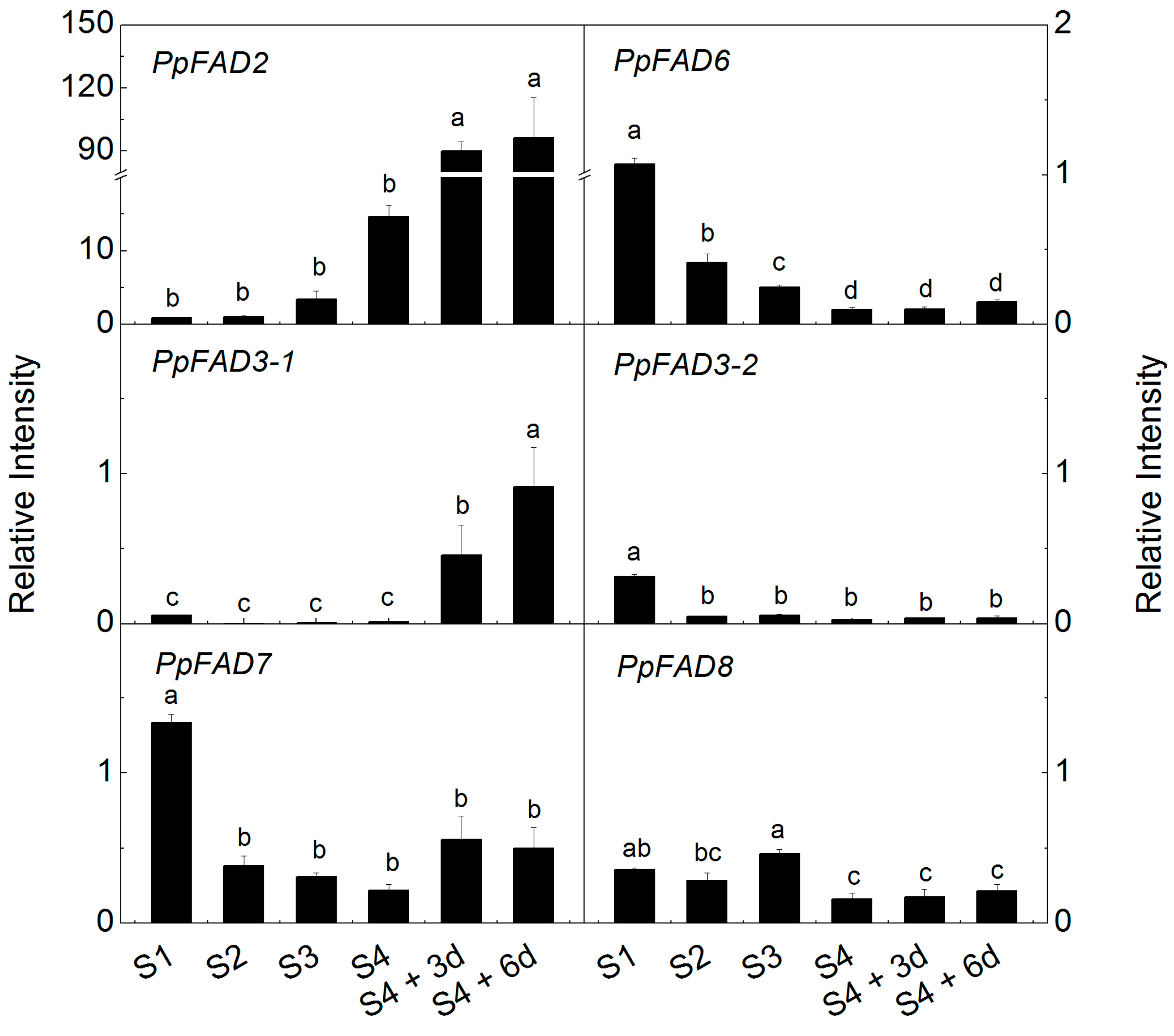
2.4. Expression of Peach FAD Genes
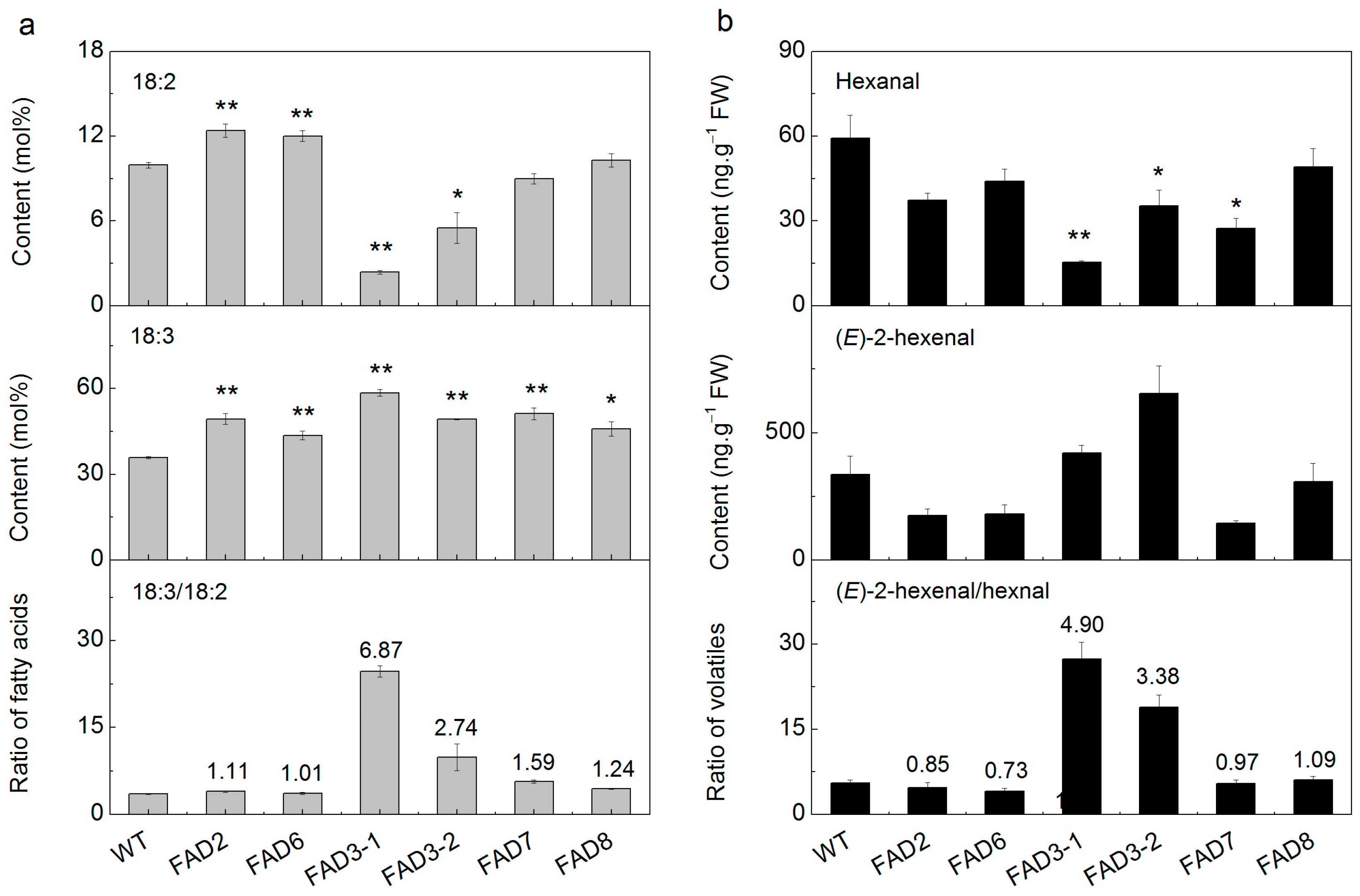
2.5. Heterologous Expression of FADs in Tobacco
3. Materials and Methods
3.1. Plant Material and Sampling
3.2. Fatty Acid Analysis
3.3. GC-MS Analysis of Volatile Compounds
3.4. Sequence Analysis of Peach FAD Genes
3.5. RNA Extraction and qPCR Analysis
3.6. Tobacco Transformation, Growth Conditions and Sampling
3.7. Experimental Design and Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sanchez, G.; Besada, C.; Luisa Badenes, M.; Jose Monforte, A.; Granell, A. A non-targeted approach unravels the volatile network in peach fruit. PLoS ONE 2012, 7, e38992. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, C.; Li, S.; Yang, L.; Wang, Y.; Zhao, J.; Jiang, Q. Volatile characteristics of 50 peaches and nectarines evaluated by HP-SPME with GC-MS. Food Chem. 2009, 116, 356–364. [Google Scholar] [CrossRef]
- Schwab, W.; Davidovich-Rikanati, R.; Lewinsohn, E. Biosynthesis of plant-derived flavor compounds. Plant J. 2008, 54, 712–732. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.P.; Zhang, B.; Liang, L.; Shen, J.Y.; Wei, W.W.; Xu, C.J.; Allan, A.C.; Ferguson, I.B.; Chen, K.S. Postharvest temperature influences volatile lactone production via regulation of Acyl-CoA oxidases in peach fruit. Plant Cell Environ. 2012, 35, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yin, X.R.; Li, X.; Yang, S.L.; Ferguson, I.B.; Chen, K.S. Lipoxygenase gene expression in ripening kiwifruit in relation to ethylene and aroma production. J. Agric. Food Chem. 2009, 57, 2875–2881. [Google Scholar] [CrossRef] [PubMed]
- Ties, P.; Barringer, S. Influence of lipid content and lipoxygenase on flavor volatiles in the tomato peel and flesh. J. Food Sci. 2012, 77, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Tao, S.; Zhang, H.; Huang, W.; Wu, J.; Xu, Y.; Zhang, S. Evolution of the aroma volatiles of pear fruits supplemented with fatty acid metabolic precursors. Molecules 2014, 19, 20183–20196. [Google Scholar] [CrossRef] [PubMed]
- Browse, J.; Warwick, N.; Somerville, C.R.; Slack, C.R. Fluxes through the prokaryotic and eukaryotic pathways of lipid-synthesis in the “16:3” plant Arabidopsis thaliana. Biochem. J. 1986, 235, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Wallis, J.G.; Browse, J. Mutants of Arabidopsis reveal many roles for membrane lipids. Prog. Lipid Res. 2002, 41, 254–278. [Google Scholar] [CrossRef]
- Zhang, M.; Barg, R.; Yin, M.G.; Gueta-Dahan, Y.; Leikin-Frenkel, A.; Salts, Y.; Shabtai, S.; Ben-Hayyim, G. Modulated fatty acid desaturation via overexpression of two distinct ω-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 2005, 44, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Fu, D.Q.; El-Habbak, M.; Navarre, D.; Ghabrial, S.; Kachroo, A. Silencing genes encoding ω-3 fatty acid desaturase alters seed size and accumulation of bean pod mottle virus in soybean. Mol. Plant. Microbe Interact. 2011, 24, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Avila, C.A.; Arevalo-Soliz, L.M.; Jia, L.; Navarre, D.A.; Chen, Z.; Howe, G.A.; Meng, Q.-W.; Smith, J.E.; Goggin, F.L. Loss of function of fatty acid desaturase7 in tomato enhances basal aphid resistance in a salicylate-dependent manner. Plant Physiol. 2012, 158, 2028–2041. [Google Scholar] [CrossRef] [PubMed]
- Bilyeu, K.D.; Palavalli, L.; Sleper, D.A.; Beuselinck, P.R. Three microsomal ω-3 fatty-acid desaturase genes contribute to soybean linolenic acid levels. Crop Sci. 2003, 43, 1833–1838. [Google Scholar] [CrossRef]
- Pham, A.T.; Lee, J.D.; Shannon, J.G.; Bilyeu, K.D. Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with the high oleic acid seed oil trait. BMC Plant Biol. 2010, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, M.L.; Mancha, M.; Martinez-Rivas, J.M. Molecular cloning and characterization of genes encoding two microsomal oleate desaturases (FAD2) from olive. Phytochemistry 2005, 66, 1417–1426. [Google Scholar] [CrossRef] [PubMed]
- Luisa Hernandez, M.; Padilla, M.N.; Mancha, M.; Martinez-Rivas, J.M. Expression analysis identifies FAD2-2 as the olive oleate desaturase gene mainly responsible for the linoleic acid content in virgin olive oil. J. Agric. Food Chem. 2009, 57, 6199–6206. [Google Scholar] [CrossRef] [PubMed]
- Matteucci, M.; D’Angeli, S.; Errico, S.; Lamanna, R.; Perrotta, G.; Altamura, M.M. Cold affects the transcription of fatty acid desaturases and oil quality in the fruit of Olea europaea L. genotypes with different cold hardiness. J. Exp. Bot. 2011, 62, 3403–3420. [Google Scholar] [CrossRef] [PubMed]
- Canoles, M.A.; Beaudry, R.M.; Li, C.Y.; Howe, G. Deficiency of linolenic acid in Lefad7 mutant tomato changes the volatile profile and sensory perception of disrupted leaf and fruit tissue. J. Am. Soc. Hortic. Sci. 2006, 131, 284–289. [Google Scholar]
- Dominguez, T.; Luisa Hernandez, M.; Pennycooke, J.C.; Jimenez, P.; Manuel Martinez-Rivas, J.; Sanz, C.; Stockinger, E.J.; Sanchez-Serrano, J.J.; Sanmartin, M. Increasing ω-3 desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol. 2010, 153, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Shen, J.Y.; Wei, W.W.; Xi, W.P.; Xu, C.J.; Ferguson, I.; Chen, K.S. Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening. J. Agric. Food Chem. 2010, 58, 6157–6165. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, G.; Venegas-Caleron, M.; Salas, J.J.; Monforte, A.; Badenes, M.L.; Granell, A. An integrative “omics” approach identifies new candidate genes to impact aroma volatiles in peach fruit. BMC Genom. 2013, 14, 343. [Google Scholar] [CrossRef] [PubMed]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Kakiuchi, N.; Ohmiya, A. Changes in the composition and content of volatile constituents in peach fruits in relation to maturity at harvest and artificial ripening. J. Jpn. Soc. Hortic. Sci. 1991, 60, 209–216. [Google Scholar] [CrossRef]
- Aubert, C.; Milhet, C. Distribution of the volatile compounds in the different parts of a white-fleshed peach (Prunus persica L. Batsch). Food Chem. 2007, 102, 375–384. [Google Scholar] [CrossRef]
- Hatanaka, A.; Harada, T. Formation of cis-3-hexenal, trans-2-hexenal and cis-3-hexenol in macerated Thea sinensis leaves. Phytochemistry 1973, 12, 2341–2346. [Google Scholar] [CrossRef]
- Hatanaka, A.; Kajiwara, T.; Matsui, K. The Biogeneration of green odour by green leaves and its physiological functions-past, present and future. Z. Naturforsch. C 1995, 50, 467–472. [Google Scholar]
- Izzo, R.; Scartazza, A.; Masia, A.; Galleschi, L.; Quartacci, M.F.; Navariizzo, F. Lipid evolution during development and ripening of peach fruits. Phytochemistry 1995, 39, 1329–1334. [Google Scholar] [CrossRef]
- Duan, Y.X.; Dong, X.Y.; Liu, B.H.; Li, P.H. Relationship of changes in the fatty acid compositions and fruit softening in peach (Prunus persica L. Batsch). Acta Physiol. Plant. 2013, 35, 707–713. [Google Scholar] [CrossRef]
- Hernández, M.L.; Sicardo, M.D.; Martínez-Rivas, J.M. Differential contribution of endoplasmic reticulum and chloroplast ω-3 fatty acid desaturase genes to the linolenic acid content of olive (Olea europaea) fruit. Plant Cell Physiol. 2016, 57, 138–151. [Google Scholar] [CrossRef] [PubMed]
- Heppard, E.P.; Kinney, A.J.; Stecca, K.L.; Miao, G.H. Developmental and growth temperature regulation of two different microsomal ω-6 desaturase genes in soybeans. Plant Physiol. 1996, 110, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Chambers, A.H.; Pillet, J.; Plotto, A.; Bai, J.; Whitaker, V.M.; Folta, K.M. Identification of a strawberry flavor gene candidate using an integrated genetic-genomic-analytical chemistry approach. BMC Genom. 2014, 15, 217. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Sevilla, J.F.; Cruz-Rus, E.; Valpuesta, V.; Botella, M.A.; Amaya, I. Deciphering gamma-decalactone biosynthesis in strawberry fruit using a combination of genetic mapping, RNA-Seq and eQTL analyses. BMC Genom. 2014, 15, 218. [Google Scholar] [CrossRef] [PubMed]
- Mehrshahi, P.; Johnny, C.; Della Penna, D. Redefining the metabolic continuity of chloroplasts and ER. Trends Plant Sci. 2014, 19, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Tong, Z.; Gao, Z.; Wang, F.; Zhou, J.; Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 2009, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Song, S.; Allan, A.C.; Liu, X.F.; Yin, X.R.; Xu, C.J.; Chen, K.S. Differential activation of anthocyanin biosynthesis in Arabidopsis and tobacco over-expressing an R2R3 MYB from Chinese bayberry. Plant Cell Tissue Organ Cult. 2013, 113, 491–499. [Google Scholar] [CrossRef]
| Genes | Forward Primers (5′–3′) | Reverse Primers (5′–3′) |
|---|---|---|
| PpFAD2 | CGGTTTTCAAGGCAATGTTC | CCTACACTCATTCGGGCAAT |
| PpFAD6 | ACGTTGCCTTTGACCAACTT | AATGACTGTGACCCCACCAC |
| PpFAD3-1 | AGTGACACAGGAGATATTGTGT | TTCGAAAGATTACGAGGATTTCA |
| PpFAD3-2 | ATTGAGATGGCAGGGATGAA | CCTTCTCAAGGTTTTTCAGCA |
| PpFAD7 | TCAGGCACAACAATTGAAGC | AAGAATGGCTGCCCATACAG |
| PpFAD8 | TGACCACAAAGACAACCTTTCA | ACTAGGGCACCACCCTTTTT |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.-J.; Liu, H.-R.; Gao, J.; Huang, Y.-J.; Zhang, B.; Chen, K.-S. Two ω-3 FADs Are Associated with Peach Fruit Volatile Formation. Int. J. Mol. Sci. 2016, 17, 464. https://doi.org/10.3390/ijms17040464
Wang J-J, Liu H-R, Gao J, Huang Y-J, Zhang B, Chen K-S. Two ω-3 FADs Are Associated with Peach Fruit Volatile Formation. International Journal of Molecular Sciences. 2016; 17(4):464. https://doi.org/10.3390/ijms17040464
Chicago/Turabian StyleWang, Jiao-Jiao, Hong-Ru Liu, Jie Gao, Yu-Ji Huang, Bo Zhang, and Kun-Song Chen. 2016. "Two ω-3 FADs Are Associated with Peach Fruit Volatile Formation" International Journal of Molecular Sciences 17, no. 4: 464. https://doi.org/10.3390/ijms17040464
APA StyleWang, J.-J., Liu, H.-R., Gao, J., Huang, Y.-J., Zhang, B., & Chen, K.-S. (2016). Two ω-3 FADs Are Associated with Peach Fruit Volatile Formation. International Journal of Molecular Sciences, 17(4), 464. https://doi.org/10.3390/ijms17040464







